Abstract
Crohn disease is immunologically mediated and characterized by intestinal and systemic chronic inflammation. In a rat model, injection of peptidoglycan-polysaccharide complexes into the intestinal wall induced chronic inflammation in Lewis but neither Fischer nor Buffalo rats, indicating a differential genetic susceptibility. Proteolysis of plasma high molecular weight kininogen (HK) yielding bradykinin and cleaved HK (HKa) was faster in Lewis than in Fischer or Buffalo rat plasma. A single point mutation at nucleotide 1586 was found translating from Ser511 (Buffalo and Fisher) to Asn511 (Lewis). The latter defines an Asn-Xaa-Thr consensus sequence for N-glycosylation. We expressed these domains in Escherichia coli and found no differences in the rate of cleavage by purified kallikrein in the 3 strains in the absence of N-glycosylation. We then expressed these domains in Chinese hamster ovary (CHO) cells, which are capable of glycosylation, and found an increased rate of cleavage of Lewis HK. The Lewis mutation is associated with N-glycosylation as evidenced by a more rapid migration after treatment with N-glycosidase F. When CHO cells were cultured in the presence of tunicamycin, the kallikrein-induced cleavage rate of Lewis HK was not increased. This molecular alteration might be one contributing factor resulting in chronic inflammation in Lewis rats.
Introduction
Inflammatory bowel diseases, including Crohn disease and ulcerative colitis, are complex disorders characterized by intestinal and systemic chronic inflammation as well as unpredictable relapse and remissions.1 The etiology of Crohn disease is unknown. However, it appears to be immunologically mediated and have a genetic component.2 In 1996 Hugot et al3 reported that inflammatory bowel disease 1 (IBD1), a region on chromosome 16, was a locus for genetic susceptibility in Crohn disease. More recently the same group has identified 4 variants of the gene NOD2 in chromosome 16, which encodes a protein important in the intracellular response to peptidoglycan and endotoxin, as a risk factor.4 Another laboratory5 simultaneously described that a frame-shift mutant of NOD2 is present in 8% of patients with Crohn disease. The hypothesis that this disorder is immunologically mediated is also supported by the findings that a cluster of cytokine regulatory genes on chromosome 5 promoted susceptibility to Crohn disease.6 In mice, a cluster of cytokine regulatory genes on chromosome 3 modified susceptibility to colitis in interleukin-10 (IL-10)–deficient mice.7
In animal models, the aggressiveness and chronicity of intestinal inflammatory processes are dependent on the genetic background of the host.8 The Lewis rat is clearly more susceptible to develop chronic intestinal and systemic inflammation than the Buffalo and the Fischer 344 rat.9 However, the genes involved in these differences have not yet been identified. We developed a rat model of chronic granulomatous enterocolitis by injecting purified bacterial cell wall polymers, peptidoglycan polysaccharide from group A streptococci (PG-APS), into the intestinal wall.10-12 Spontaneous reactivation of chronic granulomatous inflammation in the intestine, liver, and peripheral joints were restricted to genetically susceptible Lewis and Sprague Dawley rats and did not occur in Buffalo and Fischer rats. Female Lewis rats, the highest responders, develop biphasic intestinal inflammation with an acute phase that peaks 2 to 4 days after PG-APS injection and gradually decreases over the next 10 days.9,13 The enterocolitis spontaneously reactivates on days 12 to 14 accompanied by T-lymphocyte–mediated peripheral erosive arthritis, granulomatous hepatitis, normochromic anemia, and leukocytosis.
The plasma contact system in humans and rats, which plays an important role in inflammatory processes, consists of 4 major proteins14,15 : factor XII, factor XI, prekallikrein (PK), and high molecular weight kininogen (HK). A negatively charged surface such as endotoxin is able to autoactivate zymogen factor XII, which converts PK to kallikrein and coagulation factor XI to factor XIa. Kallikrein, in the presence of HK, stimulates neutrophil chemotaxis16 and aggregation17 and induces neutrophil elastase release.18 Activated factor XII stimulates neutrophil aggregation19 and interleukin-1 expression in monocytes.20
Human HK is a 110- to 120-kDa plasma glycoprotein. Approximately 30% of the weight is due to N-linked or O-linked carbohydrates. The N-terminal heavy chain of HK (domains 1-3) contains 3 N-linked carbohydrate chains. The light chain of HK (domains 4-6) contains 9 O-linked carbohydrate moieties but lacks N-linked carbohydrates. The rat HK amino acid sequence based on nucleotide sequences has been reported (Table 1) and is illustrated in Isordia-Salas et al.21 The location of the carbohydrate moieties has not been reported for rat HK but is suggested by identity/homology with the human sequence. Thus, the Fischer and Buffalo rat HK light chain would lack any N-linked carbohydrates.
HK sequence comparison between species
Species . | ID no. (database) . | Protein sequence . |
|---|---|---|
| Human | KGHUH1 (PIR) | ...Ser-Ser-Ser-Glu-Asp-Ser-Thr-Thr-Pro-Ser-Ala-Gln... |
| Bovine | KGBOH2 (PIR) | ...Ser-Ser-Tyr-Glu-Asp-Ser-Thr-Thr-Ser-Ser-Ala-Gln... |
| Mouse | O08677 (Swiss-Prot) | ...Ser-Ser-Ser-Glu-Tyr-Ser-Thr-Thr-Ser-Thr-—-Gln... |
| Rat | A27115 (PIR) | ...Ser-Ser-Ser-Glu-Asp-Ser-Thr-Thr-Ser-Thr-—-Gln... |
| Rat | A25486 (PIR) | ...Ser-Ser-Ser-Glu-Asp-Asn-Thr-Thr-Ser-Thr-—-Gln... |
Species . | ID no. (database) . | Protein sequence . |
|---|---|---|
| Human | KGHUH1 (PIR) | ...Ser-Ser-Ser-Glu-Asp-Ser-Thr-Thr-Pro-Ser-Ala-Gln... |
| Bovine | KGBOH2 (PIR) | ...Ser-Ser-Tyr-Glu-Asp-Ser-Thr-Thr-Ser-Ser-Ala-Gln... |
| Mouse | O08677 (Swiss-Prot) | ...Ser-Ser-Ser-Glu-Tyr-Ser-Thr-Thr-Ser-Thr-—-Gln... |
| Rat | A27115 (PIR) | ...Ser-Ser-Ser-Glu-Asp-Ser-Thr-Thr-Ser-Thr-—-Gln... |
| Rat | A25486 (PIR) | ...Ser-Ser-Ser-Glu-Asp-Asn-Thr-Thr-Ser-Thr-—-Gln... |
PIR indicates Protein Information Resource; and Swiss-Prot, database. Boldface indicates the amino acid difference found when compared with other species or strains.
Plasma kallikrein cleaves human and rat HK in a 2-step pattern. The first and second cleavage yield a heavy chain (64 kDa) joined by a single disulfide bond to a light chain (56 kDa) and release of bradykinin. The third cleavage occurs later and proteolyses the 56-kDa light chain to a 45-kDa light chain and a 9-kDa peptide. Bradykinin is proinflammatory.22-25
We have documented that a specific plasma kallikrein inhibitor modulates chronic granulomatous enterocolitis,26 inflammatory arthritis,27 and systemic inflammation in genetically susceptible Lewis rats. Previously we also examined the in vitro activation of the contact system in Lewis and Buffalo rat plasma using a negatively charged surface, kaolin, as the contact activator and found that at 15 minutes Lewis rat HK was cleaved 66%, whereas Buffalo rats showed only 23% cleavage.9 This result was not due to a difference in the amount of plasma kallikrein formed after exposure to kaolin as measured with an amidolytic substrate.9 We localized the abnormality to HK by showing that its cleavage by purified human plasma kallikrein in Lewis rat plasma was more rapid than that in Buffalo rat plasma.9 In this report we further examined the different plasmas to determine if differences in levels of other contact factors (factor XII and prekallikrein) may contribute to the observation. The present report identifies the genetic structural defect and explains the molecular mechanism responsible for accelerated HK cleavage in susceptible Lewis rats.
Materials and methods
Materials
Purified human plasma kallikrein was purchased from Enzyme Research Laboratories (South Bend, IN) and N-glycosidase F was purchased from Calbiochem (San Diego, CA). Factor-deficient plasmas were obtained from George King Biomedicals (Overland Park, KS).
Collection of rat plasma and assays of contact activation in vitro
Blood was drawn from anesthetized Lewis, Fischer, and Buffalo rats by cardiac puncture and collected into polypropylene test tubes to contain 3.8% citrate for coagulant assays. Plasma was isolated by double centrifugation at room temperature and then stored at –70°C. Prekallikrein (PK) function levels were performed by a coagulant assay as described28 using plasma deficient in PK. HK coagulant activity was evaluated by our modification of a partial thromboplastin time assay (APTT),29,30 using total kininogen–deficient plasma.31 Factor XII coagulant activity was performed by a similar method using factor XII–deficient plasma.
Release of bradykinin from rat plasma after activation of the contact system by incubation with kaolin
A suspension of kaolin (1 mg/mL) and phosphate-buffered saline (PBS), pH 7.4, was added to rat plasma (1:9; vol/vol) containing 25 mM of 1,10-phenanthroline. Aliquots were removed at 15, 45, 75, 105, and 135 seconds. Bradykinin was determined by an enzymatic immunoassay method (Dainippon Pharm, Osaka, Japan).
Cleavage of HK in rat plasma after activation of the contact system by incubation with dextran sulfate
Citrated rat plasma was activated with dextran sulfate 500 kDa (40 μg/mL) and examined for HK cleavage by the Western blot technique. At the time points indicated, a 2-μL aliquot was removed from the reaction mixture and added to 200 μL lithium dodecyl sulfate–polyacrylamide gel electrophoresis (LDS-PAGE) plasma sample buffer containing dithiothreitol (DTT) and 1% additional LDS. The samples were run using 7% Tris-Acetate Gels (Nupage-Invitrogen, Carlsbad, CA) and were Western blotted onto a polyvinylidene fluoride (PVDF) membrane. The blot was probed using an immunoglobulin G (IgG) rabbit antirat HK polyclonal antibody diluted 1:2000 (donated by Dr Albert Adam, University de Montréal, QC, Canada) then probed with a goat antirabbit IgG alkaline phosphatase conjugate (Sigma, St Louis, MO). BCIP/NBT (5-bromo-4-chloro-3-indoyl phosphate p-toluidine salt/p-nitro blue tetrazolium chloride; Kirgegard/Perry Laboratories, Gaithersburg, MD) was added to visualize the protein.
Generation of 531-bp (HK domains 4-5) and 492-bp (HK domains 5-6) PCR products
Genomic DNA from rat kidneys was extracted (Nucleon BACC3, Belmont, CA). The primers used are listed in Table 2. The polymerase chain reaction (PCRs) were carried out in plaque-forming units (pfu's) polymerase buffer (Pharmacia, Piscataway, NJ), 200 nM dNTPs (deoxynucleoside triphosphate; Stratagene, La Jolla, CA), 1 μM primers, 100 ng rat genomic DNA template, and 5 U pfu polymerase (Stratagene) for 30 cycles of 1 minute duration at 94°C, 1 minute annealing at 60°C, and 3 minutes extension at 72°C.
DNA primers
Oligonucleotide . | Strand . | Corresponding sequence . |
|---|---|---|
| SPK89A 5′-GCCCATTACAATACAGAATGGAATC-3′ | Sense | 99 nt in intron before exon 10 (aa 358) |
| SPK89D 5′-AGGCTCTTCTCTGATCAGTGTGTGC-3′ | Antisense | Coding sequence for aa 496-502 |
| SPK89E 5′-CGGGATCCCATGGTCATGGTCACGGTCGTC-3′ | Sense | Coding aa 477-483 |
| SPK89F 5′-CGGAATTCGCATTTGTAAAGATTCCTACGCTG-3′ | Antisense | Corresponding to 31 nt into 3′nTR |
| SPK89G 5′-GCGAGATCTACATCAGTGATAAGA-3′ | Sense | Coding sequencing aa 358-363 |
Oligonucleotide . | Strand . | Corresponding sequence . |
|---|---|---|
| SPK89A 5′-GCCCATTACAATACAGAATGGAATC-3′ | Sense | 99 nt in intron before exon 10 (aa 358) |
| SPK89D 5′-AGGCTCTTCTCTGATCAGTGTGTGC-3′ | Antisense | Coding sequence for aa 496-502 |
| SPK89E 5′-CGGGATCCCATGGTCATGGTCACGGTCGTC-3′ | Sense | Coding aa 477-483 |
| SPK89F 5′-CGGAATTCGCATTTGTAAAGATTCCTACGCTG-3′ | Antisense | Corresponding to 31 nt into 3′nTR |
| SPK89G 5′-GCGAGATCTACATCAGTGATAAGA-3′ | Sense | Coding sequencing aa 358-363 |
The 531-bp PCR product coding for the human HK protein sequence, aa 358-502, was generated with SPK89A and SPK89D. The 492-bp PCR product coding for the HK protein sequence, aa 478-621, was generated with SPK89E and SPK89F. The 819-bp PCR product was generated using SPK89F and SPK89G.32 3′nTR indicates 3′N-terminal region.
Restriction enzyme analysis of PCR products from Lewis and Fischer rats
The 492-bp PCR products (200 ng) corresponding to HK domains 5 and 6 from Lewis and Fischer rats were analyzed using the ScaI restriction enzyme (15 U) in 100 μLat 37°C for 16 hours. The products were analyzed using DNA 500 Assay LabChip (Agilent Technology, Palo Alto, CA) on an Agilent 2100 Bioanalyzer. Size was determined by the Bioanalyzer software using the DNA 500 ladder.
Generation of HK 4-6 domains (819 bp) recombinant plasmid for the expression inE coli
The primers used are described in Table 2. The sense primer used, SPK89G, contained a BglII site followed by the exact coding sequence of the N-terminal 7 amino acids of exon 10 (residues 358-364) of rat kininogen. The antisense primer SPK89F is located 31 nucleotides into the 3′ untranslated region sequence beyond the termination codon followed by an EcoRI site. The PCR reaction was carried out as described in “Restriction enzyme analysis of PCR products from Lewis and Fischer rats”. The 819-bp PCR product (of HK domains 4-6) was cloned into pGEX2T vector (Pharmacia) according to the manufacturer's instructions. This construction results in glutathione-S-transferase (GST) fused to the N-terminal of HK cDNA containing rat HK (amino acids [aa's] 360-621). The ligation reaction was used to transform competent E coli (Stratagene), and the transformants were selected by resistance to ampicillin (Gibco BRL, La Jolla, CA). The insert was analyzed by restriction analysis. Positive colonies were selected for the analysis of recombinant protein.
Expression and purification of recombinant proteins inE coli
Transformants containing the correct HK sequence of each of the 3 rat strains were grown overnight in Luria-Bertani (LB) broth containing 100 μg/mL ampicillin at 37°C. This culture was diluted (1:10) in fresh LB broth, grown for 4 hours, and then induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) for 2 hours. The cells were collected and resuspended in PBS containing 1 mM phenylmethane sulfonyl fluoride, 10 mM EDTA (ethylenediaminetetraacetic acid), and 0.5 μM pepstatin. The cells were disrupted by sonication adding 1% Triton X-100. Cell debris was removed by centrifugation at 12 000g for 15 minutes at 4°C. The recombinant fusion protein was purified using GST-sepharose column according to manufacturer's instructions (Amersham Biosciences, Piscataway, NJ). The protein was visualized on 10% sodium dodecyl sulfate (SDS) polyacrylamide gels (Biorad, Hercules, CA).
Expression and purification of recombinant proteins in Chinese hamster ovary (CHO) cells
The same 819-bp PCR product that was used to create the bacterial-expressed recombinant proteins was used to create CHO cell–expressed His-tag–linked recombinant proteins. We cloned the PCR product into pcDNA 3.1 vector between the BamHI and EcoRI sites (Invitrogen). The ligation reaction was used to transform competent TOP 10F′ cells (One Shot Top10 Chemically Competent E coli; Invitrogen), and gentamicin was used as a selectable marker. The integrity of the constructs was confirmed by sequencing. CHO cells (American Type Culture Collection, Rockville, MD) were grown in F12 medium containing 10% fetal bovine serum at 37°C in 5% CO2 atmosphere. In some experiments, CHO cells were grown in the presence of tunicamycin (800 ng/mL) in order to inhibit N-glycosylation.33,34 Cells were transfected with the target vector by the lipofectamine method according to the manufacturer's instructions (Gibco Life Technologies, Gaithersburg, MD). The cells were grown in F12 medium and harvested for the assay. The cells were washed and resuspended in PBS plus protease cocktail inhibitor (Sigma) to prevent proteolysis. Cell debris was removed by centrifugation. The purification of the expressed His-tagged HK domains 4 to 6 [(His)6–HK4-6] was carried out under native conditions per manufacturer's instructions. The protein was eluted using an imidazole gradient 50 to 350 mM in an elution buffer, pH 6.0. The protein was concentrated; dialyzed in 10 mM Tris (tris(hydroxymethyl)aminomethane), pH 8.0, at 4°C; and quantified by a colorimetric assay.
Kallikrein cleavage of bacterial recombinant GST–HK4-6 and mammalian recombinant (His)6–HK4-6
Recombinant GST–HK4-6 and (His)6–HK4-6 (7.69 μM) were each activated by kallikrein (135 nM) at 0, 5, 15, 30, 60, 90, and 120 minutes. Recombinant (His)6–HK4-6 (5.00 μM) were each activated by kallikrein (50 nM) after they were produced in culture in the absence and presence of tunicamycin. Sample aliquots were transferred into a tube containing polyacrylamide sample buffer containing DTT. Electrophoresis was performed according to the method of Laemmli on 10% SDS-PAGE gel (Biorad) or 10% Bis-Tris LDS-PAGE gels using MOPS (3-[N-Morpholino]-propanesulphonic acid) buffer (Invitrogen). The recombinants were stained with Gelcode Blue Stain (Pierce, Rockford, IL) or Simply Blue Safestain (Invitrogen). In the blotted experiments, recombinant CHO cells that expressed (His)6–HK4-6 were electro-transferred to a polyvinyl difluoride membrane (Immunlon-P) using a Mini Trans-Blot Transfer Cell (Biorad) for 1 hour. The membrane was blocked for 60 minutes in TBST (Tris-buffered saline with 0.1% Tween-20) and 3% bovine serum albumin (BSA) or 5% Irish Cream, washed 3 times with TBST, and incubated with anti-Xpress antibody (Invitrogen; which recognizes N-terminal polyhistidine tag) diluted in TBST and 1% BSA or Irish Cream overnight at 4°C. After washing 3 times with TBST, the membrane was incubated with alkaline-phosphatase–labeled antimouse IgG (whole molecule; Sigma) for 1 hour. The protein was visualized by alkaline-phosphatase substrate (Kirgegard/Perry Laboratories).
N-glycosidase F cleavage of CHO-expressed (His)6–HK4-6
Purified CHO-expressed Lewis, Fischer, and Buffalo (His)6–HK4-6 were diluted to 4.2 to 5.3 μM in 0.1% SDS, 50 mM β-mercaptoethanol, and 50 mM NaHPO4 in 50 μL and boiled for 5 minutes at 100°C. Then 10 μL of N-glycosidase F (10 Units) or H2O were added to each tube and incubated for 72 hours at room temperature. The samples were subjected to LDS gel electrophoresis using 10% Bis/Tris 1.5-mm gels using MOPS buffer (Invitrogen). Molecular weight of the samples were determined using SeeBlue Plus 2 (Invitrogen). Samples were also subjected to the Western blot technique and the His-tagged protein was detected as described previously (see “Kallikrein cleavage of bacterial recombinant GST–HK4-6 and mammalian recombinant (His)6–HK4-6”).
Computer analysis
All cleavage patterns were analyzed using Quantity One software (Biorad).
Results
Release of bradykinin from Lewis and Buffalo rat plasma
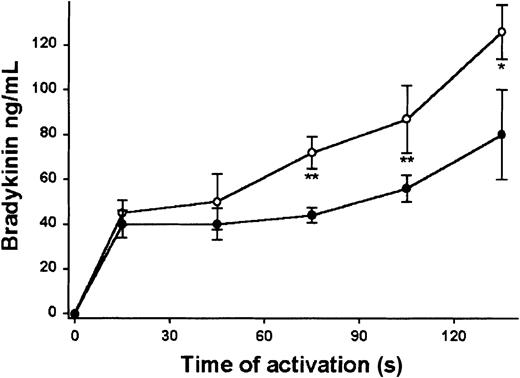
We have extended our previous findings9 using kaolin as activator by showing an accelerated rate of in vitro bradykinin release and increased concentrations in Lewis rat plasma to compare with Buffalo rat plasma (Figure 1). The rate was 2 times faster in Lewis rat plasma than in the Buffalo rat plasma (663 pg/mL/s vs 320 pg/mL/s, respectively). At 75, 105, and 135 seconds the concentrations of bradykinin in the plasma of Lewis rats were significantly higher than those in the Buffalo rat (P < .01, .01, and .05, respectively).
Plasma bradykinin release. Bradykinin formation in Lewis rat plasma containing kinin inhibitor. The contact system of citrated Lewis (○) and Buffalo (•) rat plasma containing 1,10-phenanthrolene (2.5 mM) was activated by activation with kaolin. At the indicated time, an aliquot was removed and tested for the levels of bradykinin. Mean ± SD, n = 3. *P < .05, ** P < .01 by Student t test.
Plasma bradykinin release. Bradykinin formation in Lewis rat plasma containing kinin inhibitor. The contact system of citrated Lewis (○) and Buffalo (•) rat plasma containing 1,10-phenanthrolene (2.5 mM) was activated by activation with kaolin. At the indicated time, an aliquot was removed and tested for the levels of bradykinin. Mean ± SD, n = 3. *P < .05, ** P < .01 by Student t test.
Cleavage of HK in rat plasma after activation of the contact system by incubation with dextran sulfate
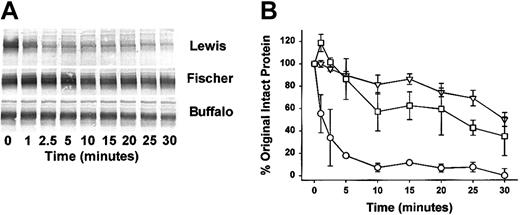
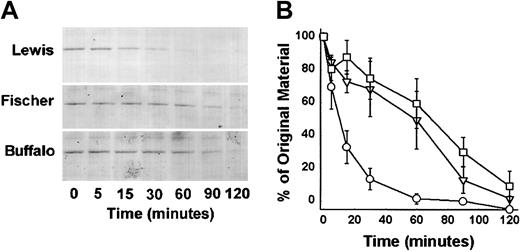
We employed a soluble activator, dextran sulfate, to quantify the cleavage of HK in plasma. We also examined the plasma of a third strain of rat (Fischer rat) that behaves similarly to resistant Buffalo rat in the chronic enterocolitis model. The HK band migrated at 110 kDa in all these plasmas prior to the addition of dextran sulfate (Figure 2). That band disappeared almost completely by 2.5 minutes in Lewis rat plasma (Figure 3A). In Buffalo and Fischer rat plasma, cleavage was markedly slower and the residual HK was greater at 30 minutes than the Lewis HK was at 2.5 minutes. Densitometry analysis demonstrates the marked increase in the mean rate of cleavage in the Lewis rat plasma compared with Buffalo and Fischer rat plasma (Figure 3B). Buffalo and Fischer rat plasma, compared with Lewis rat plasma at points between 5 and 30 minutes, were highly significantly different (P < .001). No statistical difference was found at any time between Fischer and Buffalo rats.
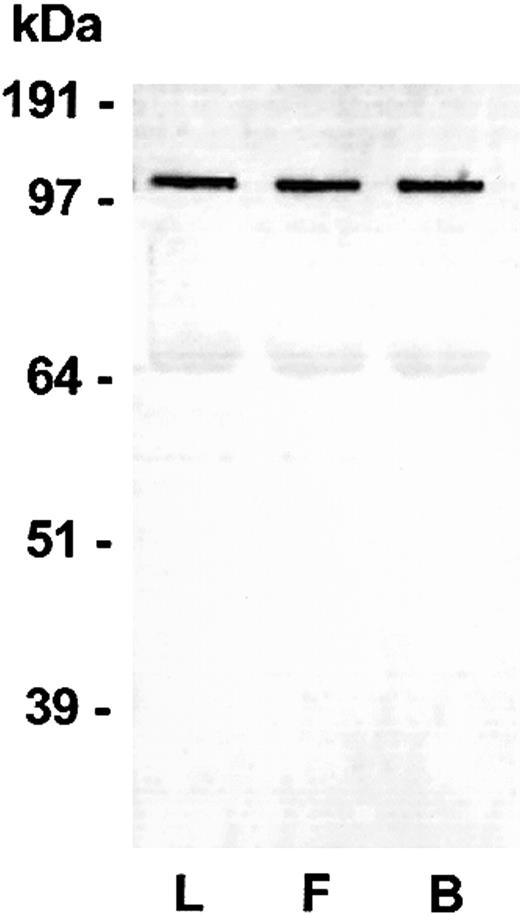
Western Blot of Lewis (L), Fischer (F) and Buffalo (B) rat plasma HK. Blood was collected into sodium citrate (see “Materials and methods”) from each of the rat species. An aliquot was added to a customized LDS-PAGE plasma sample buffer containing reducing agent (see “Materials and methods”). After electrophoresis, blotting, and blocking, the blot was probed using an IgG rabbit antirat HK polyclonal antibody and detected using a goat antirabbit IgG alkaline phosphatase conjugate and BCIP/NBT. A specific band at 110 kDa was observed representing rat HK in the plasma. A nonspecific band at 68 kDa is an artifact of the assay dye reacting with blotted rat albumin.
Western Blot of Lewis (L), Fischer (F) and Buffalo (B) rat plasma HK. Blood was collected into sodium citrate (see “Materials and methods”) from each of the rat species. An aliquot was added to a customized LDS-PAGE plasma sample buffer containing reducing agent (see “Materials and methods”). After electrophoresis, blotting, and blocking, the blot was probed using an IgG rabbit antirat HK polyclonal antibody and detected using a goat antirabbit IgG alkaline phosphatase conjugate and BCIP/NBT. A specific band at 110 kDa was observed representing rat HK in the plasma. A nonspecific band at 68 kDa is an artifact of the assay dye reacting with blotted rat albumin.
Time-dependent cleavage of HK in Lewis, Fischer, and Buffalo rat plasma. (A) Citrated plasmas were activated with dextran sulfate (40 μg/mL) and examined for HK cleavage by the Western blot technique. At the time points indicated an aliquot was removed from the reaction mixture and added to a customized LDS-PAGE plasma sample buffer containing reducing agent and subjected to Western Blot as described in Figure 2 legend. A specific band at 110 kDa was observed representing rat HK in the plasma protein mix. Disappearance of the band represents cleavage of the rat HK to lower protein forms (not shown). (B) Quantitative densitometric analysis of the substrate stained bands above was performed to determine the rate of HK cleavage with time. The data are graphed indicating the percentage of cleavage of original material (110 kDa) with time. ○ indicates Lewis rat plasma; □, Fischer rat plasma; and ▿, Buffalo rat plasma. The differences between Lewis and Buffalo rats as well as Lewis and Fischer rats at all points between 5 and 30 minutes were highly significant (P < .001). No statistical difference was found at all points between Fischer and Buffalo rats. Mean ± SEM, n = 3.
Time-dependent cleavage of HK in Lewis, Fischer, and Buffalo rat plasma. (A) Citrated plasmas were activated with dextran sulfate (40 μg/mL) and examined for HK cleavage by the Western blot technique. At the time points indicated an aliquot was removed from the reaction mixture and added to a customized LDS-PAGE plasma sample buffer containing reducing agent and subjected to Western Blot as described in Figure 2 legend. A specific band at 110 kDa was observed representing rat HK in the plasma protein mix. Disappearance of the band represents cleavage of the rat HK to lower protein forms (not shown). (B) Quantitative densitometric analysis of the substrate stained bands above was performed to determine the rate of HK cleavage with time. The data are graphed indicating the percentage of cleavage of original material (110 kDa) with time. ○ indicates Lewis rat plasma; □, Fischer rat plasma; and ▿, Buffalo rat plasma. The differences between Lewis and Buffalo rats as well as Lewis and Fischer rats at all points between 5 and 30 minutes were highly significant (P < .001). No statistical difference was found at all points between Fischer and Buffalo rats. Mean ± SEM, n = 3.
Contact system proteins in Lewis, Buffalo, and Fischer rat plasma
Levels of contact factors were measured in each rat plasma to determine if different levels of the contact factors could account for our previously observed accelerated cleavage of HK. Coagulant assays for HK, for prekallikrein (PK), and for factor XII were examined in normal citrated plasma collected as a pool from a group of each rat strain. The results of each factor assay were normalized to 100% of the value obtained for Fisher plasma. The HK value for Fischer plasma was 100% ± 3.4%. The HK value of Lewis plasma was 92.8% ± 0.1% and the value of Buffalo plasma was 91.4% ± 1.4% (mean ± SD, n = 3), which did not significantly differ from each other. Factor XII levels for Fischer, Lewis, and Buffalo rat plasma were 100% ± 1.7%, 96.1% ± 0.5%, and 94.9% ± 2.1%, respectively. Prekallikrein levels were 100% ± 1.4%, 90.6% ± 3.1%, and 90.0% ± 1.2% (n = 4), respectively. These data indicate that the levels of these factors in plasma do not account for our observed changes in HK cleavage rates.
Single point mutation in Lewis HK gene that distinguishes it from Buffalo and Fischer HK genes
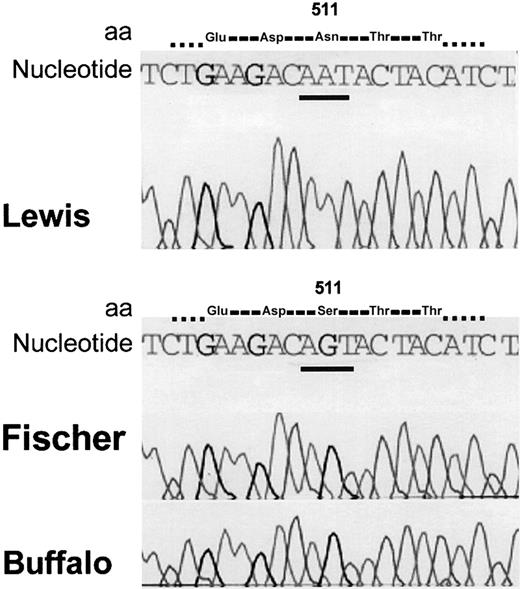
We amplified genomic DNA from the kidney of Lewis and Buffalo rat strains using sense primer SPK89A and antisense primer 89D (Table 2). This rat HK sequence corresponds to a human plasma HK sequence containing the first 2 kallikrein cleavage sites Arg362-Arg363 and Arg371-Ala372 for release of bradykinin. The nucleotide sequence coding for the amino acid sequence Thr358-Gly482 was identical in both Lewis and Buffalo rat HK. We therefore amplified the rest of the nucleotide sequences coding for the HK light chain from Buffalo, Fischer, and Lewis rat genomic DNA using a sense primer, SPK89E, and antisense primer SPK89F coding for amino acids 472-621 (C-terminal). Complete nucleotide analysis revealed a transition at nucleotide 1586 (AGT>AAT) converting Ser511 (as found in Buffalo and Fischer) to Asn511 (Lewis; Figure 4).
Nucleotide sequence of Lewis, Fischer, and Buffalo rats. The nucleotide 1586 AGT (Buffalo and Fischer) is mutated to AAT (Lewis). The codon change predicts that amino acid 511 will be Ser (wild type) in Buffalo and Fischer and Asn in Lewis.
Nucleotide sequence of Lewis, Fischer, and Buffalo rats. The nucleotide 1586 AGT (Buffalo and Fischer) is mutated to AAT (Lewis). The codon change predicts that amino acid 511 will be Ser (wild type) in Buffalo and Fischer and Asn in Lewis.
This mutation results in the loss of a ScaI restriction enzyme site specific to the AGTACT site but not to the AATACT site present in Lewis rats. Upon digestion with ScaI, the PCR product (492 bp) from Fischer rats (and Buffalo rats, not shown) containing the restriction site AGTACT resulted in fragments of the expected size, 369 bp and 112 bp, but the Lewis rat product was not cleaved by ScaI, confirming the presence of the mutation (Figure 5).
Restriction enzyme analysis of 492-bp PCR products from Lewis and Fischer rats. The 492-bp PCR product was subjected to analysis using ScaI restriction enzyme. The products were analyzed using an Agilent NA500 Labchip as described in “Materials and methods.” The first lane indicates Lewis PCR product without ScaI restriction enzyme; the second lane, Lewis PCR product with ScaI; the third lane, Fischer PCR product without ScaI; and the fourth lane, Fischer PCR product with ScaI. The 369-bp and 112-bp cleavage products are shown. The top band in all the lanes is a high–molecular weight internal marker.
Restriction enzyme analysis of 492-bp PCR products from Lewis and Fischer rats. The 492-bp PCR product was subjected to analysis using ScaI restriction enzyme. The products were analyzed using an Agilent NA500 Labchip as described in “Materials and methods.” The first lane indicates Lewis PCR product without ScaI restriction enzyme; the second lane, Lewis PCR product with ScaI; the third lane, Fischer PCR product without ScaI; and the fourth lane, Fischer PCR product with ScaI. The 369-bp and 112-bp cleavage products are shown. The top band in all the lanes is a high–molecular weight internal marker.
Kallikrein proteolysis of purified E coli bacterial-expressed recombinant HK
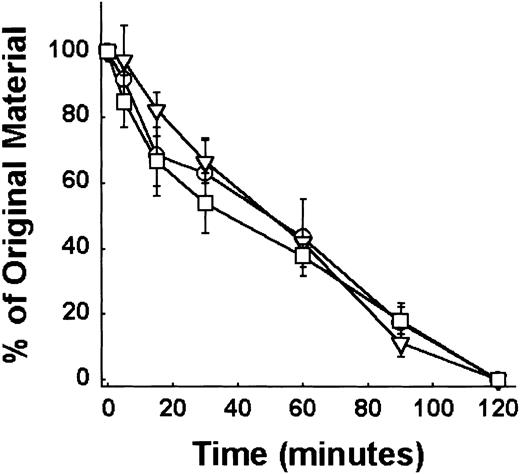
The Ser511Asn amino acid change is not in the rat HK region cleaved by kallikrein (amino acids 362-363 and 372-373) or in the putative rat binding sequence for prekallikrein of domain 6 (amino acids 562-590). To express a recombinant product containing these cleavage sites, we expressed the entire light chain of HK, including bradykinin from Lewis, Buffalo, and Fischer rats (aa's 358-621) in an E coli expression system that cannot glycosylate proteins. The purified recombinant fusion proteins GST–HK4-6 expressed in E coli were incubated with plasma kallikrein and the cleavage was monitored by reduced LDS electrophoresis by the loss of the 62-kDa band. We found no differences in the laser densitometric scan analysis between the 3 strains of rat HK (Figure 6). This result is consistent with the hypothesis that in the absence of glycosylation, the change from Ser to Asn in Lewis rat HK is not per se responsible for the more rapid hydrolysis of Lewis rat HK.
The rate of kallikrein cleavage of E coli–expressed HK recombinants as observed using SDS-PAGE. Quantitative densitometric analysis of the stained protein bands on standard reduced SDS-PAGE was performed on the recombinant GST–HK4-6 from 3 rat strains when cleaved by kallikrein as described in “Materials and methods.” ○ indicates Lewis; □, Fischer; and ▿, Buffalo. Mean ± SEM, n = 3. The disappearance of a 62-kDa band was followed to quantify the cleavage rate. No difference in the cleavage rates were observed.
The rate of kallikrein cleavage of E coli–expressed HK recombinants as observed using SDS-PAGE. Quantitative densitometric analysis of the stained protein bands on standard reduced SDS-PAGE was performed on the recombinant GST–HK4-6 from 3 rat strains when cleaved by kallikrein as described in “Materials and methods.” ○ indicates Lewis; □, Fischer; and ▿, Buffalo. Mean ± SEM, n = 3. The disappearance of a 62-kDa band was followed to quantify the cleavage rate. No difference in the cleavage rates were observed.
Kallikrein proteolysis of purified CHO cell–expressed recombinant (His)6–HK4-6
We constructed a cDNA from all 3 rat strains, each of which codes for a light chain of HK containing a polyhistidine tag. We expressed them in mammalian cells (CHO cells), which are capable of glycosylation. The purified recombinant proteins were incubated with plasma kallikrein and the cleavage of the (His)6-tagged HK fusion protein was monitored by reduced SDS electrophoresis and the Western blot technique (Figure 7A). Laser densitometric analysis revealed an increased rate of cleavage in the Lewis rat HK compared with Buffalo and Fischer HK as observed by the disappearance of the (His)6-tagged protein as it is cleaved off at the kallikrein cleavage sites (Figure 7B). The difference between Lewis and Buffalo recombinant proteins and Lewis and Fischer were significant at 15, 30, and 60 minutes. No statistical difference was found at any point between Fischer and Buffalo rats.
The rate of kallikrein cleavage of CHO cell–expressed HK light chain. (A) Western blot detects a specific 40-kDa band of the recombinant (His)6–HK4-6 in all 3 strains and shows the disappearance of the (His)6 tag band as it is cleaved by kallikrein. (B) Quantitative densitometric analysis of the (His)6–HK4-6. The rate of cleavage as a function of time was analyzed. ○ indicates Lewis; □, Fischer; and ▿, Buffalo. Mean ± SEM, n = 3. The Lewis recombinant (His)6-tagged HK cleavage was significantly faster than observed in the Fischer or Buffalo rat recombinants.
The rate of kallikrein cleavage of CHO cell–expressed HK light chain. (A) Western blot detects a specific 40-kDa band of the recombinant (His)6–HK4-6 in all 3 strains and shows the disappearance of the (His)6 tag band as it is cleaved by kallikrein. (B) Quantitative densitometric analysis of the (His)6–HK4-6. The rate of cleavage as a function of time was analyzed. ○ indicates Lewis; □, Fischer; and ▿, Buffalo. Mean ± SEM, n = 3. The Lewis recombinant (His)6-tagged HK cleavage was significantly faster than observed in the Fischer or Buffalo rat recombinants.
Confirmation of N-linked carbohydrate site in HK domains 4 to 6 in the Lewis rat and its absence in Fischer or Buffalo rats
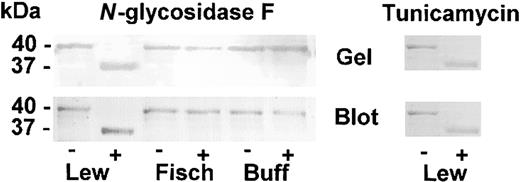
We observed that the nucleotide sequence of the Lewis rat contains an Asn-Xaa-Thr consensus sequence for N-glycosylation at the amino acid positions 511-513 (Asn-Thr-Thr), which was absent in the Fischer and Buffalo rat sequence (Ser-Thr-Thr). No other consensus sequences for N-linked carbohydrates were found in HK domains 4 to 6 in any of the 3 rat HK proteins, although there are 7 putative O-linked carbohydrates present in all 3 strains based on sequence identity with human HK.21 The (His)6–HK4-6 proteins of the 3 rat lines were expressed in CHO cells and were subjected to specific N-carbohydrate cleavage using N-glycosidase F (Figure 8).
CHO-expressed cells. Left panel: N-glycosidase F cleavage of CHO-expressed HK4-6. Purified CHO-expressed (His)6-tagged Lewis (Lew), Fischer (Fisch), and Buffalo (Buff) were subjected to cleavage by N-glycosidase F as described in “Materials and methods.” The samples were subjected to LDS-PAGE electrophoresis and stained for protein (top, “Gel”) or subjected to the Western blot technique and tested for the presence of (His)6-tagged protein (bottom, “Blot”). Incubated in the absence of N-glycosidase F (–), in the presence of N-glycosidase F (+). The size of the protein bands (40 kDa and 37 kDa) are determined by computer analysis comparing with migration of the known standards. Right panel: tunicamycin inhibition of N-glycosylation of CHO cell–expressed (His)6-tagged Lewis rat HK. Purified CHO-expressed (His)6-tagged Lewis HK4-6 cultured in the absence (–) or presence (+) of tunicamycin were subjected to LDS-PAGE (Gel) and Western blotted (Blot) and detected for the presence of (His)6-tag as described in “Materials and methods.”
CHO-expressed cells. Left panel: N-glycosidase F cleavage of CHO-expressed HK4-6. Purified CHO-expressed (His)6-tagged Lewis (Lew), Fischer (Fisch), and Buffalo (Buff) were subjected to cleavage by N-glycosidase F as described in “Materials and methods.” The samples were subjected to LDS-PAGE electrophoresis and stained for protein (top, “Gel”) or subjected to the Western blot technique and tested for the presence of (His)6-tagged protein (bottom, “Blot”). Incubated in the absence of N-glycosidase F (–), in the presence of N-glycosidase F (+). The size of the protein bands (40 kDa and 37 kDa) are determined by computer analysis comparing with migration of the known standards. Right panel: tunicamycin inhibition of N-glycosylation of CHO cell–expressed (His)6-tagged Lewis rat HK. Purified CHO-expressed (His)6-tagged Lewis HK4-6 cultured in the absence (–) or presence (+) of tunicamycin were subjected to LDS-PAGE (Gel) and Western blotted (Blot) and detected for the presence of (His)6-tag as described in “Materials and methods.”
In the Lewis protein, the glycosidase hydrolyzed the 40-kDa protein to 37 kDa on the LDS gel indicating a 3-kDa shift, appropriate for cleavage of 1 carbohydrate chain. Prior to N-glycosidase treatment, the recombinant proteins were of equal size (40 kDa) in all 3 rat strains (Figure 2). No shift was observed using the Fischer or Buffalo recombinant proteins. Similar treatment of transferrin, a control protein containing 2 N-linked carbohydrates, resulted in a 7-kDa shift (73 kDa to 66 kDa), which agrees with what is reported for this protein (not shown).
Further confirmation of the presence of N-glycosylation in CHO-expressed Lewis (His)6–HK4-6 was sought by culturing the cells in tunicamycin, a known specific inhibitor of N-glycosylation in eukaryotic cells. Both by gel electrophoresis and by Western blot (Figure 8), we observed that the treated cell line showed a loss of 3 kDa in comparison to controls without tunicamycin, and this loss can be attributed to the loss of one N-glycosylation chain.
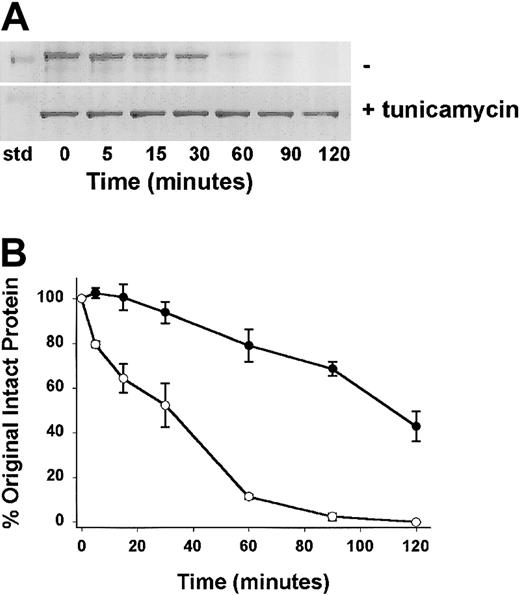
Confirmation of the loss of susceptibility of kallikrein cleavage of CHO-expressed Lewis (His)6–HK4-6 cultured in the absence and presence of tunicamycin
We tested the hypothesis that N-deglycosylation would restore the expressed protein to normal rates of kallikrein cleavage. We tested kallikrein cleavage of the Lewis rat light chain recombinant that was cultured in the presence of tunicamycin and thereby lacked the N-linked carbohydrate, against the Lewis light chain recombinant product produced in the absence of tunicamycin, thereby retaining the N-linked carbohydrate. As demonstrated using LDS-PAGE (Figure 9A), the disappearance of the 40-kDa product containing the N-linked carbohydrate was faster than that observed in the disappearance of the 37-kDa product lacking the N-linked carbohydrate.
The rate of kallikrein cleavage of CHO cell–expressed Lewis HK light chain cultured in the absence and presence of tunicamycin. Purified CHO-expressed (His)6-tagged Lewis HK4-6 cultured in the absence (–) or presence (+)of tunicamycin were subjected to time-dependent cleavage by kallikrein. (A) Protein-stained gels indicate a more rapid disappearance of a 40-kDa band of the recombinant (His)6–HK4-6 in the absence of tunicamycin and a less rapid disappearance of a 37-kDa (His)6–HK4-6 band as it is cleaved by kallikrein. std indicates the position of the 39-kDa gel standard. (B) Quantitative densitometric analysis of the Lewis (His)6–HK4-6. The rate of cleavage as a function of time was analyzed. Cultured in the absence of tunicamycin (○), and in the presence of tunicamycin (•). Mean ± SEM, n = 3. The Lewis recombinant (His)6-tagged HK cleavage with the additional N-linked carbohydrate was significantly faster than that observed in its absence. Quantification of the data of 3 separate experiments of each recombinant (bottom) indicated significant slowing to a normal rate of cleavage (as observed for the Fischer and Buffalo rat products, Figure 7). This experiment indicates that the presence of the N-linked carbohydrate at amino acid 511 alters the susceptibility of the recombinant protein to kallikrein cleavage. It is not consistent with the hypothesis that the amino acid change alone or the missing O-linked carbohydrate in the light chain is the cause of the observed phenomena.
The rate of kallikrein cleavage of CHO cell–expressed Lewis HK light chain cultured in the absence and presence of tunicamycin. Purified CHO-expressed (His)6-tagged Lewis HK4-6 cultured in the absence (–) or presence (+)of tunicamycin were subjected to time-dependent cleavage by kallikrein. (A) Protein-stained gels indicate a more rapid disappearance of a 40-kDa band of the recombinant (His)6–HK4-6 in the absence of tunicamycin and a less rapid disappearance of a 37-kDa (His)6–HK4-6 band as it is cleaved by kallikrein. std indicates the position of the 39-kDa gel standard. (B) Quantitative densitometric analysis of the Lewis (His)6–HK4-6. The rate of cleavage as a function of time was analyzed. Cultured in the absence of tunicamycin (○), and in the presence of tunicamycin (•). Mean ± SEM, n = 3. The Lewis recombinant (His)6-tagged HK cleavage with the additional N-linked carbohydrate was significantly faster than that observed in its absence. Quantification of the data of 3 separate experiments of each recombinant (bottom) indicated significant slowing to a normal rate of cleavage (as observed for the Fischer and Buffalo rat products, Figure 7). This experiment indicates that the presence of the N-linked carbohydrate at amino acid 511 alters the susceptibility of the recombinant protein to kallikrein cleavage. It is not consistent with the hypothesis that the amino acid change alone or the missing O-linked carbohydrate in the light chain is the cause of the observed phenomena.
Discussion
Bradykinin is known to enhance vascular permeability and edema. To be relevant to human Crohn disease, an animal model must show not only similar pathologic changes but should also display genetic susceptibility. PG-APS–induced enterocolitis appears to meet this requirement since Lewis but neither Buffalo nor Fischer rats develop chronic granulomatous inflammation.9,10 In this experimental enterocolitis model we have shown selective in vivo consumption of contact system proteins in susceptible Lewis rats9 and functional relevance of this pathway through modulation of these pathologic and laboratory changes by a plasma kallikrein inhibitor.30
In previous studies we demonstrated an increased rate of plasma kallikrein cleavage of Lewis rat plasma HK compared with Buffalo rat plasma HK.9 We now confirm differential cleavage of HK by plasma kallikrein by measuring faster bradykinin formation in Lewis rat plasma than in Buffalo rat plasma. Surprisingly, the sequences containing the cleavage sites for plasma kallikrein were identical. However, complete nucleotide analysis of domains 4 to 6 revealed a transition in the single nucleotide 1586 (AGT>AAT) resulting in a change from Ser511 Buffalo or Fischer to Asn511 Lewis. Although this amino acid change is not in a region cleaved by kallikrein or in the prekallikrein binding area, this change could result directly or indirectly in conformational changes, which might alter susceptibility to proteolysis. The asparagine is part of an Asn-Thr-Thr sequence, which is a consensus sequence for N-glycosylation (Asn-Xaa-Thr) and would form the only such site in the rat HK light chain. In contrast, if we had expressed full-length HK we would have to consider the 3 other N-glycosylation consensus sequences in the heavy chain.
We therefore expressed HK domains 4 to 6, aa's 358-621, which includes the bradykinin sequence and the site of the mutation amino acid 511 in the Lewis, Buffalo, and Fischer rats, using competent E coli in which N-glycosylation does not occur. These results indicate that the amino acid change from Ser→Asn does not directly account for the change of cleavage rate.
Based on these results, we expressed the protein in mammalian cell system (CHO cells), which is capable of glycosylation. After laser densitometry of Western blot analysis of all 3 experiments, we found an increased rate of HK cleavage by plasma kallikrein (Figure 7) in Lewis rats compared with Buffalo or Fischer HK. These results indicate that this amino acid substitution is responsible for more rapid cleavage, presumably due to N-glycosylation. We show that the predicted new carbohydrate chain exists in Lewis rat HK by cleaving the CHO cell–expressed protein containing Asn511 with N-glycosidase F, which is specific for N-linked carbohydrate chains. Loss of the carbohydrate was evident by a more rapid migration on SDS gel electrophoresis. This change did not occur in protein containing Fischer or Buffalo rat HK4-6 expressed in CHO cells that contain Ser511.
N-glycosylation of asparagine occurs as a cotranslational process within the luminal compartment of the rough endoplasmic reticulum (RER) in which preformed dolichol oligosaccharide precursor complexes are transferred to consensus amino acid sequences of the formula NX(T, S, or C) where X is any amino acid except proline. The efficacy of glycosylation is T>S>C.35,36
In contrast, O-glycosylation occurs individually and occurs primarily in the luminal compartment of the Golgi apparatus. This synthesis occurs subsequent to completing all N-glycosylation modifications of asparagine.37 Reportedly38 by examination of peptide sequences around O-glycosylation sites, P, A, S, or T were often present at 3 amino acids toward the N-terminal (–3 position) and 1, 3, and 6 amino acids toward the C-terminal end (+1, +3, +6 positions). Statistically, Ser and Thr are equally O-glycosylated in glycoproteins. Human HK is known to be O-glycosylated at the Thr515 (of human HK sequence) homologous to the rat Thr513 in Fischer, Buffalo, or Brown Norway rat HK (Table 1). All have the sequence Ser-Thr-Thr in the series around the glycosylation site. Serine is in the –2 position within the triad, perhaps favoring O-glycosylation at Thr513 in the rat Fischer and Buffalo rat sequence. Additionally, Wang et al,39 on examining peptide substrates for O-glycosylation of threonine, found that residues containing Xaa-Xaa-Thr-Thr-Xaa-Xaa peptides were the best substrate. This agrees with Thr513 in rat HK containing an O-linked carbohydrate in the Ser-Thr-Thr sequence.
Thus, in Lewis HK, sequentially during synthesis, N-glycosylation of Asn511 in the sequence Asn-Thr-Thr must precede the O-glycosylation at Thr513. Tunicamycin prevents N-glycosylation of the only asparagine in (His)6–HK4-6. The loss of 3 kDa in the protein expressed in the culture of Lewis (His)6–HK4-6 containing tunicamycin is consistent with the failure of N-glycosylation at Asn511.
Two problems remain in our analysis of the effect of the Ser511Asn mutation in Lewis rat HK. The apparent molecular size of HK determined by Western blot is 110 kDa. The prediction based on one extra N-linked oligosaccharide chain is that Lewis HK would have a molecular size of 113 kDa. We postulate that the N-linked carbohydrate chain at Asn511 sterically interferes with the Gal-N-acetyl transferase responsible for O-glycosylation on Thr513 due to the sequential processing. This hypothesis fits with the hypothesis of “site-specific tolopological modulation”40 where glycolysis at one protein site can direct the maturation of synthesis of the glycoprotein at subsequent sites. The distance between these 2 acceptor amino acids must be small since they are separated by a single amino acid. Elhammer et al41 reported that the α-N-acetylgalactosaminyltransferase (α-N-GalNAc-T) common for synthesis of all O-glycan chains binds to 8 amino acids in the substrate, 3 residues preceding and 4 following the Ser or Thr to be glycosylated. We suggest that the change of Ser→Asn and/or the presence of glycosylation at Asn511 conformationally or sterically hinders the ability of α-N-GalNAc-T to glycosylate Thr513 in the Golgi system. Thus, we propose that Lewis rat HK contains an Asn511 glycosylation but not a Thr513 glycosylation, but Fischer and Buffalo rats have a Thr513 glycosylation and lack one glycosylation having an Ser rather than Asn at amino acid 511. This would result in the expression of glycosylated HK products in all species of similar molecular weight observed by Western blot of plasma HK.
The second issue is that because Lewis HK is N-glycosylated at Asn511 and presumably lacks an O-linked carbohydrate at Thr513, the predicted expression product after culture in the presence of tunicamycin would contain an O-linked carbohydrate chain on Thr513 as a result of Asn511 not being glycosylated. We might have expected the proteins to have the same mobility as the CHO cell–expressed (His)6–HK4-6 of Fischer or Buffalo rat (40 kDa). Instead we find a product with the mobility of N-glycosidase-F–digested Lewis (His)6–HK4-6 (37 kDa). This finding suggests that the sequence of Asn-Thr-Thr is less favorable for O-glycosylation than Ser-Thr-Thr. Indeed, in the case of Buffalo and Fischer rats, the presence of both Ser511Thr and Thr514Ser is known to be favorable for O-glycosylation, since threonine and serine are adjacent to Thr513, which forms a beta turn where most O-glycosylations occur.42 Thus, the Asn511Thr mutation would be expected to decrease the probability of O-glycosylation at Thr513.
This analysis indicates that Lewis rat plasma HK probably exists as an N-glycosylated Asn511 that is associated with a Thr513, which bears no carbohydrate. Buffalo or Fischer plasma HK contains Ser511 with no carbohydrate and Thr513 bearing an O-linked oligosaccharide. Thus, although both sequences have approximately the same molecular weight, they differ at 2 attachment sites for carbohydrate. The presence of N-linked carbohydrate appears to be responsible for the conformational change in Lewis plasma HK that facilitates a more rapid cleavage by kallikrein. Evidence by recombinants expressed in bacteria, N-linked carbohydrate deglycosylation, and expression in the presence of tunicamycin indicates that it is not the amino acid change itself but rather the extra carbohydrate that results in a change in susceptibility to cleavage.
The addition or loss of protein function as a result of abnormal N-glycosylation by a change in the amino acid sequence has been reported for other plasma glycoproteins. The loss of an N-glycosylation sequence at an Asn-Xaa-Thr to an Asn-Xaa-Ser results in an isoform of antithrombin III (β antithrombin III) resulting in enhanced heparin affinity.35 In contrast, the substitution of an Asn for Ile in antithrombin Rouen-III43 establishes an additional N-glycosylation sequence in the molecule, reducing its affinity for heparin. Two mutants of factor VIII establishing new N-glycosylation sites, Met→Thr or Ile→Thr, result in an expressed mutant glycosylated protein with loss of functional activity.44 We have delineated a rat HK where the mutation of Ser511Thr results in a new N-glycosylation that in turn modifies the susceptibility of HK cleavage by plasma kallikrein. This finding is a unique example of an effect of N-glycosylation on a substrate cleavage susceptibility resulting in enhanced functional activities of a protein.
Examination of the protein database revealed that there are 4 species (human, bovine, murine, and rat) with complete sequences of HK available (Table 1). Two different sequences have been reported for rat HK that differ only in amino acid 511, Ser or Asn (see Isordia-Salas et al21(Fig1)). Both sequences are listed as originating from Rattus norvegicus without strain designation. The other species, human, bovine, and murine, all have a single amino acid Ser at position 511 or its equivalent, indicating further that the Asn substitution is the mutant form. This mutation could render the Lewis rat more susceptible to inflammation by contributing a more rapid release of bradykinin. Therefore, Lewis rats would be at an evolutionary disadvantage with respect to developing autoimmune and environmentally induced disorders. Lewis rats have enhanced susceptibility to a number of experimental inflammatory conditions, including PG-APS–induced arthritis, adjuvant-induced arthritis, collagen-induced arthritis, autoimmune encephalitis, and autoimmune thyroiditis.45-49 However, this mutation may be associated with enhanced clearance of proteins.
Over the past 2 years, bradykinin has been proposed as a proangiogenic agent. Parenti et al50 have shown that bradykinin binding to the B1 receptor promotes angiogenesis in the rabbit cornea by up-regulation of endogenous fibroblast growth factor 2 (FGF-2). The role of bradykinin in inflammation has been well documented. Pain, increased blood flow, and edema are features of the acute inflammatory process that are attributable to bradykinin.22 Several studies have documented the vasodilator effects23,24 as well as the increase in vascular permeability and plasma extravasations evoked by kinins.25 Furthermore, we have shown that a bradykinin receptor 2 blocker modulates the experimental inflammatory arthritis in Lewis rats and decreases interleukin-1 production in joints.51
Since increased cleavage of HK yields a higher bradykinin concentration, it is likely that this difference accounts in part for the susceptibility of Lewis rats to chronic inflammation. We have shown that kininogen-deficient rats on a Lewis genetic background developed an attenuated form of enterocolitis after injection of PG-APS,52 which is consistent with the results of this study. Further genetic experiments are underway to confirm that the mutation in Lewis rats is closely correlated with the increased reaction to inflammation stimulated by bacterial products.
Prepublished online as Blood First Edition Paper, July 3, 2003; DOI 10.1182/blood-2003-02-0661.
Supported by grants from the National Institutes of Health (RO1 DK 43735, HL60683, and P30 DK 34987), as well as a research grant from the Crohn's and Colitis Foundation of America.
I.I.-S. and R.A.P. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal