Abstract
The first hematopoietic and endothelial progenitors are derived from a common embryonic precursor termed the hemangioblast. The genetic cascades that regulate the differentiation of the hemangioblast to hematopoietic and endothelial cells are largely unknown. In general, much of embryonic development is coordinately regulated by temporal and spatial expression of transcription factors, such as the Homeobox (Hox) gene family. We and others isolated a divergent homeobox gene termed Hex (or Prh) that is preferentially expressed in hematopoietic and endothelial cells. Using in vitro Hex-/- embryonic stem (ES) cell differentiation, in vivo yolk sac hematopoietic progenitor assays, and chimeric mouse analysis, we found that Hex is required for differentiation of the hemangioblast to definitive embryonic hematopoietic progenitors and to a lesser extent endothelial cells. Therefore, Hex is a novel regulator of hemangioblast differentiation to hematopoietic and endothelial cells. (Blood. 2003;102:2428-2435)
Introduction
In embryonic development the first hematopoietic and endothelial cells are derived from a common mesodermal precursor termed the hemangioblast.1,2 The hemangioblast requires the functional presence of the transcription factors Scl and Runx1 (also AML1), and the vascular endothelial growth factor (VEGF) receptor Flk-1, for proper formation.3-5 Beyond those fundamental studies, little is known about the cascades of gene expression that regulate either initial hemangioblast formation or its subsequent decisions to differentiate. The hemangioblast gives rise to the 2 types of embryonic hematopoiesis, primitive and definitive.1-5 Primitive hematopoiesis produces nucleated erythrocytes with embryonic globins, and monocytes. Primitive hematopoietic progenitors cannot fully recapitulate hematopoiesis in an adult who underwent transplantation. In contrast, definitive embryonic hematopoietic progenitors can restore hematopoiesis in an adult who underwent transplantation. Definitive hematopoiesis produces enucleated erythrocytes with fetal and adult globins, and produces all hematopoietic lineages. Recent evidence suggests that there may be distinct hemangioblasts for primitive and definitive hematopoiesis.4 This hypothesis came from the finding that murine embryonic stem cells that have Runx1 homozygously deleted can form hemangioblasts that produce primitive but not definitive hematopoietic cells.
Much of embryonic development is coordinately regulated by temporal and spatial expression of transcription factors, such as the Homeobox (Hox) gene family.6 Hox proteins are known to play critical roles in adult hematopoiesis.7,8 We and several others cloned an orphan homeobox gene termed Hex (also Prh) that in the adult was preferentially expressed in hematopoietic cells.9-11 It is generally down-regulated during terminal differentiation of hematopoietic cells.9-12 It is located on chromosome 10, near the Hox11 locus, indicating that one may have arisen from the other by endoduplication.9 Hex is thought to mediate anterior-posterior organizer inducing signals and endodermal organ formation during vertebrate embryonic development.13 Whole mount in situ hybridization studies in early chicken embryos found Hex transcripts expressed in all cells of the early endoderm, and then later in both endothelial and hematopoietic cells.14 These findings were generally duplicated in the developing mouse embryo.15
We found that homozygous deletion of murine Hex resulted in embryonic lethality at days 11 to 13 from anterior truncation of the forebrain, and failure of the liver and thyroid to develop.16 Consistent with the knock-out data, Hex has been found to regulate gene expression in anterior endodermal structures such as the thyroid or liver.17,18 A second group also reported that Hex deletion produced embryonic lethality around embryonic day 11 with a similar phenotype as above.19 They also described a decrease in liver macrophage formation. Otherwise, the role of Hex in the regulation of hematopoiesis or angiogenesis was not characterized in these models. However, studies in xenopus and zebrafish indicated that Hex may play a role not just in endodermal organ formation but also in the origin of mesodermal structures such as blood or endothelium.20,21
In this study we used in vitro Hex-/- murine embryonic stem cell differentiation, in vivo murine yolk sac hematopoietic progenitor analysis, and chimeric mouse analysis to test whether the lack of Hex reduces the ability of the hemangioblast to form embryonic hematopoietic progenitors and endothelial tubules. We found that the lack of Hex was detrimental to the differentiation of the hemangioblast to hematopoietic progenitors and to a lesser extent, to endothelial cells.
Materials and methods
Hemangioblast assay
Hex+/- and Hex-/- murine E14 embryonic stem (ES) cells were generated using homologous recombination and elevated G418 as previously described.16 This deleted the entire coding sequence of Hex. The hemangioblast assay was performed as previously described.3 Briefly, day-3 embryoid bodies' (EBs) disaggregated cells were plated at 12 500 cells/mL in the presence of 10% lot-tested fetal calf serum (differentiation serum; Stem Cell Technologies, Vancouver, BC, Canada), 25% D4T cell-conditioned media (D4T cells kindly provided by Dr Gordon Keller, Mt Sinai School of Medicine, New York, NY), 100 ng/mL stem cell factor, and 5 ng/mL vascular endothelial growth factor. Hemangioblast colonies (termed BL-CFCs) are distinguished after 4 days of culture from transitional colonies and secondary EBs by morphology, high expression of both Flk-1 and Runx1 but low expression of Brachyury, and their ability to form both hematopoietic and endothelial cells when plucked and placed in secondary cultures as described.1,3,4
Primitive and definitive hematopoietic progenitor assays
Disaggregated day-6 EB cells were plated at 1 to 2.5 × 105 cells/mL in 0.9% methylcellulose-based media as we described22,23 and counted after 7 days of culture. Colony-forming units-megakaryocyte (CFU-Meg) colonies were scored based on their colony morphology and CD41 expression as assessed by in situ immunohistology. Each experiment was performed at least 3 times in triplicate. For analysis of in vivo yolk sac hematopoietic progenitors, timed pregnant day-8.5 Hex+/- mice (mated to Hex+/- males) were killed, individual embryos were dissected free of the uterus, and the yolk sac and the embryo proper were separated using an inverted microscope as we previously described.22 DNA was isolated from the embryo proper and used for genotyping as we previously described.16
Microarray analysis
Total RNA was extracted from 4 distinct cultures of day-6 Hex-/- and Hex+/+ EBs using the RNAeasy kit according to the manufacturer's instructions (Qiagen, Valencia, CA). First-strand cDNA was synthesized from the DNA-free total RNA using a dT primer that has the bacterial T7 promoter sequence coupled to it. After second-strand synthesis, biotinylated cRNA is synthesized from the T7 promoter, and then fragmented to an average size of 300 nucleotides. The cRNA is spiked with control-labeled cRNA to standardize the assay. After hybridization to an Affymetrix murine GeneChip and washing, the chips were scanned in a dedicated GeneArray scanner, using Affymetrix software (Santa Clara, CA) as the Indiana University Center for Medical Genomics previously described.24
Vascular sprout formation assay
Aliquots of 200 μL Matrigel (BD Pharmingen, San Diego, CA) were added to each precooled well of 24-well culture plates and incubated at 37°C until gelation occurred as described.25 Then 40 day-6 Hex-/-, Hex+/-, and Hex+/+ EBs were suspended in 200 μL α-modified essential medium containing 15% fetal bovine serum, 4.5 × 10-5 M 2-mercaptoethanol, 2 mM glutamine, 5 ng/mL recombinant murine vascular endothelial growth factor, 100 ng/mL recombinant human β-fibroblast growth factor, and 50 μg/mL endothelial cell growth supplement (BD Pharmingen), and were seeded on the Matrigel. The EBs were incubated at 37°C in a 5% CO2, 2% O2, and 85% N2 humidified environment and sprout formation was assessed. EBs were scored in blinded fashion according to 4 standard classes based on vascular sprout formation: I, no sprout formation; II, few sprouts; III, many sprouts but no network; and IV, many sprouts with network.25 To ensure that the sprouts formed were derived from endothelial cells the colonies were immunohistologically stained in situ with anti-CD31.
Hex-/- chimeric mouse analysis
Hex-/- ES cells were injected into C57/Bl6 blastocysts to provide a 50:50 ratio of Hex-/- to Hex+/+ cells. These blastocysts were placed in utero in pseudopregnant females. Chimeric mice were killed at 3 months of age, and DNA was isolated from specific organs of each mouse. Marrow and spleen cells were flow cytometrically sorted into hematopoietic subpopulations using cell surface markers (Ter119, CD19, and Gr-1; BD Pharmingen), and genomic DNA was isolated. Analysis of the tissue contribution of the Hex-/- ES cells was performed by densitometrically comparing the ratio of Hex with neomycin phosphotransferase (neo) DNA using normalized multiplex log-phase polymerase chain reaction (PCR) as we previously described.16 If Hex-/- ES cells contributed equally well to the generation of a given organ or lineage, then the ratio of neo to Hex sequences would be 1:1. If Hex-/- ES cells contributed poorly to a given organ or lineage then the ratio of neo to Hex would be significantly less than 1.0.
Results
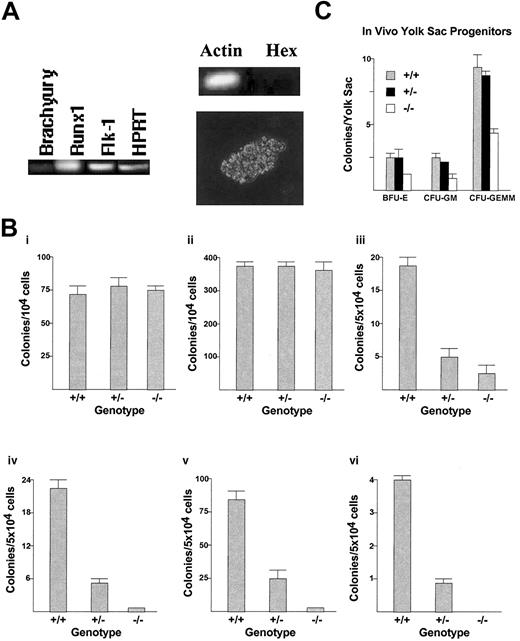
To test the effect of deleting the homeobox gene Hex on hemangioblast formation, in vitro hemangioblast assays (BL-CFC assays) were performed using Hex+/+, Hex+/-, and Hex-/- embryoid bodies (EBs) derived from differentiating ES cells (Figure 1A-B). There was no statistical distinction in the ability to form hemangioblasts between the Hex+/+, Hex+/-, or Hex-/- EBs. Interestingly, whereas the BL-CFCs express Runx1 and Flk-1, with a low level of expression of Brachyury, as previously described,4 we found that they do not appreciably express Hex (Figure 1A). Next, the capability of Hex wild-type (wt) or knock-out EBs to form primitive erythroid progenitors (EryPs) was analyzed using colony-formation assays (Figure 1B). There was no statistical difference between the ability of the Hex cell types to form EryP colonies. Therefore, the lack of Hex did not affect the formation of BL-CFC hemangioblasts or EryPs from differentiating EBs.
The effect ofHexdeletion on embryonic hematopoiesis. (A) RT-PCR and morphology of BL-CFC hemangioblast colonies. In vitro formation of definitive hemangioblasts from day-3 EBs expresses low levels of Brachyury, but high levels of Runx1 and Flk-1, and has characteristic BL-CFC colony morphology.1,4 Hex is not expressed in these BL-CFCs by RT-PCR. BL-CFC colony morphology is shown at × 40 magnification. (B) Primitive and definitive hematopoietic progenitor assays from in vitro differentiation of Hex-/-, Hex+/-, and Hex+/+ EBs. There was no effect of the deletion of the homeoprotein Hex on in vitro formation in day-3 EB hemangioblasts (i) and day-6 EB EryPs (ii). However, there was a dose-dependent requirement for the presence of Hex for the proper development of in vitro EB definitive BFU-E (iii), CFU-GEMM (iv), CFU-GM (v), and CFU-Meg (vi) hematopoietic progenitors. All assays were performed 3 times in triplicate. (C) Measurement of in vivo day-8.5 yolk sac hematopoietic progenitor formation. Both primitive and definitive hematopoietic progenitors are present at this stage and are measured together in this assay. The lack of Hex also reduces the ability of yolk sacs to develop in vivo in hematopoietic progenitors (n = 41 embryos). Error bars represent SEM.
The effect ofHexdeletion on embryonic hematopoiesis. (A) RT-PCR and morphology of BL-CFC hemangioblast colonies. In vitro formation of definitive hemangioblasts from day-3 EBs expresses low levels of Brachyury, but high levels of Runx1 and Flk-1, and has characteristic BL-CFC colony morphology.1,4 Hex is not expressed in these BL-CFCs by RT-PCR. BL-CFC colony morphology is shown at × 40 magnification. (B) Primitive and definitive hematopoietic progenitor assays from in vitro differentiation of Hex-/-, Hex+/-, and Hex+/+ EBs. There was no effect of the deletion of the homeoprotein Hex on in vitro formation in day-3 EB hemangioblasts (i) and day-6 EB EryPs (ii). However, there was a dose-dependent requirement for the presence of Hex for the proper development of in vitro EB definitive BFU-E (iii), CFU-GEMM (iv), CFU-GM (v), and CFU-Meg (vi) hematopoietic progenitors. All assays were performed 3 times in triplicate. (C) Measurement of in vivo day-8.5 yolk sac hematopoietic progenitor formation. Both primitive and definitive hematopoietic progenitors are present at this stage and are measured together in this assay. The lack of Hex also reduces the ability of yolk sacs to develop in vivo in hematopoietic progenitors (n = 41 embryos). Error bars represent SEM.
There is evidence that there is a distinct hemangioblast population for primitive compared with definitive hematopoiesis.4 The lack of Hex might have different effects on primitive versus definitive embryonic hematopoiesis. Therefore, the effect of deleting Hex on the formation of definitive hematopoietic progenitors was analyzed (Figure 1B). Hex+/+ EB cells had 2.5-fold more definitive colony-forming units-granulocyte/macrophage (CFUGMs) than Hex+/- EB cells, and 96-fold more CFU-GMs than Hex-/- EBs. Hex+/+ EB cells had 4.4-fold more definitive colony-forming units-granulocyte/erythroid/macrophage/megakaryocyte (CFU-GEMMs) than Hex+/- EB cells, and 55-fold more CFUGEMMs than Hex-/- EB cells. Hex+/+ EB cells had 5.1-fold more definitive blast-forming units-erythroid (BFU-Es) than Hex+/- EB cells, and 9.5-fold more definitive BFU-Es than Hex-/- EB cells. Hex+/+ EB cells had 5.7-fold more colony-forming units-megakaryocyte (CFU-Meg's) than Hex+/- EB cells. No CFUMeg's developed from the Hex-/- EB cells. Thus, the lack of one Hex allele statistically significantly decreased the ability of EBs to form definitive hematopoietic progenitors, whereas the lack of both alleles almost completely abrogated the ability of EBs to form such progenitors.
To assess whether the hematopoietic defect observed with Hex deletion in vitro was also present in vivo, the affect of Hex deletion on yolk sac hematopoietic progenitor formation was tested (Figure 1C). Yolk sacs from day-8.5 pregnant Hex+/- female mice were microdissected and hematopoietic progenitors quantified by colony-formation analysis. By day 8.5, both primitive and definitive hematopoietic progenitors are present in the yolk sac, and both will be counted in these colony-formation assays. There was no statistical difference between the number of Hex+/+ and Hex+/- yolk sac hematopoietic progenitors. However, Hex+/+ yolk sacs had 1.7-fold more BFU-Es, 2.4-fold more CFU-GMs, and 2.1-fold more CFU-GEMMs than did Hex-/- yolk sacs. Heterogeneity was noted in the number of Hex-/- yolk sac progenitors, where 8 of 12 Hex-/- yolk sacs had essentially no progenitors and 4 of 12 had near normal numbers. On average, these data demonstrated that Hex deletion also decreased in vivo yolk sac hematopoietic progenitor development in addition to in vitro EBs.
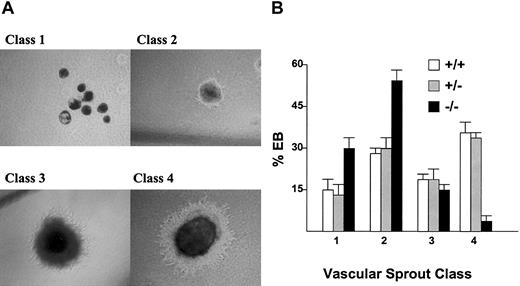
Since the hemangioblast gives rise to endothelial cells as well as hematopoietic cells, the effect of Hex deletion on vascular sprout formation was assessed using in vitro EB differentiation (Figure 2A). Hex+/+ EBs cultured in conditions that stimulated vascular sprout formation had 33.6% of the EBs form mature class IV vascular sprouts (abundant networked tubules), whereas Hex+/- cells had 32.8% and Hex-/- cells had 6.7% of the EBs form class IV vascular sprouts (Figure 2B).
The effect ofHexdeletion on in vitro vascular sprout formation in differentiating EBs. (A) The 4 classes of vascular sprout development in EBs, as described in “Materials and methods.” Original magnification, × 10. (B) The percent of EBs in each class by Hex genotype. There were fewer CD31+ mature vascular tubules in the Hex-/- EBs. Assays were preformed 3 times in triplicate. Error bars represent SEM.
The effect ofHexdeletion on in vitro vascular sprout formation in differentiating EBs. (A) The 4 classes of vascular sprout development in EBs, as described in “Materials and methods.” Original magnification, × 10. (B) The percent of EBs in each class by Hex genotype. There were fewer CD31+ mature vascular tubules in the Hex-/- EBs. Assays were preformed 3 times in triplicate. Error bars represent SEM.
Flow cytometric analysis of Hex-/- differentiating day-6 EBs demonstrated decreased vascular endothelial (VE)-Cadherin-positive endothelial cells and decreased Gr-1+ myeloid cells compared with Hex+/+ EBs. Hex-/- EBs had an average of 2.9% VE-Cadherin-positive cells, whereas Hex+/+ EBs had an average of 5.6%. Hex-/- EBs had an average of 1.2% Gr-1-positive cells, whereas Hex+/+ EBs had an average of 8.9% Gr-1-positive cells.
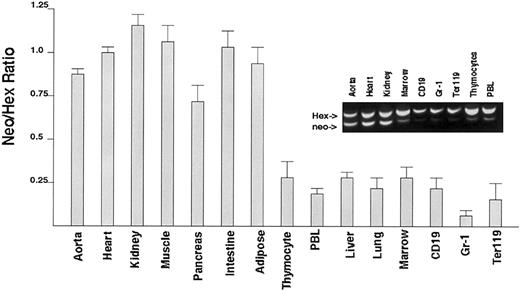
To assess whether Hex-/- ES cells could contribute to hematopoietic tissue in an adult mouse, Hex+/+ and Hex-/- chimeric mice were generated by blastocyst mixing. The percent contribution of Hex-/- ES cells to specific hematopoietic tissues was characterized by the ratio of neo (from Hex-/- cells) to Hex (from Hex+/+ cells) DNA (Figure 3). Carefully isolated marrow, lung, liver, total peripheral blood leukocytes, and thymocytes all had significantly decreased contribution of Hex-/- ES cells as demonstrated by a lack of neo sequences (Hex deletion) compared with the presence of normal Hex DNA. In addition, Hex-/- cells contributed very little to the flow sorted CD19+ B cells, Gr-1+ mature granulocytes, and Ter119+ erythroid cells. The unexpected finding that the lung also had few cells that did not contain Hex indicates that it also may require Hex for development. Hex-/- mice die before the lung is fully developed, and therefore the role of Hex in lung organogenesis has not been defined. The finding that Hex-/- ES cells could not contribute to both lymphoid as well as myeloid lineages implicates a requirement by the hematopoietic stem cell (HSC) for Hex. Hex-/- contribution to endothelial cells could not be accurately assessed because of contamination with other tissues, especially smooth muscle, which does not require Hex.
Analysis of the ability ofHex-/-ES cells to contribute hematopoietic tissues in chimeric mice. The ratio of Hex-/- DNA to Hex+/+ DNA in specific organs or flow cytometrically sorted individual hematopoietic cell types was obtained using normalized multiplex PCR. The expected ratio would be 1.0 if both Hex-/- and Hex+/+ cells contributed without bias to tissue development. There were only rare contributions from Hex-/- ES cells to any hematopoietic cell type. Aorta will contain smooth muscle, which does not require Hex. Each ratio is the averaged value from 4 distinct mice analyzed individually. Error bars represent SEM. Inset shows genomic PCR of Hex chimeric mice.
Analysis of the ability ofHex-/-ES cells to contribute hematopoietic tissues in chimeric mice. The ratio of Hex-/- DNA to Hex+/+ DNA in specific organs or flow cytometrically sorted individual hematopoietic cell types was obtained using normalized multiplex PCR. The expected ratio would be 1.0 if both Hex-/- and Hex+/+ cells contributed without bias to tissue development. There were only rare contributions from Hex-/- ES cells to any hematopoietic cell type. Aorta will contain smooth muscle, which does not require Hex. Each ratio is the averaged value from 4 distinct mice analyzed individually. Error bars represent SEM. Inset shows genomic PCR of Hex chimeric mice.
To validate the cell biology findings above, and to identify genes that Hex might regulate, the gene expression patterns of the day-6 Hex+/+ EBs were compared with the expression patterns of Hex-/- EBs using microarray analysis (Tables 1, 2). This analysis found that a number of hematopoietic- and endothelial-specific genes demonstrated decreased expression in day-6 EBs when Hex was deleted, and several mesodermal or hemangioblast genes were increased. For example, definitive erythroid-specific proteins such as alpha- and beta-globin, Band 4.2, glycophorin A, and T-cell surface markers such as cytotoxic T-lymphocyte antigen 4 (CTLA4) were decreased when Hex was absent. Transcription factors important in hematopoiesis such as EKLF, Gata-1, NF-AT, and PPARgamma were also decreased. The liver hematopoietic stem/progenitor cell marker AA4 was decreased, but there was no change in the levels of Flk-1, Runx1, or Scl transcripts. This is consistent with the lack of a change in Flk-1 expression in the Hex-/- day-9 embryos.16 However, BMP-4, an essential inducer of hematopoietic mesoderm, was increased. The endothelial marker VE-Cadherin was decreased when Hex was absent, consistent with the flow cytometry data. In addition, the endothelial genes Tie1, tissue factor pathway inhibitor (TFPI), and lipoprotein lipase were also decreased in the microarray analysis.
Down-regulated genes in day-6 Hex−/− embryoid bodies
P . | Fold change . | Gene . |
|---|---|---|
| .002653 | −3.78 | X04480: Insulin-like growth factor 1 |
| .010953 | −3.75 | X02677: Solute carrier family 4 (anion exchanger) |
| .000017 | −3.56 | X58196: H19 fetal liver |
| .001568 | −2.78 | AF081789: Cell surface antigen AA4 |
| .031296 | −2.57 | U82758: Claudin 5 |
| .000067 | −2.52 | Y17851: Ganglioside-induced differentiation-associated protein 2 |
| .001432 | −2.40 | AF057527: Erythrocyte membrane protein Rh50 (Rhag) |
| .001569 | −2.37 | AW060556: Stabilin 1 (Stab1) |
| .004042 | −2.34 | D49544: Kinesin family member C1 (Kifc1) |
| .003058 | −2.29 | AI853217: VE-cadherin |
| .001761 | −2.28 | J04696: Mouse glutathione S-transferase class mu (GST5-5) |
| .000048 | −2.23 | L17076: HnRNP-associated with lethal yellow RNA-binding protein mRNA |
| .000672 | −2.18 | V00722: Beta-1-globin |
| .004949 | −2.12 | AW046391: Hypothetical protein |
| .000210 | −2.10 | J00413: Hemoglobin, beta adult major chain |
| .002405 | −2.02 | AF087644: Coagulation factor X |
| .032201 | −2.01 | U04055: Erythrocyte protein band 4.2 |
| .018315 | −1.98 | AV303514: Phosphatidylinositol-4-phosphate 5-kinase, type II |
| .000977 | −1.98 | D16215: Flavin-containing monooxygenase |
| .008922 | −1.96 | AW048779: hypothetical gene supported by BC027302 |
| .003111 | −1.94 | X15591: Cytotoxic T lymphocyte-associated protein 2 alpha |
| .017759 | −1.89 | AV266785: Complement component 1 subunit r |
| .010371 | −1.88 | M26385: Glycophorin A |
| .000978 | −1.88 | AJ250490: Receptor activity modifying protein 2 (Ramp2 gene) |
| .015505 | −1.85 | AA690483: Unknown Mus musculus adult male liver tumor cDNA |
| .025388 | −1.83 | AA673486: Unknown Mus musculus 4 days neonate thymus and hematopoietic stem cell cDNA |
| .011468 | −1.81 | AW049647: ADP ribosylation factor 6 interacting protein 5 |
| .000226 | −1.78 | U19604: DNA ligase I, ATP dependent |
| .009390 | −1.78 | AI845915: POL 1 and transcript release factor |
| .023288 | −1.77 | M15501: Actin, alpha, cardiac |
| .018349 | −1.75 | AV235418: Tyrosine kinase receptor 1 (Tie 1) |
| .011321 | −1.74 | X14061: Hemoglobin beta, pseudogene bh3 |
| .008537 | −1.73 | AF017275: Growth factor-independent 1B |
| .017719 | −1.73 | AF110520: NADH oxidoreductase |
| .017687 | −1.71 | X15763: GATA-1 |
| .003072 | −1.70 | U95736: Frataxin |
| .015075 | −1.70 | AF004833: Tissue factor pathway inhibitor (TFPI) |
| .000928 | −1.69 | M27938: Male enhanced antigen 1 |
| .026540 | −1.68 | M97200: Erythroid Kruppel-like factor (EKLF) |
| .013354 | −1.66 | AA285978: ADP-ribosylation-like factor 6 interacting protein 5 (Arl6ip5) |
| .006031 | −1.66 | AF084524: EIA-stimulated genes CREG |
| .013729 | −1.64 | AF647239: Unknown adult male kidney cDNA |
| .009494 | −1.62 | AA726364: Lipoprotein lipase (Lpl) |
| .034037 | −1.61 | AW124226: Unknown adult male hippocampus cDNA |
| .004018 | −1.60 | AF087434: NF-ATc isoform a (NF-ATc) |
| .021581 | −1.60 | U20619: Serine-rich RNA polymerase suppressor protein 1 (SRP1) |
| .020747 | −1.60 | AW124340: Unknown Mus musculus 2 days neonate thymic cells cDNA, similar to zinc transporter |
| .012994 | −1.59 | AI840130: Src activating and signaling molecule (Srcasm) |
| .019533 | −1.59 | M35153: Lamin B1 |
| .006731 | −1.56 | AW125390: Unknown Mus musculus 18-day embryo whole body cDNA |
| .012535 | −1.56 | AB011665: BAZF, complete cds/cds = (24,1448) |
| .001342 | −1.56 | AI844357: Immature colon carcinoma transcript 1 |
| .017822 | −1.55 | V00714: Hemoglobin alpha, adult chain 1 |
| .043322 | −1.55 | AA755260: Deleted in polyposis 1-like 1 |
| .029021 | −1.55 | AA726223: Meltrin beta metalloproteinase |
| .009957 | −1.55 | AI849082: Goliath-related zinc finger protein |
| .013799 | −1.55 | AI197161: ELL-related RNA polymerase II, elongation factor |
| .002423 | −1.54 | AF000294: Peroxisome proliferator-activated receptor gamma binding protein (PPARgamma) |
| .013278 | −1.54 | U34691: Uroporphyrinogen decarboxylase |
| .009814 | −1.53 | AJ010045: Rho guanine nucleotide-exchange factor |
| .006577 | −1.51 | AI836162: SRY-box containing gene 18 (Sox18) |
| .028268 | −1.51 | AI155192: BIRC 5 (Baculovirus IAP-repeat containing 5) |
| .005933 | −1.50 | L35528: Manganese superoxide dismutase (MnSOD) gene |
| .021623 | −1.50 | AI841295: Glutathione S-transferase subunit 13 |
| .003194 | −1.50 | X77952: Endoglin |
| .007243 | −1.50 | L07508: Golli-mpb |
| .003337 | −1.49 | AW050240: Casein kinase II, alpha I-related sequence 4 |
| .007000 | −1.49 | AB007136: Protease (prosome, macropain) 28 subunit, alpha |
| .011080 | −1.48 | Y17343: Protein-tyrosine-phosphatase IF2P |
| .002985 | −1.48 | AW122685: Similar to TMP21 vesicular transport protein |
| .000312 | −1.47 | AF069051: Pituitary tumor transforming gene protein (PTTG) |
| .014099 | −1.47 | AF004326: Angiopoietin 2 |
| .003649 | −1.47 | AI842600: Pre-T cell RS21-C6 (Tdrg-TL1) protein |
| .000485 | −1.47 | AI836509: Unknown 12 days embryo male wolffian duct |
| .012981 | −1.47 | D85818: RNA polymerase II subuunit RPB14 |
| .004562 | −1.47 | AI848107: Unknown Mus musculus 18-day embryo whole body cDNA |
| .023514 | −1.46 | J04181: A-X actin mRNA |
P . | Fold change . | Gene . |
|---|---|---|
| .002653 | −3.78 | X04480: Insulin-like growth factor 1 |
| .010953 | −3.75 | X02677: Solute carrier family 4 (anion exchanger) |
| .000017 | −3.56 | X58196: H19 fetal liver |
| .001568 | −2.78 | AF081789: Cell surface antigen AA4 |
| .031296 | −2.57 | U82758: Claudin 5 |
| .000067 | −2.52 | Y17851: Ganglioside-induced differentiation-associated protein 2 |
| .001432 | −2.40 | AF057527: Erythrocyte membrane protein Rh50 (Rhag) |
| .001569 | −2.37 | AW060556: Stabilin 1 (Stab1) |
| .004042 | −2.34 | D49544: Kinesin family member C1 (Kifc1) |
| .003058 | −2.29 | AI853217: VE-cadherin |
| .001761 | −2.28 | J04696: Mouse glutathione S-transferase class mu (GST5-5) |
| .000048 | −2.23 | L17076: HnRNP-associated with lethal yellow RNA-binding protein mRNA |
| .000672 | −2.18 | V00722: Beta-1-globin |
| .004949 | −2.12 | AW046391: Hypothetical protein |
| .000210 | −2.10 | J00413: Hemoglobin, beta adult major chain |
| .002405 | −2.02 | AF087644: Coagulation factor X |
| .032201 | −2.01 | U04055: Erythrocyte protein band 4.2 |
| .018315 | −1.98 | AV303514: Phosphatidylinositol-4-phosphate 5-kinase, type II |
| .000977 | −1.98 | D16215: Flavin-containing monooxygenase |
| .008922 | −1.96 | AW048779: hypothetical gene supported by BC027302 |
| .003111 | −1.94 | X15591: Cytotoxic T lymphocyte-associated protein 2 alpha |
| .017759 | −1.89 | AV266785: Complement component 1 subunit r |
| .010371 | −1.88 | M26385: Glycophorin A |
| .000978 | −1.88 | AJ250490: Receptor activity modifying protein 2 (Ramp2 gene) |
| .015505 | −1.85 | AA690483: Unknown Mus musculus adult male liver tumor cDNA |
| .025388 | −1.83 | AA673486: Unknown Mus musculus 4 days neonate thymus and hematopoietic stem cell cDNA |
| .011468 | −1.81 | AW049647: ADP ribosylation factor 6 interacting protein 5 |
| .000226 | −1.78 | U19604: DNA ligase I, ATP dependent |
| .009390 | −1.78 | AI845915: POL 1 and transcript release factor |
| .023288 | −1.77 | M15501: Actin, alpha, cardiac |
| .018349 | −1.75 | AV235418: Tyrosine kinase receptor 1 (Tie 1) |
| .011321 | −1.74 | X14061: Hemoglobin beta, pseudogene bh3 |
| .008537 | −1.73 | AF017275: Growth factor-independent 1B |
| .017719 | −1.73 | AF110520: NADH oxidoreductase |
| .017687 | −1.71 | X15763: GATA-1 |
| .003072 | −1.70 | U95736: Frataxin |
| .015075 | −1.70 | AF004833: Tissue factor pathway inhibitor (TFPI) |
| .000928 | −1.69 | M27938: Male enhanced antigen 1 |
| .026540 | −1.68 | M97200: Erythroid Kruppel-like factor (EKLF) |
| .013354 | −1.66 | AA285978: ADP-ribosylation-like factor 6 interacting protein 5 (Arl6ip5) |
| .006031 | −1.66 | AF084524: EIA-stimulated genes CREG |
| .013729 | −1.64 | AF647239: Unknown adult male kidney cDNA |
| .009494 | −1.62 | AA726364: Lipoprotein lipase (Lpl) |
| .034037 | −1.61 | AW124226: Unknown adult male hippocampus cDNA |
| .004018 | −1.60 | AF087434: NF-ATc isoform a (NF-ATc) |
| .021581 | −1.60 | U20619: Serine-rich RNA polymerase suppressor protein 1 (SRP1) |
| .020747 | −1.60 | AW124340: Unknown Mus musculus 2 days neonate thymic cells cDNA, similar to zinc transporter |
| .012994 | −1.59 | AI840130: Src activating and signaling molecule (Srcasm) |
| .019533 | −1.59 | M35153: Lamin B1 |
| .006731 | −1.56 | AW125390: Unknown Mus musculus 18-day embryo whole body cDNA |
| .012535 | −1.56 | AB011665: BAZF, complete cds/cds = (24,1448) |
| .001342 | −1.56 | AI844357: Immature colon carcinoma transcript 1 |
| .017822 | −1.55 | V00714: Hemoglobin alpha, adult chain 1 |
| .043322 | −1.55 | AA755260: Deleted in polyposis 1-like 1 |
| .029021 | −1.55 | AA726223: Meltrin beta metalloproteinase |
| .009957 | −1.55 | AI849082: Goliath-related zinc finger protein |
| .013799 | −1.55 | AI197161: ELL-related RNA polymerase II, elongation factor |
| .002423 | −1.54 | AF000294: Peroxisome proliferator-activated receptor gamma binding protein (PPARgamma) |
| .013278 | −1.54 | U34691: Uroporphyrinogen decarboxylase |
| .009814 | −1.53 | AJ010045: Rho guanine nucleotide-exchange factor |
| .006577 | −1.51 | AI836162: SRY-box containing gene 18 (Sox18) |
| .028268 | −1.51 | AI155192: BIRC 5 (Baculovirus IAP-repeat containing 5) |
| .005933 | −1.50 | L35528: Manganese superoxide dismutase (MnSOD) gene |
| .021623 | −1.50 | AI841295: Glutathione S-transferase subunit 13 |
| .003194 | −1.50 | X77952: Endoglin |
| .007243 | −1.50 | L07508: Golli-mpb |
| .003337 | −1.49 | AW050240: Casein kinase II, alpha I-related sequence 4 |
| .007000 | −1.49 | AB007136: Protease (prosome, macropain) 28 subunit, alpha |
| .011080 | −1.48 | Y17343: Protein-tyrosine-phosphatase IF2P |
| .002985 | −1.48 | AW122685: Similar to TMP21 vesicular transport protein |
| .000312 | −1.47 | AF069051: Pituitary tumor transforming gene protein (PTTG) |
| .014099 | −1.47 | AF004326: Angiopoietin 2 |
| .003649 | −1.47 | AI842600: Pre-T cell RS21-C6 (Tdrg-TL1) protein |
| .000485 | −1.47 | AI836509: Unknown 12 days embryo male wolffian duct |
| .012981 | −1.47 | D85818: RNA polymerase II subuunit RPB14 |
| .004562 | −1.47 | AI848107: Unknown Mus musculus 18-day embryo whole body cDNA |
| .023514 | −1.46 | J04181: A-X actin mRNA |
Microarray analysis of gene expression patterns in Hex+/+ versus Hex−/− EBs. Note that a number of hematopoietic- and endothelial-specific genes exhibit decreased expression. These data represent averages and statistical analysis from 4 distinct Hex+/+ and 4 distinct Hex−/− day-6 EB experiments performed using individual gene chips, for each experiment (total 8 chips).
Up-regulated genes in day-6 Hex−/− embryoid bodies
P . | Fold change . | Gene . |
|---|---|---|
| .019536 | 1.50 | X70764: ELKL motif kinase |
| .025304 | 1.50 | S78454: Metal response element binding transcription factor 2 |
| .101605 | 1.51 | D26046: AT motif binding factor 1 |
| .009346 | 1.51 | X71978: Fused toes |
| .008006 | 1.51 | X80171: Placental growth factor |
| .017930 | 1.53 | Y15910: Dia protein NAP-22 membrane attached signal protein 1 |
| .031838 | 1.53 | L47480: BMP-4 gene |
| .024428 | 1.53 | AF069953: G protein gamma 3 subunit (Gng3) |
| .015906 | 1.54 | AA733868: CD2 antigen (cytoplasmic tail) binding protein 2 (Cd2bp2) |
| .012801 | 1.54 | AW123115: Reticulon 1 (Rtn1) |
| .001579 | 1.55 | AF041472: Ataxin-2 |
| .001456 | 1.56 | AV276058: SCA2 |
| .022530 | 1.56 | U92885: LERK-3 (Epl3) |
| .022782 | 1.56 | M87802: Homeobox D |
| .003225 | 1.57 | AJ002366: Transcription factor TFIIH |
| .102759 | 1.57 | AA600647: Rag D |
| .005952 | 1.58 | AA795486: HIV-1 Rev binding protein |
| .010549 | 1.59 | AW048233: Ets2 repressor factor |
| .027050 | 1.59 | AW125031: Chemokine-like super family 6 |
| .017595 | 1.59 | AI848841: Unknown Mus musculus cDNA |
| .003082 | 1.60 | D76440: Necdin |
| .009446 | 1.61 | AA189677: Similar to Luc7 homolog (S cerevisiae) |
| .000965 | 1.62 | AV171460: Unknown Mus musculus 13 days embryo forelimb cDNA |
| .058823 | 1.63 | U36778: Tal1 interrupting locus (Sil) |
| .003858 | 1.63 | AB020886: SSeCKS |
| .000481 | 1.64 | AJ001166: PXMP1-L (PMP69, P70R) |
| .000044 | 1.65 | AA268823: CD59B |
| .023294 | 1.66 | AI838398: Capicua (Cic) |
| .010314 | 1.66 | AI848050: Kruppel-like factor 9 (Klf9) |
| .008523 | 1.67 | AF039021: Pericentriolar material 1 (Pcm1) |
| .016667 | 1.67 | AW121328: T-Box transcription factor 3 |
| .013826 | 1.67 | AA981581: Unknown thymus cDNA |
| .027554 | 1.67 | U05252: Nuclear matrix attachment DNA-binding protein SATB1 |
| .012993 | 1.68 | U88567: Secreted frizzled-related protein sFRP-2 (Sfrp2) |
| .001160 | 1.69 | D83966: Protein tyrosine phosphatase, nonreceptor type 13 |
| .022100 | 1.73 | X68363: POU domain, class 2, transcription factor 1 |
| .000117 | 1.73 | X96639: Exostoses-1 |
| .011775 | 1.77 | AI042802: Solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (Slc6a6) |
| .001983 | 1.77 | AW061234: Unknown Mus musculus cDNA |
| .002101 | 1.77 | AW121960: Unknown Mus musculus adult male aorta cDNA |
| .007887 | 1.77 | U13837: Vacuolar adenosine triphosphatase subunit A |
| .029744 | 1.78 | X73230: Arylsulfatase A |
| .007617 | 1.78 | AJ002730: Ubiquitously transcribed tetratricopeptide repeat |
| .003410 | 1.78 | AW046101: Unknown protein, contains gaunylate kinase domain |
| .013035 | 1.80 | X75285: Fibulin 2 |
| .013581 | 1.80 | AB024538: Islr (immunoglobulin superfamily containing leucine-rich repeat) |
| .006885 | 1.81 | AW046038: Transcriptional coactivator (Taz gene) |
| .000209 | 1.82 | U06944: Praja1 |
| .000066 | 1.86 | AF022432: Mus musculus matrix metalloproteinase-14 (Mmp14) |
| .001276 | 1.87 | X68363: POU domain, class 2, transcription factor 1 |
| .009344 | 1.89 | D10011: Glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| .001754 | 1.89 | U22325: Faciogenital dysplasia homolog |
| .009816 | 1.90 | X78989: Testin |
| .007261 | 1.91 | M19380: Calmodulin 3 |
| .000211 | 1.93 | X83569: Neuronatin |
| .030738 | 1.94 | AJ243608: Ns7 protein, ortholog of the human nM15 gene, melanoma antigen, family L, 2 (Magel2) |
| .008798 | 1.95 | AW122897: Frizzled homolog 4 (Drosophila) |
| .021436 | 2.00 | U02098: Purine-rich element binding protein A (Pura) |
| .029488 | 2.01 | AA711516: Similar to splicing factor, arginine/serine-rich 7 |
| .017521 | 2.02 | AI839004: Long-chain fatty-acyl elongase (Lce) |
| .008565 | 2.04 | AI852838: Dlk1 gene for delta-like and Gtl2 gene |
| .024052 | 2.05 | AW121323: Similar to yeast URM1: ubiquitin-like protein |
| .008120 | 2.07 | U14941: ELF-1 |
| .002046 | 2.08 | X16672: Type IIB intracisternal A-particle element encoding integrase and gag |
| .013533 | 2.08 | M97590: Protein tyrosine phosphatase-1 (PTP-1) |
| .003222 | 2.08 | AF056187: Insulin-like growth factor I receptor |
| .003911 | 2.15 | AI851048: Rab6, RAS family member |
| .005097 | 2.17 | X65138: Eph receptor A4 |
| .006860 | 2.18 | AA759910: Similar to e(y)2 homolog, 12 days embryo male wolffian duct |
| .009977 | 2.26 | AW260482: NMDA receptor-regulated gene 1 (Narg1) |
| .031771 | 2.31 | AW048581: Ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| .008367 | 2.37 | AI642417: Unknown Mus musculus cDNA, 3 end |
| .001441 | 2.49 | AW107922: SRY-box containing gene 11 (Sox11) |
| .030771 | 2.49 | U82122: Ubiquitin conjugating enzyme E3A |
| .022656 | 2.49 | M95200: Vascular endothelial growth factor (VEGF) |
| .004646 | 2.53 | D50086: Neuropilin |
| .007256 | 2.62 | AI645050: Sartine nexin 5 |
| .015276 | 2.62 | AA795284: Hypothetical protein, containing Tol transport and WD40 domains |
| .004607 | 2.75 | X60136: Trans-acting transcription factor 1 |
| .004792 | 2.77 | AI503821: Zinc finger protein 144 (Zfp144) |
| .001863 | 2.88 | U92437: Phosphatase and tensin homolog |
| .003833 | 2.93 | U85614: SRG3 |
| .012519 | 2.97 | AW209098: IQ motif containing GTPase activating protein 1 |
| .031953 | 3.13 | AW212708: Unknown Mus musculus 10, 11 days embryo whole body cDNA, contains FOG zinc fingers |
| .032497 | 3.26 | X84037: E-selectin ligand-1 |
| .003827 | 3.28 | AI551087: Mus musculus hypothetical gene supported by BC020078, containing elF4 domains |
| .013229 | 3.38 | AI449034: RE1-silencing transcription factor (Rest) |
| .006480 | 3.39 | AI853712: Kruppel-like factor 7 (Klf-7) |
| .001798 | 3.45 | AI843739: Transducin beta-like 1X protein |
| .000640 | 3.46 | AW123697: CD99 (E2 antigen, Mic2 protein) |
| .025527 | 3.52 | C80249: Unknown Mus musculus blastocyst cDNA |
| .006833 | 4.32 | AV299153: DEAD/H box polypeptide 36 protein (Ddx36) |
| .001673 | 4.70 | C79248: Unknown Mus musculus adult pancreas islet cells and blastocyst cDNA |
| .007836 | 5.08 | AI854614: Bromodomain-containing protein BRD4 |
P . | Fold change . | Gene . |
|---|---|---|
| .019536 | 1.50 | X70764: ELKL motif kinase |
| .025304 | 1.50 | S78454: Metal response element binding transcription factor 2 |
| .101605 | 1.51 | D26046: AT motif binding factor 1 |
| .009346 | 1.51 | X71978: Fused toes |
| .008006 | 1.51 | X80171: Placental growth factor |
| .017930 | 1.53 | Y15910: Dia protein NAP-22 membrane attached signal protein 1 |
| .031838 | 1.53 | L47480: BMP-4 gene |
| .024428 | 1.53 | AF069953: G protein gamma 3 subunit (Gng3) |
| .015906 | 1.54 | AA733868: CD2 antigen (cytoplasmic tail) binding protein 2 (Cd2bp2) |
| .012801 | 1.54 | AW123115: Reticulon 1 (Rtn1) |
| .001579 | 1.55 | AF041472: Ataxin-2 |
| .001456 | 1.56 | AV276058: SCA2 |
| .022530 | 1.56 | U92885: LERK-3 (Epl3) |
| .022782 | 1.56 | M87802: Homeobox D |
| .003225 | 1.57 | AJ002366: Transcription factor TFIIH |
| .102759 | 1.57 | AA600647: Rag D |
| .005952 | 1.58 | AA795486: HIV-1 Rev binding protein |
| .010549 | 1.59 | AW048233: Ets2 repressor factor |
| .027050 | 1.59 | AW125031: Chemokine-like super family 6 |
| .017595 | 1.59 | AI848841: Unknown Mus musculus cDNA |
| .003082 | 1.60 | D76440: Necdin |
| .009446 | 1.61 | AA189677: Similar to Luc7 homolog (S cerevisiae) |
| .000965 | 1.62 | AV171460: Unknown Mus musculus 13 days embryo forelimb cDNA |
| .058823 | 1.63 | U36778: Tal1 interrupting locus (Sil) |
| .003858 | 1.63 | AB020886: SSeCKS |
| .000481 | 1.64 | AJ001166: PXMP1-L (PMP69, P70R) |
| .000044 | 1.65 | AA268823: CD59B |
| .023294 | 1.66 | AI838398: Capicua (Cic) |
| .010314 | 1.66 | AI848050: Kruppel-like factor 9 (Klf9) |
| .008523 | 1.67 | AF039021: Pericentriolar material 1 (Pcm1) |
| .016667 | 1.67 | AW121328: T-Box transcription factor 3 |
| .013826 | 1.67 | AA981581: Unknown thymus cDNA |
| .027554 | 1.67 | U05252: Nuclear matrix attachment DNA-binding protein SATB1 |
| .012993 | 1.68 | U88567: Secreted frizzled-related protein sFRP-2 (Sfrp2) |
| .001160 | 1.69 | D83966: Protein tyrosine phosphatase, nonreceptor type 13 |
| .022100 | 1.73 | X68363: POU domain, class 2, transcription factor 1 |
| .000117 | 1.73 | X96639: Exostoses-1 |
| .011775 | 1.77 | AI042802: Solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (Slc6a6) |
| .001983 | 1.77 | AW061234: Unknown Mus musculus cDNA |
| .002101 | 1.77 | AW121960: Unknown Mus musculus adult male aorta cDNA |
| .007887 | 1.77 | U13837: Vacuolar adenosine triphosphatase subunit A |
| .029744 | 1.78 | X73230: Arylsulfatase A |
| .007617 | 1.78 | AJ002730: Ubiquitously transcribed tetratricopeptide repeat |
| .003410 | 1.78 | AW046101: Unknown protein, contains gaunylate kinase domain |
| .013035 | 1.80 | X75285: Fibulin 2 |
| .013581 | 1.80 | AB024538: Islr (immunoglobulin superfamily containing leucine-rich repeat) |
| .006885 | 1.81 | AW046038: Transcriptional coactivator (Taz gene) |
| .000209 | 1.82 | U06944: Praja1 |
| .000066 | 1.86 | AF022432: Mus musculus matrix metalloproteinase-14 (Mmp14) |
| .001276 | 1.87 | X68363: POU domain, class 2, transcription factor 1 |
| .009344 | 1.89 | D10011: Glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| .001754 | 1.89 | U22325: Faciogenital dysplasia homolog |
| .009816 | 1.90 | X78989: Testin |
| .007261 | 1.91 | M19380: Calmodulin 3 |
| .000211 | 1.93 | X83569: Neuronatin |
| .030738 | 1.94 | AJ243608: Ns7 protein, ortholog of the human nM15 gene, melanoma antigen, family L, 2 (Magel2) |
| .008798 | 1.95 | AW122897: Frizzled homolog 4 (Drosophila) |
| .021436 | 2.00 | U02098: Purine-rich element binding protein A (Pura) |
| .029488 | 2.01 | AA711516: Similar to splicing factor, arginine/serine-rich 7 |
| .017521 | 2.02 | AI839004: Long-chain fatty-acyl elongase (Lce) |
| .008565 | 2.04 | AI852838: Dlk1 gene for delta-like and Gtl2 gene |
| .024052 | 2.05 | AW121323: Similar to yeast URM1: ubiquitin-like protein |
| .008120 | 2.07 | U14941: ELF-1 |
| .002046 | 2.08 | X16672: Type IIB intracisternal A-particle element encoding integrase and gag |
| .013533 | 2.08 | M97590: Protein tyrosine phosphatase-1 (PTP-1) |
| .003222 | 2.08 | AF056187: Insulin-like growth factor I receptor |
| .003911 | 2.15 | AI851048: Rab6, RAS family member |
| .005097 | 2.17 | X65138: Eph receptor A4 |
| .006860 | 2.18 | AA759910: Similar to e(y)2 homolog, 12 days embryo male wolffian duct |
| .009977 | 2.26 | AW260482: NMDA receptor-regulated gene 1 (Narg1) |
| .031771 | 2.31 | AW048581: Ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| .008367 | 2.37 | AI642417: Unknown Mus musculus cDNA, 3 end |
| .001441 | 2.49 | AW107922: SRY-box containing gene 11 (Sox11) |
| .030771 | 2.49 | U82122: Ubiquitin conjugating enzyme E3A |
| .022656 | 2.49 | M95200: Vascular endothelial growth factor (VEGF) |
| .004646 | 2.53 | D50086: Neuropilin |
| .007256 | 2.62 | AI645050: Sartine nexin 5 |
| .015276 | 2.62 | AA795284: Hypothetical protein, containing Tol transport and WD40 domains |
| .004607 | 2.75 | X60136: Trans-acting transcription factor 1 |
| .004792 | 2.77 | AI503821: Zinc finger protein 144 (Zfp144) |
| .001863 | 2.88 | U92437: Phosphatase and tensin homolog |
| .003833 | 2.93 | U85614: SRG3 |
| .012519 | 2.97 | AW209098: IQ motif containing GTPase activating protein 1 |
| .031953 | 3.13 | AW212708: Unknown Mus musculus 10, 11 days embryo whole body cDNA, contains FOG zinc fingers |
| .032497 | 3.26 | X84037: E-selectin ligand-1 |
| .003827 | 3.28 | AI551087: Mus musculus hypothetical gene supported by BC020078, containing elF4 domains |
| .013229 | 3.38 | AI449034: RE1-silencing transcription factor (Rest) |
| .006480 | 3.39 | AI853712: Kruppel-like factor 7 (Klf-7) |
| .001798 | 3.45 | AI843739: Transducin beta-like 1X protein |
| .000640 | 3.46 | AW123697: CD99 (E2 antigen, Mic2 protein) |
| .025527 | 3.52 | C80249: Unknown Mus musculus blastocyst cDNA |
| .006833 | 4.32 | AV299153: DEAD/H box polypeptide 36 protein (Ddx36) |
| .001673 | 4.70 | C79248: Unknown Mus musculus adult pancreas islet cells and blastocyst cDNA |
| .007836 | 5.08 | AI854614: Bromodomain-containing protein BRD4 |
See footnote to Table 1.
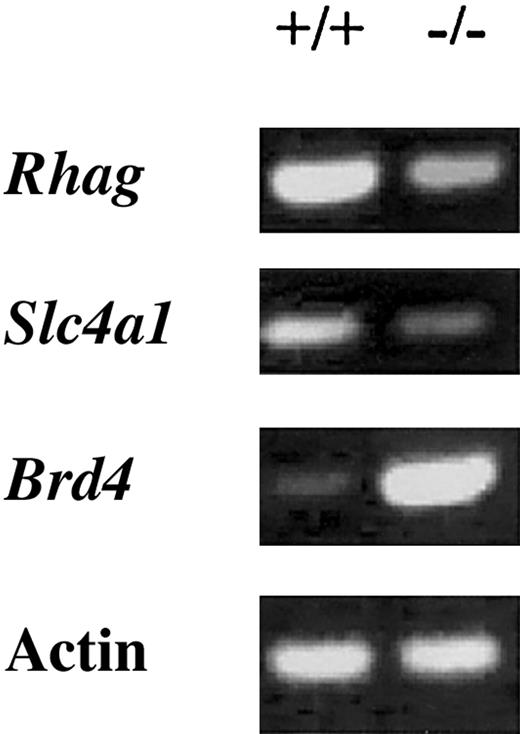
To validate the microarray findings, we performed reverse transcription (RT)-PCR expression analysis on 3 genes that were found to be significantly regulated by Hex in the microarray analysis (Figure 4). Genes for 2 erythrocyte proteins, erythrocyte membrane protein Rh50 (Rhag) and solute carrier family 4 (Slc4a1), were found to be significantly decreased in the Hex-/- day-6 EBs by microarray analysis. Rhag50 was decreased by 2.5-fold and Slc4a by 2.8-fold, very consistent with the microarray data shown in Tables 1, 2. Brd4, a bromodomain-containing protein important in mitosis, was increased 6.7-fold in this RT-PCR analysis, again consistent with the microarray data.
RT-PCR expression analysis of 3 genes found to be regulated byHexin the microarray experiments shown in Tables1,2. The expression of Rhag and Slc4a1 are decreased, whereas Brd4 is increased. These are the same results seen in the microarray analysis, validating the microarray findings.
Discussion
The lack of Hex does not affect hemangioblast formation as assessed by the BL-CFC assay. This is consistent with the finding that Hex is not expressed in the BL-CFC colonies. The Hex-/- defect is limited to definitive embryonic hematopoiesis, with primitive embryonic hematopoiesis being unaffected. This distinction is made more clear by the microarray analysis, which shows that the embryonic betaH1- and ery2-globins do not appear to be decreased, in contrast to the adult (alpha and beta1) globins, which are decreased in the Hex-/- EBs.
This primitive versus definitive defect is similar to what is seen with Runx1 homozygous deletion, where hemangioblasts that give rise to definitive hematopoietic cells are decreased, but hemangioblasts that give rise to primitive hematopoiesis seem to be maintained.4 However, that report indicates that Runx1 would function before Hex is required, since total hemangioblast function was not affected by Hex deletion.
Although the Runx1 deletion studies indicate that there may be 2 distinct hemangioblasts, arising from developing mesoderm, the data here imply that there are distinct pathways of differentiation by the hemangioblast after it is formed: one for primitive and one for definitive hematopoiesis. The data also lend weight to the hypothesis that there are distinct HSCs for each type of embryonic hematopoiesis.1,4 Thus, if there are distinct primitive versus definitive hemangioblasts each giving rise to distinct primitive versus definitive HSCs, respectively, it appears that Hex is required for only the transition from the definitive hemangioblast to a definitive HSC, and to a somewhat lesser extent, endothelial cells. Hex-/- EBs did not form endothelial tubules as well as wt EBs. However, there was not a complete lack of endothelial tubule formation, consistent with the microscopic observation that Hex-/- embryos can form some vessels before they expire at day 12. Thus, the inhibition of angiogenesis from the differentiating hemangioblast appears less severe than the inhibition of definitive hematopoietic progenitor development. This raises the question of whether there are primitive versus definitive endothelial progenitors, since the data here show a partial defect in Hex-/- endothelial sprout formation, yet Hex-/- embryos show minimal vessel disruption at death.16
Although a dose effect for Hex is seen in the analysis of the definitive hematopoietic progenitors, Hex+/- mice survive to adulthood without obvious defects. Therefore, it is likely that there is a threshold below which Hex expression must fall before there is enough phenotypic damage to the definitive HSC to prevent its development or expansion. In addition, environmental signals within the developing embryo/yolk sac/placenta complex in vivo may compensate for the lack of one Hex allele, perhaps stimulating its overexpression. Such signaling would not be present in the in vitro EB. The microarray data provide some candidates for this, such as BMP-4. Also, it may be the persistence of primitive erythropoiesis in the Hex-/- embryos that allows them to mature to day 12 where the failure of endodermal organ development becomes lethal.
Thus, the in vitro ES cell differentiation experiments, the in vivo yolk sac progenitor assay, the chimeric mice analysis, and the microarray data all lend credence to the hypothesis that Hex is not needed for initial hemangioblast formation, but is required for proper differentiation of the hemangioblast once it is formed. The finding that Hex-/- cells could not contribute significantly to the formation of either lymphoid or myeloid tissue implicates a role for Hex in the generation of hematopoietic stem cells.
As mentioned, the finding that primitive erythropoiesis was normal implies that there is a distinct HSC for primitive versus definitive hematopoiesis, and that this stem cell does not require Hex. Therefore, the transcriptional regulation of gene expression cascades is distinct in the primitive and the definitive HSC. These data, especially the ES cell phenotypes, and the microarray experiments, allow for hypothetical placement of Hex in the cascade of transcriptional regulators and cytokines expressed during hemangioblast formation. Based on these data, one would hypothesize that BMP-4, Flk-1, Runx1, and Scl are epistatically above or at least parallel to Hex. IGF-1, Gata-1, EKLF, NF-AT, and PPARgamma require Hex for expression, and therefore may be epistatically below Hex. In summary, this study provides evidence that Hex may have an essential role in hematopoietic stem cell origins and begins to define that role in the context of other known factors.
Prepublished online as Blood First Edition Paper, June 5, 2003; DOI 10.1182/blood-2003-02-0634.
Supported by National Institutes of Health (NIH) grants to K.Z. (NIH GM36477), R.C. (NRSA CA84677), M.Y. (NIH HL63169), and R.H. (NIH HL66308). The microarray studies were performed by the Indiana University Center for Medical Genomics, which is supported by the 21st Century Indiana Research Technology Fund and the Indiana Genomics Initiative (Lilly Endowment).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We gratefully acknowledge the critical role the late Dr Rosa Beddington played in this project.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal