Abstract
The transcription factor homeobox B4 (HOXB4) is preferentially expressed in immature hematopoietic cells and implicated in the transition from primitive hematopoiesis to definitive hematopoiesis as well as in immature hematopoietic cell proliferation and differentiation. We previously identified Hox response element 1 (HxRE-1) and HxRE-2/E-box as 2 critical DNA-binding sites of the HOXB4 promoter active in hematopoietic cells and demonstrated that upstream stimulating factor 1 and 2 (USF1/2) activate HOXB4 transcription through their binding to the E-box site. Here we report that the trimeric regulatory complex nuclear factor Y (NF-Y) is the factor that recognizes HxRE-1 and activates the HOXB4 promoter in hematopoietic cells. We further show that NF-Y interacts biochemically with USF1/2 on the HOXB4 promoter, and that the formation of this NF-Y/USF1/2 complex is required for the full activity of the HOXB4 promoter. Most important, NF-Ya subunit protein levels are found to be lower in c-Kit-Gr-1+ granulocytic bone marrow (BM) cells than in c-Kit+ immature BM cells, in parallel with a reduction of NF-Y occupancy on the HOXB4 promoter as shown by chromatin immunoprecipitation (ChIP) assay. These results suggest that NF-Y is a developmentally regulated inducer of the HOXB4 gene in hematopoietic cells. (Blood. 2003;102:2420-2427)
Introduction
The homeobox (HOX) genes encode a family of evolutionarily conserved transcription factors that control embryonic body patterning and organogenesis.1 Their involvement in regulating blood cell development was first suggested by studies demonstrating that the expression of certain homeobox genes varies with hematopoietic stem cell (HSC) differentiation.2-4 Of particular interest is HOXB4, which is abundantly expressed in primitive hematopoietic cells, but then declines with lineage-specific terminal differentiation. Enforced expression of HOXB4 in mouse bone marrow (BM) cells induces a net expansion of HSCs that retain the ability to repopulate full lymphoid-myeloid lineages in vivo.5,6 A recent study reported that the overexpression of HOXB4 confers on precirculation yolk sac hematopoietic progenitors the ability to rescue both myeloid and lymphoid reconstitution in lethally irradiated adult mice, suggesting a possible role of HOXB4 involving the transition from primitive to definitive hematopoiesis during embryonic development.7 Interestingly, however, the hematopoietic effect of HOXB4 overexpression may be dose dependent, as Schiedlmeier et al8 have recently found that high-level overexpression of HOXB4 in human CD34+ umbilical cord blood cells impaired their differentiation to myeloid cells and B lymphocytes in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice, in addition to expanding the number of CD34+ cells. Taken together, these studies suggest that the molecular regulations of homeobox gene expression may be among the critical nodal points controlling HSC self-renewal and differentiation.
HOXB4 may also regulate later stages in hematopoietic differentiation. HOXB4 expression, which declines as HSCs begin to differentiate, was shown to rise again in monocytic maturation.9 Moreover, Vitamin D3-induced terminal monocytic differentiation of leukemic myeloid HL-60 cell in vitro is inhibited by the treatment with antisense HOXB4 deoxyoligonucleotides, perhaps through preventing HOXB4′s inhibition of the elongation of c-Myc gene transcription.10
Despite these consistent observations supporting a key role of HOXB4 levels in the early phase of hematopoiesis as well as an involvement in terminal monocytic differentiation, the upstream regulatory mechanisms governing HOXB4 gene transcription in hematopoietic tissues are largely unknown.11 Previously, we determined that HOXB4 is transcriptionally regulated in both normal and malignant human primitive hematopoietic cells, and HOXB4 transcription is induced by activation of a 99-base pair (bp) HOXB4 promoter.12 Mutations within this promoter revealed 2 critical DNA-binding sites: an inverted CCAAT box (Hox response element 1 [HxRE-1]) and an E-box (HxRE-2). Yeast one-hybrid assays evaluating BM and leukemia K562 libraries for HxRE-2 recognition identified upstream stimulating factor-2 (USF2) and microphthalmia transcription factor (MITF), and electrophoresis mobility shift assays (EMSAs) with K562 cell nuclear extract (NE) confirmed that these proteins, along with USF1, bind to the HOXB4 promoter in vitro. Cotransfection assays in both K562 and normal BM CD34+ cells showed that USF1 and USF2, but not MITF, induced the HOXB4 promoter in response to signals stimulating mitogen-activated protein kinase (MAPK) pathway. However, variations in USF1 and/or USF2 levels with differentiation in either K562 or CD34+ cells were not detected, so the mechanisms by which HOXB4 expression is modulated in hematopoiesis were not apparent from these studies.
Here we report that the trimeric transcriptional regulatory complex nuclear factor Y (NF-Y) is the factor that binds to the HxRE-1 site within the HOXB4 promoter. We find that NF-Y protein that binds to the HxRE-1 site complexes with USF1/2 bound to HxRE-2, activating the transcription of the HOXB4 gene. Protein levels of one subunit of NF-Y, NF-Ya, as well as its occupancy of the HOXB4 promoter, vary with normal hematopoietic differentiation. These results indicate that modulation of NF-Y activity, through its inducible component NF-Ya, regulates the expression of HOXB4 in hematopoietic cells via its interaction with USF1/2 on the HOXB4 promoter, and that the loss of NF-Y occupancy on the HOXB4 promoter parallels HOXB4 mRNA decay with stem cell differentiation.
Materials and methods
HOXB4/luc mutants and NF-Y cDNAs
The 99-bp HOXB4 promoter sequence in plasmid pGL-3 (Promega, Madison, WI) was digested in the E-box by PML1 and re-ligated via double-strand DNA oligonucleotides with a sense sequence of GCTCGGAGGATCAC to produce HxRE-INS mutant. The expected insertion of this synthesized oligo was confirmed by DNA sequencing. The cDNAs for human NF-Ya (short form), NF-Yb, and NF-Yc were amplified from human BM cell RNA by Pfu DNA polymerase (Stratagene, La Jolla, CA), and these sequences were confirmed by DNA sequencing. A mutated human NF-Ya cDNA (referred to as DN-NF-Ya) was constructed with the site-directed mutagenesis method (Stratagene), by introducing 3 point mutations into its DNA binding domain as in NF-YA29.13 Murine NF-YA29-expressing plasmid was kindly provided by Dr Roberto Mantovani of Universita di Milano (Milan, Italy). The MigR1 retroviral vector was obtained from Dr Warren Pear, Department of Pathology, University of Pennsylvania (Philadelphia, PA).14 NF-Ya and DN-NF-Ya cDNAs were directionally cloned into MigR1 vector by EcoR1 and XhoI sites to form plasmid MigR1-NF-Ya and MigR1-DN-NF-Ya.
Integration of human HOXB4 promoter-coupled luciferase into the murine HOXB4 locus of ES cells
The gene targeting vector pflexneo, containing the phosphoglycerate kinase-thymidine kinase (PGK-TK1) and neomycin resistance cassettes, was provided by Dr Edward Scott, University of Florida (Gainesville, FL). Approximately 1 kilobase (kb) 5′ human HOXB4 promoter sequence (nucleotides [nt] -1139 to -55) was ligated to the coding sequence for luciferase, and then cloned into targeting vector between the PGK-TK1 and neomycinr cassettes. The remaining coding and 3′ sequence of the HOXB4 gene (3 kb, +40 to +3420 nt) was inserted into the cloning sites downstream of the neomycinr cassette. Then, 25 μg of the constructed plasmid was electroporated into 1 × 107 embryonic stem (ES) cells in 0.8 mL Dulbecco minimum essential medium (DMEM) at 250 V and 500 microfarads (500 μF) by a gene pulser (Bio-Rad, Hercules, CA). Cells were selected in complete medium containing 0.4 mg/mL G418 and 2 μM ganciclovir for 5 days. At 7 days after electroporation, colonies were picked, trypsinized, and replated for integration examination.
Transient transfection and luciferase assay
All reporter-gene assays based on transient transfection were performed according to our previous work.12 Briefly described, 1 × 107 cells were mixed with 3 to 10 μg reporter plasmid, 1 μg Renilla plasmid (Promega), and 1 to 10 μg plasmids expressing the genes of interest, or empty-vector plasmid to compensate for the DNA amount. The mixture was then electroporated once at 200 V and 960 μF. At 48 hours after transfection, the cells were collected, and both luciferase activity and Renilla activity of 20 μL cell lysate were measured by a luminometer (Monolight 2010; Analytical Luminescence Laboratories, San Diego, CA).
Electrophoresis mobility shift assays
The 99-bp HOXB4 core promoter, as well as its mutants, was radiolabeled by [α-32P]-deoxycytidine triphosphate (dCTP) and Klenow fragment. For in vitro binding reaction, 2 μL NE derived from 1 × 105 K562 cells were incubated with the radiolabeled probes at 30°C for 30 minutes in 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) pH 7.9, 50 mM KCl, 1 to 5 mM dithiothreitol (DTT), 0.5 mM EDTA (ethylenediaminetetraacetic acid), 10% glycerol, 0.1% Nonidet P-40 (NP-40), 1 mg/mL bovine serum albumin (BSA), and 0.1 μg/mL poly dI-C (5 mM MgCl2, optional). For competition or supershift experiment, the NEs were treated with 20 × molar cold probes or 1 μg antibody for 30 minutes at room temperature prior to the addition of the radiolabeled probes. The reaction products were then loaded onto 4% to 5% polyacrylamide gel electrophoresis (PAGE) and run at 100 to 200 V in 0.5 × Tris (tris(hydroxymethyl)aminomethane)-boreate-EDTA (TBE) for 5 to 7 hours before gel-drying and autoradiography. The antibodies against USF1, USF2, and 3 subunits of NF-Y were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Rockland Immunochemicals (Gilbertsville, PA).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitate (ChIP) assays were performed by following the protocols proposed by Kuo and Allis15 and Orlando.16 Briefly, K562 cells, primary BM, or spleen cells were treated with 1% formaldehyde for 10 minutes at room temperature for cross-linking, which was stopped by the addition of glycine. The cell pellets were treated with hypotonic buffer to release cytoplasmic proteins, and then the nuclear pellets were lysed in 0.5 to 1 mL of 1% sodium dodecyl sulfate (SDS) lysis buffer for 10 minutes on ice and immediately sonicated. The sonicated supernatants were diluted into the dilution buffer and precleared with an agorse slurry of single-stranded DNA (ssDNA)/protein. Transcription factors along with bound DNA were then immunoprecipitated with 1 to 5 μg antibody against either USF1, USF2, or NF-Ya, or a control immunoglobulin G (IgG), and immune complexes were recovered by 30 μL ssDNA/protein A-agarose slurry. After 6 washings in the dilution buffer and Tris-EDTA (TE) buffer, the protein-DNA complexes were released from the beads by proteinase K treatment, and the cross-linking was then dissociated by 65°C heating. The solutions were extracted by phenol/chloroform and precipitated with ethanol. The DNA pellets were dissolved in 20 to 50 μL TE. Then, 5 μL this final solution was used as the template for polymerase chain reaction (PCR), with primer pairs spanning 363 bp of the HOXB4 promoter, centered with both HxRE-1 and E-box (primers, 5′CCCCGCAGGAGCCCTATGTA3′ -274 to -253 nt and 5′GTAGGTAATCGCTCTGTGAATA3′ +68 to +89 nt). For ChIP experiments on stably transfected K562 cells, primers derived from pGL-3 sequence surrounding the inserted HOXB4 promoter, forward primer CAAACTAGCAAAATAGGCTGTC and reverse primer CTTTATGTTTTTGGCGTCTTCC, were used to identify these transfected DNA sequences.
Western blotting, coimmunoprecipitation, and glutathione S-transferase fusion protein pull-down assays
For Western blotting, 3 × 107 32Dc13 cells were washed twice in cold phosphate-buffered saline (PBS) and resuspended in 300 μL lysing buffer (radioimmunoprecipetation assay [RIPA] buffer) containing 50 mM Tris pH 7.9, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% NP-40, 10% glycerol, and 0.2 mM phenylmethyl sulfonic fluoride (PMSF), which was exposed to mild sonication before centrifugation. For primary hematopoietic cells, 5 to 10 × 106 cells were incubated with 0.4 mL A buffer containing 10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA and EGTA (ethylene glycol tetraacetic acid), 1 mM DTT, and 0.5 mM PMSF on ice for 15 minutes. The suspension was then conditioned with 25 μL 10% NP-40, and vigorously vortexed for 10 seconds to release cytoplasmic proteins. The centrifuge nuclear pellet was then dissolved in 50 μL buffer C (20 mM HEPES pH 8.0; 0.4 M NaCl; and 1 mM each of EDTA, EGTA, DTT, and PMSF) and rocked at 4°C for 15 minutes. The supernatant was collected as nuclear extract, and the protein level was measured by Dc Protein Assay (Bio-Rad). Next, 10 μg lysate was loaded onto a 12% SDS-PAGE for separation and then blotted to Immunobilon-P (Millipore, Billerica, MA). The membrane was sequentially stained with primary and secondary antibodies (Santa Cruz Biotechnology; and Rockland Immunochemicals) before signals were detected by electrochemiluminescence (ECL) kit (Amersham, Arlington Heights, IL).
For coimmunoprecipitation assays, 2 to 5 × 107 K562 cells were collected, washed twice in cold-PBS, resuspended in 3 mL cold RIPA buffer/0.5% NP-40, and incubated on ice for 30 minutes with occasional mixings. The cell lysate was spun at 10 000g for 10 minutes at 4°C to pellet cellular debris. The supernatant was precleared by incubation with 1 μg control IgG (Santa Cruz Biotechnology) and 20 μL protein A-agarose beads for 30 minutes, and the cleared supernatant was incubated with 1 μg antibody against either USF1 or USF2 for 2 hours. Finally, we added 20 μL protein A-agarose beads for overnight incubation. The bead-antibody pellets were washed with RIPA containing 0.05% NP-40 for 5 times, resuspended in 1 × SDS loading buffer, and analyzed by 12% SDS-PAGE and Western blotting.
Glutathione S-transferase (GST)-NF-Ya, GST-NF-Yb, and GST-NF-Yc fusion proteins were produced in B21 Escherichia coli cells and purified by affinity binding to glutathione sepharose 4B beads (Amersham Pharmacia Biotech, Piscataway, NJ). For GST pull-down experiments, 1 μg GST-X fusion protein was washed with binding buffer containing 50 mM Tris pH 7.5, 40 mM NaCI, 1% glycerol, 1 mM PMSF, and 0.5 μg/mL BSA, and resuspended in 100 μL binding buffer. Then, 50 μg K562 NE was added to each sample, and the sepharose-fusion protein/NE mixture was incubated at room temperature for 30 minutes, washed 5 times with washing buffer, and suspended in 1 × SDS loading buffer. Dissociated supernatants were analyzed by Western blotting with antibodies to USF1 or USF2.
Primary murine BM and spleen cell isolation and differentiation induction of 32D cells
The primary murine hematopoietic cells were freshly obtained from the tibia and femurs as well as spleens of C57BL/6J mice at 6 to 12 weeks of age. The c-Kit+ BM cells were isolated by sequentially incubating the BM cells with biotin-conjugated CD117 antibody (2B8; Pharmingen, San Diego, CA) and streptavidin-linked microbeads (magnetic-activated cell sorting [MACS]). Then the labeled BM cell solution was passed through MACS separation columns to separate c-Kit+ from c-Kit- cells. The resulting c-Kit- cells then went through the same course, but starting with Ly-6G antibody (RB6-8C5) to obtain granulocytic antigen (Gr)-1+ BM cells. Splenic macrophages were isolated by means of the same protocol, but anti-CD11b antibody (M1/70) was substituted. The identities of the isolated cells were confirmed by morphologic examination. Granulocytic differentiation induction of 32D cells (American Type Culture Collection [ATCC], Manassas, VA) was achieved by replacing 1 ng/mL interleukin 3 (IL-3) with 10 ng/mL granulocyte colony-stimulating factor (G-CSF) in culture medium for indicated times.
RT-PCR analysis
The total RNAs from 5 × 106 freshly isolated c-Kit+ BM cells and c-Kit-Gr-1+ granulocytic BM cells were isolated by Trizol (Life Technologies, Bethesda, MD) and quantified by DU640B Spectrophotometer (Beckman Coulter, Fullerton, CA). Around 50 ng RNA was used as the template in each PCR reaction to measure the expression levels of NF-Y target genes, with a one-step reverse-transcription PCR (RT-PCR) kit (Qiagen, Valencia, CA). Mouse HOXB4 RT-PCR primers were as follows: sense 5′GGAGTTTCACTACAACCGCTACCTG3′, antisense 5′CTACCCCCCTTCTCTGTGTTTATTC3′, 29 cycles; 18s rRNA RT-PCR, 11cycles. Transduced K562 cells infected by MigR1-DN-NF-Ya or MigR1-NF-Ya were separated from nontransduced cells by green fluorescent protein-fluorescence-activated cell sorting (GFP-FACS). Total RNA isolated from 5 × 106 transduced cells in each group was prepared and quantified by both spectrometer and RNA electrophoresis. Then, 3 μg RNA from each sample was primed with 150 ng random hexamer in 40 μL RT reaction, and 2 μL RT product was used as the template for each PCR amplification. Real-time PCR reactions were performed with Taqman universal PCR master mix on iCycler (Bio-Rad), with the use of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PCR as the internal control. The PCR primers for HOXB4 mRNA span an intron, with the sequences as follows: sense primer CCCTGGATGCGCAAAGTT and antisense primer GCGGTTGTAGTGAAATTCCTTCTC. The HOXB4 probe sequence is CCTACACGCGCCAGCAGGTCTTG.
Results
Both HxRE-1 and HxRE-2/E-box sites are required for HOXB4 promoter activity in vivo
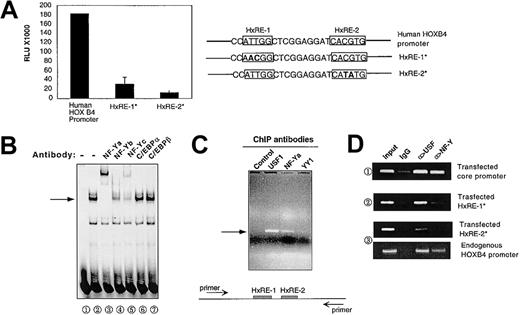
Previous studies using transient transfection of reporter constructs into K562 cells and normal human CD34+ cells identified Hox response elements 1 and 2 (HxRE-1, HxRE-2) as candidates for key HOXB4 regulatory sites. To confirm that HxRE-1 and HxRE-2 are essential for HOXB4 activation in situ, we used homologous recombination to target intact or mutated HOXB4 5′ promoter-coupled luciferase constructs into the murine HOXB4 locus on chromosome 11 in ES cells. As shown in Figure 1A, luciferase expression from these knock-in constructs was highly dependent on the presence of intact HxRE-1 and HxRE-2/E-box sites, similarly to previous results from transient transfection studies.12 HxRE-1 mutation reduced the expression of luciferase by approximately 85%, and E-box mutation reduced the expression by approximately 95%. Thus, both HxRE-1 and E-box are required for the activity of the HOXB4 promoter in vivo in the context of chromatin.
NF-Y binding to the HOXB4 promoter in vitro and in situ. (A) The HxRE-1 site and E-box are essential for HOXB4 promoter activity. A luciferase gene, coupled with a wild-type human HOXB4 5′ regulatory sequence or the mutated form of this sequence at either HxRE-1 (HxRE-1*) or HxRE-2 (HxRE-2*), was incorporated into the endogenous HOXB4 locus of murine ES cells. Luciferase activities of cell lysates from equal amounts of ES cells in each genetic modification were measured in triplicate as described in “Materials and methods.” The data presented are means ± SD. (B) NF-Y binds to the probe containing HxRE-1 site in EMSA. K562 nuclear extract (NE) was incubated with a 32P-labeled short probe containing the HxRE-1 site, and the reaction mixture was then run on a 5% nondenaturing PAGE to detect the specific retarded migrating band (arrow, lane 2). In lanes 3 through 7, K562 NE was pretreated with the indicated antibodies before being incubated with the probe. (C) NF-Y binds to HOXB4 promoter in situ. In ChIP assay, the chemically cross-linked and fragmented chromatins were precipitated by antibodies against NF-Ya, USF1, and Yin Yang protein 1 (YY1), as well as by a control IgG. The presence of HOXB4 promoter within those final precipitates was examined by PCR amplification of a specific 363-bp sequence spanning HxRE-1 and HxRE-2. The PCR primers are depicted as arrows in diagram below. (D) NF-Y and USF specifically bind to HxRE-1 and HxRE-2 sites in situ. Chemically linked chromatin from the K562 cells stably transfected with either pGL-3-HOXB4 core promoter (1), pGL-3-HOXB4 core promoter/mutated HxRE-1 (2), mutated HxRE-2 (3) were precipitated with the antibodies as indicated. Transfected HOXB4 promoter DNA bound within the immunoprecipitates were amplified with primers derived from pGL-3 vector sequence surrounding the inserted wild-type or mutated HOXB4 core promoters.
NF-Y binding to the HOXB4 promoter in vitro and in situ. (A) The HxRE-1 site and E-box are essential for HOXB4 promoter activity. A luciferase gene, coupled with a wild-type human HOXB4 5′ regulatory sequence or the mutated form of this sequence at either HxRE-1 (HxRE-1*) or HxRE-2 (HxRE-2*), was incorporated into the endogenous HOXB4 locus of murine ES cells. Luciferase activities of cell lysates from equal amounts of ES cells in each genetic modification were measured in triplicate as described in “Materials and methods.” The data presented are means ± SD. (B) NF-Y binds to the probe containing HxRE-1 site in EMSA. K562 nuclear extract (NE) was incubated with a 32P-labeled short probe containing the HxRE-1 site, and the reaction mixture was then run on a 5% nondenaturing PAGE to detect the specific retarded migrating band (arrow, lane 2). In lanes 3 through 7, K562 NE was pretreated with the indicated antibodies before being incubated with the probe. (C) NF-Y binds to HOXB4 promoter in situ. In ChIP assay, the chemically cross-linked and fragmented chromatins were precipitated by antibodies against NF-Ya, USF1, and Yin Yang protein 1 (YY1), as well as by a control IgG. The presence of HOXB4 promoter within those final precipitates was examined by PCR amplification of a specific 363-bp sequence spanning HxRE-1 and HxRE-2. The PCR primers are depicted as arrows in diagram below. (D) NF-Y and USF specifically bind to HxRE-1 and HxRE-2 sites in situ. Chemically linked chromatin from the K562 cells stably transfected with either pGL-3-HOXB4 core promoter (1), pGL-3-HOXB4 core promoter/mutated HxRE-1 (2), mutated HxRE-2 (3) were precipitated with the antibodies as indicated. Transfected HOXB4 promoter DNA bound within the immunoprecipitates were amplified with primers derived from pGL-3 vector sequence surrounding the inserted wild-type or mutated HOXB4 core promoters.
The HOXB4 promoter is bound by NF-Y both in vitro and in vivo
Sequence inspection suggested that the point mutations defining the HxRE-1 site are located within an inverted CCAAT motif (Figure 1Aii), which previous studies have identified as a potential binding site for numerous transcription factors, including CCAAT enhancer-binding protein (C/EBP) isoforms and the trimeric transcriptional regulatory complex NF-Y.17,18 We therefore investigated whether one or more of these CCAAT-binding proteins might bind to the HxRE-1 site in the HOXB4 promoter, by employing a short probe derived from the HOXB4 core promoter sequence that retains the HxRE-1 in EMSA studies with K562 cell nuclear extract (NE). As shown in Figure 1B, a distinctive band was detected (arrow), and its formation was either supershifted or disrupted by the antibodies against 3 subunits of NF-Y (NF-Ya, NF-Yb, and NF-Yc), but not by the antibody against either C/EBPα or C/EBPβ. Thus, K562 NE contains proteins detected by anti-NF-Ya, anti-NF-Yb, and anti-NF-Yc antibodies, which can bind to the HOXB4 promoter in vitro.
Then we directly performed ChIP to verify the binding of NF-Y proteins to the endogenous HOXB4 promoter in K562 cells. As shown in Figure 1C, the antibody directed against NF-Ya (the DNA-binding subunit of NF-Y) and anti-USF1 each effectively precipitated chromatin fragments containing HOXB4 promoter. In contrast, the antibody against YY1, whose binding consensus sequence CCAT also partially overlaps the HxRE-1 site, and a control IgG failed to demonstrate the ability to precipitate the target proteins tied with the HOXB4 promoter in vivo.
To determine whether specific binding of NF-Y and USF to HxRE-1 and HxRE-2 sites in situ was absolutely required for these positive ChIP signals, we performed ChIP assays on K562 cells stably transfected with either (1) wild-type HOXB4 core promoter or (2) the promoter mutated at either the HxRE-1 or the HxRE-2 site inserted in pGL-3 plasmid.12 As shown in Figure 1D, both USF and NF-Ya antibodies effectively precipitated the integrated core promoter fragments with wild-type HxRE-1 and HxRE-2. In contrast, NF-Ya antibody failed to precipitate protein binding to mutated HxRE-1, while neither anti-USF nor NF-Ya precipitated complexes containing chromatin fragments with mutated HxRE-2. As a positive control, both USF and NF-Ya antibodies effectively precipitated chromatin fragments containing the endogenous HOXB4 promoter from the same lysate from which they failed to precipitate transfected HxRE-2-mutated HOXB4 promoter. These data clearly show that NF-Y and USF specifically bind to HxRE-1 and HxRE-2 sites in situ. In addition, they suggest that binding of USF isoforms to HxRE-2 might be required for stable binding of NF-Y to HxRE-1.
NF-Y specifically recognizes the HxRE-1 site within the HOXB4 promoter, forming a complex with USF1/2
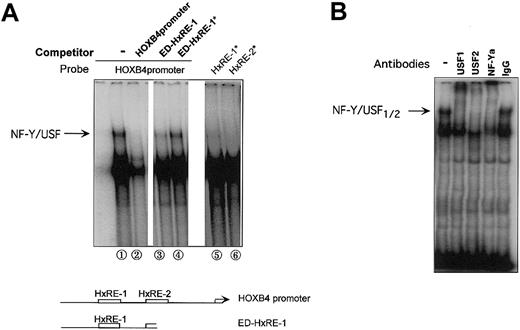
We next used EMSA to ask whether NF-Y protein bound to the HxRE-1 site interacts with USF1/2 bound to the adjacent HxRE-2 site. Using the complete 99-bp HOXB4 core promoter as probe, we detected a slowly migrating band (Figure 2A; arrow), whose formation was blocked by inclusion of either excess unlabeled probe or excess truncated probe containing HxRE-1 (ED-HxRE-1 indicates E-box deleted), but not by competition with this truncated probe containing mutated HxRE-1 (ED-HxRE-1*). The formation of this band, however, was dependent on the integrity of both HxRE-1 and the adjacent HxRE-2 site. An EMSA with HOXB4 promoter probe containing either an HxRE-1 mutation (HxRE-1*) or an E box mutation (HxRE-2*) failed to produce this HxRE-1-specific band. Likewise, inclusion of antibody against either USF1, USF2, or NF-Ya into the intact promoter/NE reaction disrupted the formation of this band (Figure 2B). Therefore, the DNA-binding protein complex responsible for this retarded mobility band contains not only NF-Y but also USF1/USF2, and these proteins bind specifically to HxRE-1 and HxRE-2, respectively.
Interaction of NF-Y and USF1/2 in binding to the HOXB4 promoter through the HxRE-1 and HxRE-2 sites. (A) Note a slowly migrating band (arrow) detected when the intact HOXB4 core promoter (99 bp) was used as the probe in an EMSA with K652 cell nuclear extracts (NE) (lane 1). In lanes 2-4, 20 × molar excess of cold wild-type probe, of a truncated HOXB4 probe containing only HxRE-1, or of the truncated form containing the neutralizing HOXB4 mutation HxRE-1* (as depicted in diagram below) was preincubated with K562 NE. In lanes 5 and 6, the HOXB4 core promoter containing either mutated HxRE-1* or HxRE-2* was used as the probe, in the absence of competing probe. (B) EMSA supershift, in which K562 NEs were pretreated with specific antibodies prior to being incubated with HOXB4 promoter probe. The upper EMSA band identified in lane 2 of panel A, which required both intact HxRE-1 and HxRE-2 sites, was disrupted by the antibody against either NF-Ya, USF1, or USF2, but not by the control IgG.
Interaction of NF-Y and USF1/2 in binding to the HOXB4 promoter through the HxRE-1 and HxRE-2 sites. (A) Note a slowly migrating band (arrow) detected when the intact HOXB4 core promoter (99 bp) was used as the probe in an EMSA with K652 cell nuclear extracts (NE) (lane 1). In lanes 2-4, 20 × molar excess of cold wild-type probe, of a truncated HOXB4 probe containing only HxRE-1, or of the truncated form containing the neutralizing HOXB4 mutation HxRE-1* (as depicted in diagram below) was preincubated with K562 NE. In lanes 5 and 6, the HOXB4 core promoter containing either mutated HxRE-1* or HxRE-2* was used as the probe, in the absence of competing probe. (B) EMSA supershift, in which K562 NEs were pretreated with specific antibodies prior to being incubated with HOXB4 promoter probe. The upper EMSA band identified in lane 2 of panel A, which required both intact HxRE-1 and HxRE-2 sites, was disrupted by the antibody against either NF-Ya, USF1, or USF2, but not by the control IgG.
NF-Y activates HOXB4 promoter reporter constructs in K562 cells, in collaboration with USF1/2
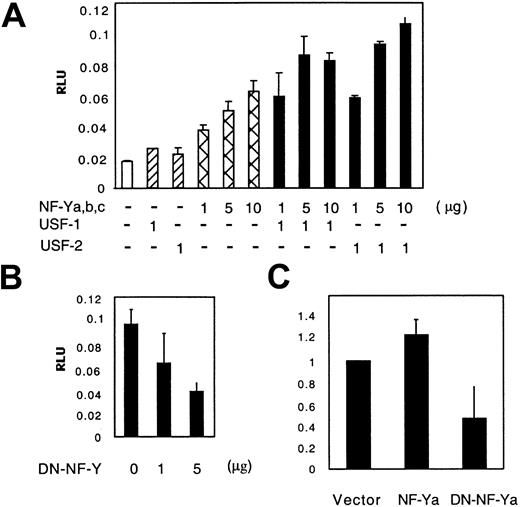
The observation that HxRE-1 mutation inhibited both HOXB4 promoter activity and NF-Y binding suggested an activating role of NF-Y on the HOXB4 promoter. To test this prediction, we cotransfected equal molar quantities of NF-Ya-, NF-Yb-, and NF-Yc-expressing plasmids, with or without plasmids that encoded USF1 or USF2, into K562 cells, and measured luciferase protein expression produced from cotransfected HOXB4 promoter-coupled luciferase reporter constructs (HOXB4/luc). As shown in Figure 3A, HOXB4/luc was activated by the overexpression of NF-Y in a dosage-dependent manner; in the presence of either USF1- or USF2-expressing plasmid, this activating effect was further enhanced. Conversely, when a dominant-negative NF-Y expression plasmid NF-YA29 (DN-NF-Y)13 was introduced into K562 cells, HOXB4/luc activity was reduced in a dosage-dependent manner (Figure 3B). To confirm that changes in NF-Y activity influence the endogenous expression of HOXB4, we transfected K562 cells with MigR1-NF-Ya and MigR1-DN-NF-Ya, and isolated the retrovirally transduced cells by GFP-FACS. HOXB4 mRNA sequences from each isolated cell population were detected and quantitated by means of real-time PCR. As shown in Figure 3C, the DN-NF-Ya overexpression resulted in 50% reduction in endogenous HOXB4 mRNA expression (P < .05). Stably infected K562 cells overexpressing NF-Ya were recovered only at modest transduction levels (as determined by GFP expression). HOXB4 mRNA levels in these moderate NF-Ya/GFP+ cells were increased by 30% versus controls.
Activation of the HOXB4 promoter by transient transfection with NF-Y, in cooperation with USF1/2. (A) NF-Y activates the HOXB4 promoter. K562 cells were transfected with 3 μg HOXB4 core promoter-coupled luciferase reporter plasmid and varying quantities (equimolar mixtures) of NF-Ya/b/c-expressing plasmids, with or without USF1- or USF2-expressing plasmids. Luciferase activities were measured 48 hours after transfection and normalized by Renilla reporter activities. The data presented are means ± SDs of triplicate measurements from 1 of 4 similar experiments. (B) HOXB4 promoter activity in the presence of DN-NF-Y. K562 cells were cotransfected with 10 μg HOXB4 promoter reporter plasmid along with varying doses of NF-YA29-expressing plasmid. (C) Endogenous HOXB4 expression in K562 cells is down-regulated by overexpression of DN-NF-Ya. The mRNA levels in transduced K562 cells were measured by real-time PCR in triplicate, as described in “Materials and methods.” The number of HOXB4 mRNA copies in the empty vector group was arbitrarily determined as 1. The data presented are means ± SDs (B-C).
Activation of the HOXB4 promoter by transient transfection with NF-Y, in cooperation with USF1/2. (A) NF-Y activates the HOXB4 promoter. K562 cells were transfected with 3 μg HOXB4 core promoter-coupled luciferase reporter plasmid and varying quantities (equimolar mixtures) of NF-Ya/b/c-expressing plasmids, with or without USF1- or USF2-expressing plasmids. Luciferase activities were measured 48 hours after transfection and normalized by Renilla reporter activities. The data presented are means ± SDs of triplicate measurements from 1 of 4 similar experiments. (B) HOXB4 promoter activity in the presence of DN-NF-Y. K562 cells were cotransfected with 10 μg HOXB4 promoter reporter plasmid along with varying doses of NF-YA29-expressing plasmid. (C) Endogenous HOXB4 expression in K562 cells is down-regulated by overexpression of DN-NF-Ya. The mRNA levels in transduced K562 cells were measured by real-time PCR in triplicate, as described in “Materials and methods.” The number of HOXB4 mRNA copies in the empty vector group was arbitrarily determined as 1. The data presented are means ± SDs (B-C).
NF-Y and HOXB4 interact biochemically and functionally to activate the HOXB4 promoter
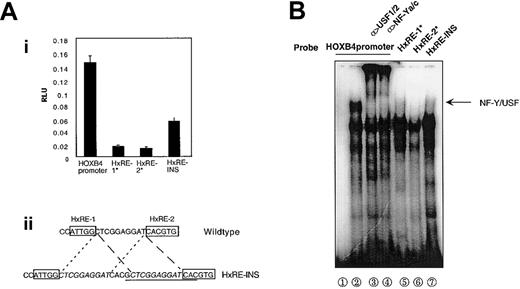
NF-Y and USF1/2 were detected in the same band in EMSA, and the mutation at either the HxRE-1 or the HxRE-2 site greatly inhibited HOXB4 promoter activity, suggesting a critical role of the NF-Y/USF1/2 complex in binding and activating the HOXB4 promoter. To further examine the relationship between NF-Y/USF interaction and HOXB4 activation, we performed both EMSA and transient transfections by using a promoter construct in which an additional 14-bp sequence was duplicated between the HxRE-1 and HxRE-2 sites in the 99-bp HOXB4 promoter. Increasing the distance between the NF-Y- and USF-binding sites without disturbing their surrounding sequences in HOXB4 promoter luciferase reporter constructs resulted in 60% reduction in reporter activity after transient transfection into K562 cells (Figure 4A). Addition of this same 14-bp sequence to HOXB4 promoter oligonucleotides probes greatly decreased the formation of the retarded mobility band representing most of promoter-bound NF-Y and USF1/2 signals as detected by antibodies to NF-Ya, NF-Yc, or USF1/2 (Figure 4B). Taken together, these results suggest that full functioning of the HOXB4 promoter requires the formation of the protein complex including both NF-Y and USF1/2 on the promoter.
Correlation of NF-Y-USF1/2 multiple protein complex with HOXB4 promoter activity. (A) Proper spacing between HxRE-1 and HxRE-2 is required for full HOXB4 promoter activity. K562 cells were transfected with 10 μg each indicated reporter plasmid, and luciferase activities were measured 48 hours later (i). (ii) HxRE-INS was constructed by inserting a 14-bp sequence (underlined) within the interval between HxRE-1 and HxRE-2, without altering the flanking sequences (italicized). The data presented are means ± SDs of triplicate measurements, from 1 of 3 similar experiments. (B) This insertion mutation between HxRE-1 and HxRE-2 (HxRE-INS) disrupts the cooperative in vitro binding of USF1/2 and NF-Y to the HOXB4 promoter, as detected by EMSA. Wild-type HOXB4 promoter probe was used in lanes 1 through 4; HOXB4 promoter with a mutation at either the HxRE-1 or the HxRE-2 site was used in lanes 5 and 6; and HxRE-INS was used in lane 7. K562 NE was added in lanes 2 through 7. In lanes 3 and 4, the NEs were pretreated with USF1/USF2 antibodies and NF-Ya/NF-Yc antibodies, respectively.
Correlation of NF-Y-USF1/2 multiple protein complex with HOXB4 promoter activity. (A) Proper spacing between HxRE-1 and HxRE-2 is required for full HOXB4 promoter activity. K562 cells were transfected with 10 μg each indicated reporter plasmid, and luciferase activities were measured 48 hours later (i). (ii) HxRE-INS was constructed by inserting a 14-bp sequence (underlined) within the interval between HxRE-1 and HxRE-2, without altering the flanking sequences (italicized). The data presented are means ± SDs of triplicate measurements, from 1 of 3 similar experiments. (B) This insertion mutation between HxRE-1 and HxRE-2 (HxRE-INS) disrupts the cooperative in vitro binding of USF1/2 and NF-Y to the HOXB4 promoter, as detected by EMSA. Wild-type HOXB4 promoter probe was used in lanes 1 through 4; HOXB4 promoter with a mutation at either the HxRE-1 or the HxRE-2 site was used in lanes 5 and 6; and HxRE-INS was used in lane 7. K562 NE was added in lanes 2 through 7. In lanes 3 and 4, the NEs were pretreated with USF1/USF2 antibodies and NF-Ya/NF-Yc antibodies, respectively.
Disruption of the NF-Y/USF1/2 complex on the HOXB4 promoter by this 14-bp insertion suggested that direct physical interaction between NF-Y and USF1/2 might be required for promoter activation. To test this, we performed coimmunoprecipitation and GST pull-down experiments with reagents detecting USF and NF-Y subunits. Immunoprecipitation of K562 lysates with antibodies against USF1 and USF2 identified protein complexes that also contained NF-Yc as detected by Western blotting (Figure 5A). Conversely, pull-down products from K562 NE identified by sepharose bead-linked GST-NF-Yc fusion protein contained protein detected by anti-USF2 and to, a lesser extent, by anti-USF1 antibodies (Figure 5B). These data suggest that NF-Yc and USF1/2 associate physically, thereby contributing to the formation of a multiple protein complex on the HOXB4 core promoter.
Complexes of NF-Yc with USF2 and USF1 in K562 cell lysates. (A) K562 cell lysate was immunoprecipitated by the antibody against either USF1 or USF2, and the resulting precipitates were probed with the antibodies against 3 subunits of NF-Y. No signals were detected with anti-NF-Ya or anti-NF-Yb. (B) K562 NE was incubated with sepharose bead-linked GST-NF-Ya, GST-NF-Yb, or GST-NF-Yc fusion protein individually. After extensive washing, the bound products were eluted and then probed by the antibody against either USF1 (top blot) or USF2 (bottom blot) in Western blotting.
Complexes of NF-Yc with USF2 and USF1 in K562 cell lysates. (A) K562 cell lysate was immunoprecipitated by the antibody against either USF1 or USF2, and the resulting precipitates were probed with the antibodies against 3 subunits of NF-Y. No signals were detected with anti-NF-Ya or anti-NF-Yb. (B) K562 NE was incubated with sepharose bead-linked GST-NF-Ya, GST-NF-Yb, or GST-NF-Yc fusion protein individually. After extensive washing, the bound products were eluted and then probed by the antibody against either USF1 (top blot) or USF2 (bottom blot) in Western blotting.
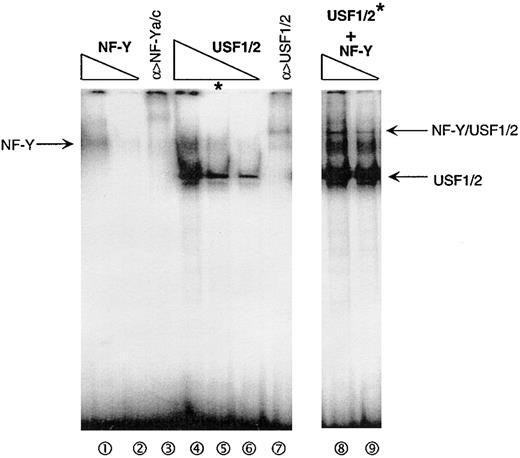
The findings that NF-Y and USF can physically associate in solution and interact on the HOXB4 promoter suggest that their association could be important in some aspect of their binding to the promoter or subsequent activation events. One of the simplest explanations would be if the binding of NF-Y to the HxRE-1 site were stabilized by the binding of USF1/2 to HxRE-2, or vice versa. To test this hypothesis, we incubated serial dilutions of purified GST-NF-Y and/or GST-USF1/2 proteins together with the HOXB4 promoter probe, and measured the strength of their binding reactions in EMSA. As shown in Figure 6 (lanes 1 and 2), adding low concentrations of stoichiometric ratios of NF-Y trimeric subcomponents resulted in the formation of a weak NF-Y band (arrow; lanes 1 and 2) in a dose-dependent manner. The formation of this band was blocked and altered by inclusion of antibodies to NF-Ya and NF-Yc, as expected (lane 3). Similarly, low concentrations of purified recombinant USF1/2 bound to the HOXB4 promoter probe to form a specific major band (lanes 4, 5, and 6). Interestingly, when the same concentrations of purified NF-Y as applied in lane 1 and 2 were added with the same concentration of USF1/2 as used in lane 5 (asterisk) before being incubated with the HOXB4 promoter (lanes 8 and 9), the EMSA was greatly augmented. Not only did a new, distinctive band appear, suggesting cooperative binding of NF-Y/USF1/2 to the HOXB4 promoter, but also the bands corresponding to NF-Y binding alone and USF1/2 binding alone were also greatly enhanced (lanes 2, 5, and 9). These data demonstrate that NF-Y and USF1/2 amplify each other's ability to bind to the HOXB4 promoter.
Cooperation of NF-Y and USF1/2 in binding to the HOXB4 promoter in vitro. Varying concentrations of recombinant GST-NF-Y or GST-USF1/2 protein, alone or in combination, were incubated with HOXB4 core promoter, and their interactions were tested in vitro by EMSA. Lanes 1 and 2 show the incubation of 2 and 0.7 ng GST-NF-Ya/b/c with 32P-labeled HOXB4 promoter probe; in lane 3, GST-NF-Ya/b/c used in lane1 plus 1 μg NF-Ya/c antibodies was added to the reaction. Lanes 4, 5, and 6 show the addition of 6, 2, and 0.7 ng GST-USF1/2, respectively, in the reaction; in lane 7, 2 ng GST-USF1/2 and 1 μg USF1/2 antibodies were added. In lanes 8 and 9, the same amounts of GST-NF-Ya/b/c protein were added to reaction as in lanes 1 and 2, together with 2 ng GST-USF1/2.
Cooperation of NF-Y and USF1/2 in binding to the HOXB4 promoter in vitro. Varying concentrations of recombinant GST-NF-Y or GST-USF1/2 protein, alone or in combination, were incubated with HOXB4 core promoter, and their interactions were tested in vitro by EMSA. Lanes 1 and 2 show the incubation of 2 and 0.7 ng GST-NF-Ya/b/c with 32P-labeled HOXB4 promoter probe; in lane 3, GST-NF-Ya/b/c used in lane1 plus 1 μg NF-Ya/c antibodies was added to the reaction. Lanes 4, 5, and 6 show the addition of 6, 2, and 0.7 ng GST-USF1/2, respectively, in the reaction; in lane 7, 2 ng GST-USF1/2 and 1 μg USF1/2 antibodies were added. In lanes 8 and 9, the same amounts of GST-NF-Ya/b/c protein were added to reaction as in lanes 1 and 2, together with 2 ng GST-USF1/2.
NF-Ya nuclear protein levels and NF-Ya occupancy on the HOXB4 promoter are down-regulated with normal hematopoietic differentiation
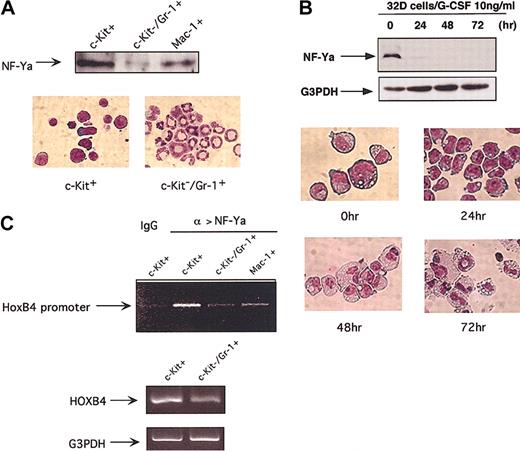
We then asked whether NF-Y interaction with the HOXB4 gene is regulated during normal hematopoiesis. In diverse cell types, NF-Ya has been shown to be the DNA-binding and regulatory subunit of NF-Y trimer, while NF-Yb and NF-Yc are constitutively expressed.17,19 To determine whether NF-Ya levels change during hematopoietic differentiation, we measured nuclear NF-Ya protein levels in primary c-Kit+ immature BM cells (0.5% to 1% of nucleated BM cells), primary c-Kit-Gr-1+ BM cells and spleen macrophage 1-positive (Mac-1+) monocytes/macrophages by Western blotting. The expression of NF-Ya was approximately 17-fold higher in immature c-Kit+ BM cells than in mature c-Kit-Gr-1+ granulocytes, and 3-fold higher than in mature splenic monocytes/macrophages (Figure 7A). Similarly, cells of the murine progenitor cell line 32D expressed easily detectable NF-Ya protein when maintained as immature blasts in IL-3, but rapidly down-regulated their levels within 24 hours of G-CSF-induced granulocytic differentiation (Figure 7B), suggesting an early reduction in NF-Ya level upon granulocytic differentiation.
Down-regulation of NF-Ya nuclear protein levels and its regulation on HOXB4 expression with hematopoietic differentiation. (A) NF-Ya protein levels decrease with terminal myeloid differentiation. Western blotting was used to measure 10 μg nuclear protein extracted from primary c-Kit+ and c-Kit-Gr-1+ BM cells as well as spleen Mac-1+ cells. (B) Primitive 32D cells maintained in IL-3 were washed and induced to granulocytic differentiation by 10 ng/mL G-CSF, and NF-Ya levels were measured by Western blotting at indicated time points. The morphology of 32D cells during differentiation is shown (original magnification, × 1000). Note that the level of NF-Ya declines rapidly and precedes the differentiation to mature granulocytes. (C) NF-Ya/HOXB4 ChIP assays in c-Kit+ premature BM cells versus mature granulocyte and monocytes/macrophages. Cross-linked chromatin fragments from 5 × 106 indicated cells were precipitated with NF-Ya antibody, and the abundance of HOXB4 promoter sequence in final precipitates was assessed by PCR amplification as described in Figure 1C. One of 2 experiments with similar PCR results is shown. The HOXB4 mRNA levels in indicated cells were examined in parallel by RT-PCR (bottom lane).
Down-regulation of NF-Ya nuclear protein levels and its regulation on HOXB4 expression with hematopoietic differentiation. (A) NF-Ya protein levels decrease with terminal myeloid differentiation. Western blotting was used to measure 10 μg nuclear protein extracted from primary c-Kit+ and c-Kit-Gr-1+ BM cells as well as spleen Mac-1+ cells. (B) Primitive 32D cells maintained in IL-3 were washed and induced to granulocytic differentiation by 10 ng/mL G-CSF, and NF-Ya levels were measured by Western blotting at indicated time points. The morphology of 32D cells during differentiation is shown (original magnification, × 1000). Note that the level of NF-Ya declines rapidly and precedes the differentiation to mature granulocytes. (C) NF-Ya/HOXB4 ChIP assays in c-Kit+ premature BM cells versus mature granulocyte and monocytes/macrophages. Cross-linked chromatin fragments from 5 × 106 indicated cells were precipitated with NF-Ya antibody, and the abundance of HOXB4 promoter sequence in final precipitates was assessed by PCR amplification as described in Figure 1C. One of 2 experiments with similar PCR results is shown. The HOXB4 mRNA levels in indicated cells were examined in parallel by RT-PCR (bottom lane).
Variation in nuclear NF-Ya levels does not necessarily accurately reflect altered NF-Y binding to the HOXB4 promoter, as the regulation of promoter occupancy is likely to be complex, involving multiple cofactors and chromatin-modeling events. We therefore used ChIP assay to directly evaluate the in vivo occupancy of NF-Y on the endogenous HOXB4 promoter in primitive and mature blood cells in vivo. As shown in Figure 7C, binding of NF-Y to the HOXB4 promoter decreased in parallel with the decline in NF-Ya protein, from immature c-Kit+ BM cells to mature c-Kit-Gr-1+ granulocytes, accompanied by the decrease in HOXB4 mRNA levels. Thus, NF-Ya levels, and its binding to the HOXB4 promoter, are significantly down-regulated with granulocytic differentiation.
Discussion
In this study, the trimeric transcription factor NF-Y was identified as an activator of the hematopoietic expression of HOXB4, through its binding to the HxRE-1 site on the HOXB4 promoter in cooperation with the binding of USF1/2 to the E-box/HxRE-2 site. NF-Y binding to the HOXB4 promoter was found to decline with myeloid differentiation, especially differentiation to mature granulocytes.
Embryonic stem cell knock-in experiments confirmed the importance of both the HxRE-1 and HxRE-2 sites for HOXB4 promoter activity within the HOX locus in the context of chromatin in vivo. HxRE-1 overlaps an inverted CCAAT box (Y box), which is located -74 bp to the identified transcription start sites in this TATA-less promoter.12 Therefore, this complete consensus sequence, inverted orientation, and location (within -60 to -100 bp), as well as its flanking regions, together represent a potential binding site for NF-Y.17,20 The NF-Y complex consists of 3 subunits encoded by 3 different genes. Current models suggest that NF-Yb and NF-Yc first form heterodimers, which then serve as a platform to recruit NF-Ya proteins. Once recruited to this trimeric complex, the DNA-binding domain of NF-Ya can specifically bind to the CCAAT box, with physical contact between chromatin histones and NF-Yb histone fold motif α1 supporting and stabilizing this specific recognition.21 Consistent with this model, our EMSA studies showed the presence of all 3 subunits of NF-Y on the HOXB4 promoter in the in vitro binding test. Furthermore, NF-Ya-linked ChIP assays confirmed NF-Y binding to the endogenous HOXB4 promoter in K562 cells, along with USF1/2 in a context of chromatin.
EMSA, coimmunoprecipitation, and GST pull-down experiments demonstrate that NF-Y and USF1/2 interact biochemically on the HOXB4 promoter when NF-Y and USF1/2 bind to HxRE-1/Y box and HxRE-2/E-box. The cotransfection studies extend these results, showing that increasing concentrations of NF-Y trimer activate HOXB4 promoter constructs, in concert with USF1 or USF2, and most importantly, HOXB4 promoter activity in K562 cells without overexpression of either NF-Y or USF1/2 appears to depend on the formation of a functional transcriptional regulatory complex including NF-Y and USF1/2 (Figure 4). These results support and extend previous transcriptional assays of NF-Y, which suggest that the regulatory roles of NF-Y may depend on the transcriptional regulatory partners to which it binds in a given genomic and cellular context.20 For example, the promoter of the human hematopoietic stem/progenitor cell antigen CD34 has been indicated to be positively regulated by NF-Y, in concert with an adjacent c-Myb-binding site.22 In B lymphocytes, it was recently shown that physical interaction of NF-Y with the transcription factor regulatory-factor X (RFX) is required for establishing a platform for the assembly of major histocompatibility complex class II (MHC-II) enhanceosomes.23 Our prior and present studies support and extend this notion12 and reveal a critical role for a novel interaction between NF-Y and USF1/2 binding to adjacent Y-box and E-box sites within the HOXB4 promoter. In contrast, in MCF7 cells, NF-Y binding to a proximal Y box within the SHP-1 gene represses target gene transcription. This SHP-1 repression by NF-Y is dependent on the integrity of a neighboring CAGTGCC motif whose DNA-binding protein is unknown.24
Interestingly, Gilthorpe et al25 recently described a TCGCCATT motif within HOXB4 intron C1 region that can act as a second potential binding site for NF-Y in mouse embryo and neuro 2A nuclear extracts in vitro. Our current data do not preclude the possibility that the regulation of HOXB4 expression in hematopoietic cells might be influenced by this nonclassic CCAAT-box site, although we have not found these intronic sequences to be essential or active in reporter assays in hematopoietic cells in vitro. Of note, we did not observe that the transcription factor YY1, which Gilthrope et al also showed capable of binding to the intronic NF-Y site by EMSA, could be detected by ChIP assays on the endogenous HOXB4 promoter in vivo (Figure 1C), nor could we detect any functional activity in cotransfection assays in hematopoietic cells in vitro (data not shown).
Sauvageau et al3 first showed that HOXB4 is preferentially expressed in the most primitive compartments of human CD 34+ BM cells. Later, observations made by Pan and Simpson9 suggested that HOXB4 is expressed and thus required for the maturation of monocytes. Recently the expression of HOXB4 in HSCs and lineage-committed progenitors ([LCPs] in adult mouse BM), along with that of HOXA9, has been detected.26
Of the 3 subunits of NF-Y, NF-Ya has previously been shown to be the regulatory subunit, and the alteration in its protein level along with hematopoiesis was indicated.19,27 To investigate whether the decline in HOXB4 mRNA level with hematopoietic differentiation is possibly caused by a corresponding changes in NF-Ya protein level, we isolated primary c-Kit+ immature BM cells and mature c-Kit-Gr-1+ granulocytic BM cells and compared their nuclear NF-Ya protein level, NF-Y occupancy on HOXB4 promoter, and HOXB4 mRNA level. We found a tight association of NF-Ya level with its occupancy on HOXB4 promoter and mRNA levels of HOXB4 in host cells, confirming that altered NF-Ya expression along with hematopoietic differentiation participates in the regulation of HOXB4 expression through its binding to HOXB4 promoter.
In contrast to the striking effect of HOXB4 overexpression on primitive hematopoietic cell behavior, HOXB4 knockout mice show only minimal hematopoietic phenotype.28 Although the reason for this discrepancy is unknown, one possible explanation could be that other HOX gene members, including perhaps paralogs of HOXB4, might compensate for HOXB4 deficiency during development. Previous studies have indicated that HOX paralog genes may overlap in their functions as well as expression patterns. HOXC4 exists in human CD34+ cells, and its overexpression induces in vitro expansion of committed as well as very early hematopoietic progenitors.29,30 Interestingly, genome sequence inspection confirms the presence of tandem potential HxRE-1 and HxRE-2 sites within the immediate 5′ noncoding sequences of the human HOXA4, HOXC4, and HOXD4 genes, with identical spacing between these sites in HOXB4, HOXC4, and HOXD4. In preliminary ChIP and RT-PCR studies, we have indeed confirmed that NF-Y and USF can bind to these sites in the HOXC4 and HOXD4 locus in K562 cells, and that both NF-Ya and DN-NF-Ya overexpression regulate their mRNA levels as well (data not shown). These data suggest the possibility that NF-Y might act as a hematopoietic regulator of both HOXB4 and its paralogs, and that this concerted action might be important in normal stem cell activity in vivo. Of note, a recent study suggested that a Drosophila gene with homology to NF-Y may act as a master gene controlling multiple target genes that are critical for dorsal-ventral patterning.31
In summary, we have identified the trimeric transcription factor NF-Y as an activator of the hematopoietic expression of HOXB4, through its binding to the HxRE-1 site on the HOXB4 promoter and interaction with USF1/2, which binds to the adjacent E-box/HxRE-2 site. NF-Y binding to the HOXB4 promoter declines with myeloid differentiation. These observations suggest that altered NF-Y activity may be a critical mediator of the differential expression of HOXB4 during hematopoiesis, through its cooperative interaction with USF1/2 on the HOXB4 promoter. In light of these data, it will be very interesting to further investigate the role of NF-Y itself in the differentiation of stem cells into hematopoietic and nonhematopoietic lineages, as well as to determine the range of NF-Y promoter targets in primitive hematopoietic cells.
Prepublished online as Blood First Edition Paper, June 5, 2003; DOI 10.1182/blood-2003-01-0251.
Supported by Public Health Service grant RO1 CA90833 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We greatly appreciate the technical support given by Gerard Joe and Dr Shaomin Zou.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal