Abstract
The cellular and molecular mechanisms underlying the blunted allo-responsiveness of umbilical cord blood (UCB) T cells have not been fully elucidated. Protein expression of NFATc2 (nuclear factor of activated T cells c2), a critical transcription factor necessary for up-regulation of multiple cytokines known to amplify T-cell allogeneic responses, is reduced in UCB T cells. Affymetrix oligonucleotide microarrays were used to compare gene expression of primary purified CD4+ UCB T cells to adult peripheral blood CD4+ T cells (AB) at baseline, 6, and 16 hours of primary stimulation. NFAT-regulated genes exhibited lower expression in UCB CD4+ T cells including the following: granulocyte-macrophage colony-stimulating factor (GM-CSF), interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), interleukin 3 (IL-3), IL-4, IL-5, IL-13, IL-2 receptor α (IL-2Rα; CD25), CD40L, and macrophage inflammatory protein 1 α (MIP-1α). Transcription factors involved in the NFAT pathway including C/EBPβ, JunB, and Fosl1 (Fra-1), as well as Th1- and Th2-related transcription factors STAT4 (signal transducers and activators of transcription 4), T-bet, and c-maf showed reduced expression in UCB compared with AB during primary stimulation. Reduced cytokine, chemokine, and receptor expression was also found in UCB. Gene array data were confirmed using RNase protection assays, flow cytometry, and quantitative multiplexed cytokine measurements. Reduced global expression of NFAT-associated genes, as well as cytokines and chemokines, in UCB CD4+ T cells may contribute to the decreased graft-versus-host disease (GVHD) observed after UCB transplantation. (Blood. 2003;102:4608-4617)
Introduction
Graft-versus-host disease (GVHD) occurring after allogeneic stem cell transplantation is mediated by donor T-cell alloreactivity against major and minor histocompatibility antigens presented by recipient antigen-presenting cells.1 Umbilical cord blood (UCB) has been shown to elicit reduced incidence and severity of GVHD.2-4 While the majority of these studies have been performed in children, recent studies have demonstrated low rates of GVHD in adults after UCB transplantation.5,6
Potential cellular and molecular mechanisms that underlie the clinical observations of low rates of GVHD in UCB recipients have been investigated. It has been shown that activated UCB lymphocytes produce lower amounts of cytokines after primary and secondary stimulation compared with activated adult T lymphocytes, including interferon-γ (IFN-γ)7-12 and tumor necrosis factor-α (TNF-α),7,10-12 which play an important role in clinical GVHD. Additionally, production of the cytokines interleukin-2 (IL-2)7,9,10 and IL-47 by activated UCB T cells is reduced compared with the response observed in adult blood (AB) T cells. Moreover, although UCB T lymphocytes proliferate at rates equivalent to adult T lymphocytes in response to primary stimulation, alloantigen-specific cytotoxicity is reduced in UCB T lymphocytes when compared with adult controls after secondary stimulation.12-14
The NFAT (nuclear factor of activated T cells) family of transcription factors contains 5 distinct NFAT proteins: NFATc1 (NFAT2 or NFATc), NFATc2 (NFAT1 or NFATp), NFATc3 (NFAT4 or NFATx), NFATc4 (NFAT3), and NFAT5 (tonicity enhancer binding protein [TonEBP]).15,16 NFATc1-NFATc4 proteins are defined by a common regulatory domain that is the target of calcineurin, a calcium/calmodulin-dependent protein phosphatase.16 Increased intracellular calcium occurring after T-cell activation results in dephosphorylation and thus activation of NFAT proteins with consequent translocation of the NFAT proteins from the cytoplasm to the nucleus and transcriptional activation of genes involved in cellular development, activation, and differentiation.16 Cyclosporin A (CsA) and FK506, effective prophylactic and treatment agents against GVHD, have been shown to inhibit calcineurin as well as dephosphorylation and translocation of NFAT proteins.17-19 NFATc2 interacts with the Fos-Jun heterodimer activator protein-1 (AP-1) for the transcriptional regulation of many genes including IL-2, IL-3, IL-4, macrophage inflammatory protein 1 α (MIP-1α), and FasL.16,20-22 Additionally, it has been shown that NFATc2 is necessary for the transcription of multiple cytokines and inflammatory proteins including cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4), IL-13, IFN-γ, CD40L, granulocyte-macrophage colony-stimulating factor (GM-CSF), and TNF-α.21-23 Reduced constitutive expression of NFATc2 protein in unstimulated UCB T lymphocytes, as well as an attenuated up-regulation of NFATc2 during primary stimulation, has been previously reported.11 Moreover, activated UCB T lymphocytes produce reduced amounts of NFATc2-regulated IL-4, IFN-γ, TNF-α, GM-CSF, and have reduced surface expression of CD40L.7,24-26 Therefore, reduced constitutive NFATc2 expression in UCB T lymphocytes and attenuated up-regulation during primary stimulation may be one underlying molecular mechanism for blunted UCB allo-responsiveness.
These observations taken together suggest that UCB differs from adult cells in both cytokine profile and effector function. To test this hypothesis we performed direct comparative gene expression profiling of purified CD4+ T-cell populations derived from adult and UCB during primary stimulation. We noted profound differences between both populations suggestive of specific signaling events that may underlie the decreased GVHD allo-reactivity observed after UCB transplantation. Our results reveal an attenuated increase in expression of NFAT family, NFAT-related, and NFAT-dependent genes, as well as globally reduced expression of cytokine and chemokine genes in UCB T lymphocytes during primary stimulation. In addition, expression of Th1 and Th2 transcription factors were reduced in UCB CD4+ T lymphocytes during primary stimulation.
Materials and methods
Cells
With informed consent, human umbilical cord blood (UCB) from scheduled cesarean sections and adult peripheral blood (AB) were collected and mononuclear cells (MNCs) were purified as previously described.11,23 For gene expression analysis using the HG-U133A&B GeneChip (Affymetrix, Santa Clara, CA), MNCs from 7 independent UCB units and 7 control adults were depleted of CD14+ cells using CD14 microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany) and CD4+ T cells were selected using CD4 microbeads (Miltenyi Biotech). All cell separations were performed using AutoMACS magnetic cell sorter (Miltenyi Biotech). For hybridization to the HG-U95A GeneChip, MNCs from 5 independent UCB units and 5 control adults were of depleted CD14+ cells using Dynabeads (Dynal, Lake Success, NY) followed by positive selection of CD4+ T cells using magnetic beads and manual columns (Miltenyi Biotech). Approval for these studies was obtained from the Case Western Reserve University/University Hospitals institutional review board (Cleveland, OH). Informed consent was provided according to the Declaration of Helsinki.
T-cell stimulation
For the HG-U133A&B gene array analysis, purified T cells from each individual were divided into 3 equal populations. One population (0 h) was extracted for RNA immediately, while the remaining 2 populations were stimulated with plate-bound anti-CD3 monoclonal antibody (mAb; Hit3a; Pharmingen, San Diego, CA) at 5 μg/mL in phosphate-buffered saline (PBS) and with soluble anti-CD28 mAb (CD28.2; Pharmingen) at 5 μg/mL in RPMI-1640 (Gibco-BRL, Gaithersburg, MD) with 10% fetal bovine serum (FBS). Cells were stimulated in 48-well plates with 1 × 106 cells per well. Stimulated cells were harvested at 6 and 16 hours and RNA extracted immediately. For studies using the HG-U95A GeneChip, cells were extracted at 0 hour or stimulated as described above for 16 hours and RNA extracted immediately.
RNA extraction and pool preparation
At the indicated time points, total RNA was isolated using TRIzol reagent per manufacturer's protocol (Gibco-BRL). Purity was verified using electrophoresis and spectophotometry and samples were stored at -80°C. To obtain sufficient mRNA and reduce interindividual genetic variability, equal amounts of RNA obtained from multiple individual UCB and AB donors was pooled for study at each time point. Initially, RNA from 5 UCB and 5 control adults were used for hybridization to the U95A microarray. This experiment was then repeated using RNA pools from additional 7 UCB and 7 adult donors, with the exception of the adult 0-hour RNA pool that contained RNA from 6 adults. In the replicate experiment, gene expression in the pooled samples was interrogated on the U133A&B microarray, and the same RNA samples were employed to validate observed changes in expression by RNase protection assays.
Purity assessment and confirmation of surface expression by flow cytometry
For purity assessment, cells were surface stained with fluorochrome-conjugated mAbs including CD3, CD4, CD8, CD14, CD19, CD56 (Pharmingen), and corresponding isotype controls. Confirmatory studies of surface expression were performed with mAbs against CD40L (Immunotech, Marseille, France), CXCR4, and CTLA-4 (Pharmingen). In each experiment, purified T cells from 1 UCB and 1 adult control were stimulated as described above and stained for CXCR4, CD40L, and CTLA-4 expression at 20 hours. This was repeated 4 times. Fluorescence of more than 10 000 events was acquired on an LSR flow cytometer (Coulter, Miami, FL) and data were analyzed using WinList (Verity Software House, Topsham, MN). Purity of CD4+/CD3+ ranged from 95% to 99% with the majority more than 97% (data not shown).
Gene expression analysis
Gene expression was interrogated at the indicated time points using Affymetrix HG-U95A and HG-U133A&B expression microarrays (Affymetrix). In the initial experiment, RNA pools from 5 adult and 5 UCBs were hybridized to U95A microarrays. In the second experiment, we used the newly released HG-U133A&B chip set, which contains 44 763 oligonucleotide probe sets for approximately 33 000 genes, to interrogate expression in an additional 7 UCB units and 7 adult controls. It was decided to switch from HG-U95A to HG-U133A&B microarrays, since the HGU133A&B array set contains refined, more robust probe pairs and since probe sets to interrogate NFATc2 expression, lacking on the U95A chip, were added to this new generation of microarrays.
Pooled total RNA from each time point was used to prepare biotinylated target cRNA, with minor modifications from the manufacturer's recommendations (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Briefly, 8 μg of total RNA was used to generate first-strand cDNA by reverse transcription using a T7-linked oligo(dT) primer. After second-strand cDNA synthesis, in vitro transcription was performed with biotinylated UTP (uridine 5′-triphosphate) and CTP (cytidine triphosphate; Enzo Diagnostics, Farmingdale, NY), resulting in the generation of biotinylated cRNA that is approximately 100-fold amplified above the initial quantity of starting material. The target biotinylated cRNA generated from each time point was processed as per manufacturer's recommendation using an Affymetrix GeneChip Instrument System. Briefly, spike transcript controls and 15 μg of fragmented cRNA were added to a hybridization cocktail. This mixture was hybridized to the expression microarray by incubation at 45°C overnight. Arrays were then washed and stained with streptavidin-phycoerythrin before being scanned on an Affymetrix GeneChip scanner. After scanning, array images were assessed by eye to confirm scanner alignment and the absence of significant bubbles or scratches on the chip surface. The 3′/5′ ratios for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and beta-actin were confirmed to be within acceptable limits (0.82-1.07), and BioB spike controls were found to be present on all chips with BioC, BioD, and CreX also present in increasing intensity. Finally, each image was scaled to a target intensity of 1500 and the scaling factors for all arrays were confirmed to be within acceptable limits (4.6-7) as were background and noise.
Data analysis
For both the HG-U95A and HG-U133A&B arrays, the fluorescent intensity of each probe was quantified using Microarray Analysis Suite version 5.0 (MAS 5.0) software (Affymetrix). The expression level of a single mRNA, defined as the signal, was determined by the MAS 5.0 software, which uses a weighted average fluorescence intensity difference obtained among the 11 to 20 probe pairs that interrogate the expression of each individual gene. This software also makes a detection call (present [P], marginal [M], or absent [A]) for each gene or probe set, based on the consistency of the performance of the individual probe pairs, the hybridization above background, and the signal-to-noise ratio.
Two-way comparisons of the microarray data were also performed using the MAS 5.0 software. Specifically, changes in gene expression between UCB and adult samples were evaluated at each time point (ie, UCB vs AB at each time point). These comparisons in MAS 5.0 provided additional data including the signal log ratio (fold change presented in logarithmic form) and the “change call” (increased [I], decreased [D], marginally increased [MI], marginally decreased [MD], or no change [NC]) for each gene being interrogated. The data were then imported into a Microsoft Excel (Redmond, WA) spreadsheet.
To identify genes that exhibited differences in expression between UCB and adult, the data sets were trimmed in Excel using the following inclusion criteria. For a gene or a probe set to be included in this trimmed data set it had to display (1) a present call (P) in at least one of the compared samples, (2) a change call other than no change (NC), and (3) at least a 2-fold difference in expression between the 2 compared samples. Additional annotation data were incorporated into the data set using the Affymetrix web-based analysis tool NetAffx.
GeneSpring analysis
The signals displayed for the genes in each sample included in the HG-U133A&B-trimmed data set were imported into GeneSpring software version 5.1 (Silicon Genetics, San Carlos, CA). To present the relative expression for a given gene or probe set in each sample, the measured signal for each probe set was divided by the median of its measurements in all samples. If the median of the raw values was below 10, then each measurement for that gene or expressed sequence tag (EST) was divided by 10.
One-way hierarchical clustering with Pearson correlation analyses and minimum distance of 0.001 was employed to order genes in the trimmed data set for the time course carried out in the UCB and adult samples. The resulting gene tree incorporated all the genes/ESTs that had met inclusion criteria from the UCB versus AB comparisons at any of the 3 time points (0, 6, and 16 hours primary stimulation).
RNase protection assay
To confirm the array data, expression of selected genes was analyzed in the same RNA pools that were hybridized to the HG-U133A&B arrays, using the RiboQuant multiprobe RNase protection assay (RPA) kits (Pharmingen). RPA kits included the following: hCK1, hStress, and hCK2b. Replicate assays were performed according to the manufacturer's protocol using 32P deoxyuridine triphosphate (dUTP), on 5 ug RNA (2 times each for the hCK1 and hStress probe sets and 3 times for the hCK2b probe set).
Multiplexed cytokine measurements
CD4+ T cells were purified and each 1 UCB and AB sample was stimulated as described above for 20 hours. Supernatants were collected and analyzed in duplicate by a multiplexed cytokine array using fluorescent microspheres27 at the Roswell Park Cancer Institute Flow Cytometry laboratory (Buffalo, NY) for the following cytokines and chemokines: IL-1β, IL-2, IL-3, IL-4, IL-5, IL-8, IL-10, IL-13, GM-CSF, monokine induced by gamma interferon (MIG), MIP-1α, and RANTES (regulated on activation, normal T cells expressed and secreted). This experimental setup was repeated twice.
Results
Consistency of microarray expression data
Two independent studies were carried out. The first study examined gene expression in UCB and AB CD4+ T cells at 0 hour and 16 hours of primary stimulation with anti-CD3 and anti-CD28, using the HG-U95A microarray (data not shown). The second study assessed gene expression at 0, 6, and 16 hours of stimulation using the HG-U133A&B microarray set. Data obtained from the 2 experiments was independently trimmed and analyzed using the same software and same inclusion criteria. After data analysis and trimming, the HG-U133A&B microarray data were compared with the data set generated by the initial HG-U95A array experiment.
The 2 array data sets were queried as follows: first, each data set was queried for similar gene name, as annotated using the Netaffx annotations for each chip. At 0 hour, 129 probe sets had the same gene name. Of these, only 4 probes (3.1%) showed conflicting data (ie, called decreased on U95A and increased on U133A&B). At 16 hours, 178 probe sets had the same gene name of which only 1 probe (0.56%) showed conflicting data. Next, the data set was queried for similar Affymetrix probe identification numbers. This query revealed that at both 0 and 16 hours, 100% showed corroboration of data between the U95A and U133A&B arrays. While data from the new HG-U133 microarray set cannot be directly compared with the HG-U95 microarray, our comparison however strongly suggests excellent reproducibility between these 2 microarray generations and thus robustness of the new HG-U133A&B microarrays. Further data mining was therefore carried out with the data from the HG-U133A&B microarrays.
Results of data mining
Genes or probe sets meeting the inclusion criteria of at least a 2-fold difference between comparison groups at any of the 3 time points were subjected to one-way hierarchical clustering (Figure 1A). This analysis revealed that UCB and AB exhibit distinct gene expression profiles at each time point, with more genes and ESTs exhibiting reduced expression in UCB than AB at each time point.
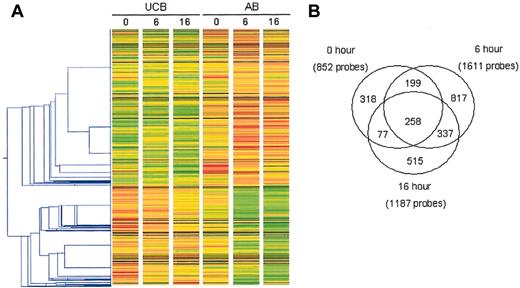
Analysis of changes in gene expression observed in unstimulated and stimulated CD4+ T cells isolated from UCB and AB. The changes in gene expression were assessed using the HG-U133A&B microarray. One-way hierarchical clustering (A) was performed on the trimmed data set using GeneSpring v5.1 following per gene normalizations of the signal. Each gene or probe set was normalized to the median of its signal across all arrays. The gene tree was constructed using the 2521 probe sets which met the inclusion criteria* in at least 1 of the 3 time points. Red and green indicate increased and decreased expression relative to the median, respectively. The Pearson correlation with a minimum distance of 0.001 was used in constructing the gene tree. (B) A Venn diagram depicts the total number of probe sets meeting inclusion criteria* at each time point. *At least one present (P) call and FC of at least 2 and change not equal to no change (NC).
Analysis of changes in gene expression observed in unstimulated and stimulated CD4+ T cells isolated from UCB and AB. The changes in gene expression were assessed using the HG-U133A&B microarray. One-way hierarchical clustering (A) was performed on the trimmed data set using GeneSpring v5.1 following per gene normalizations of the signal. Each gene or probe set was normalized to the median of its signal across all arrays. The gene tree was constructed using the 2521 probe sets which met the inclusion criteria* in at least 1 of the 3 time points. Red and green indicate increased and decreased expression relative to the median, respectively. The Pearson correlation with a minimum distance of 0.001 was used in constructing the gene tree. (B) A Venn diagram depicts the total number of probe sets meeting inclusion criteria* at each time point. *At least one present (P) call and FC of at least 2 and change not equal to no change (NC).
The final numbers of probe sets remaining after the inclusion analyses are shown in Figure 1B. At 0 hour, 852 probe sets met the criteria for inclusion, while at 6 and 16 hours 1611 probe sets and 1187 probe sets, respectively, met the criteria. Importantly, the number of differentially expressed probe sets increased after stimulation, with the largest difference at 6 hours. The Venn diagram in Figure 1B shows the overlap in these probe sets. Two hundred fifty-eight probe sets met the criteria for inclusion at all 3 time points. One hundred forty-one probe sets met inclusion criteria at 0 and 6 hours and 337 met the criteria at 6 and 16 hours, whereas only 77 probe sets were determined to be significant at 0 and 16 hours but not 6 hours.
NFAT-dependent gene expression
The NFAT pathway is crucial for expression of inflammatory cytokines and other immunomodulatory proteins as evidenced by NFATc2 gene-deleted mice.22 As NFATc2 is expressed at reduced levels in UCB,11 we queried the microarray data set for genes known to be dependent on NFATc2 and genes involved in the NFAT pathway (Tables 1, 2).15,16,21,22,28,29 All known NFATc2-dependent genes on the HG-U133A&B microarray that met inclusion criteria exhibited lower expression in UCB than AB CD4+ T lymphocytes, with the exception of the cyclins A2 and E2, which exhibited higher expression in UCB than AB (Table 1). The majority of differences were seen during stimulation, with the exception of CD25 (IL-2 receptor α [IL-2Rα]), which only showed differential expression at 0 hour. NFATc2-dependent genes including IL-3, IL-4, IL-5, IL-13, GM-CSF, IFN-γ, TNF-α, CD40L, and MIP-1α demonstrated lower expression in UCB T lymphocytes after 6 and 16 hours of stimulation. CTLA-4 expression was lower in UCB than in AB at all 3 time points. Several cyclins including A2, B1, E, and F, have been shown to be overexpressed in NFATc2 gene-deleted murine lymphocytes after stimulation.29 Consistent with this report, our data set revealed that cyclin A2 and cyclin E2 show greater expression in UCB than AB at 16 hours of stimulation.
NFAT-dependent genes with differential expression in UCB versus AB CD4+ T cells
Gene symbol . | Accession no. . | Gene name . | 0 h . | 6 h . | 16 h . |
|---|---|---|---|---|---|
| p21 | NM_000389 | Cyclin-dependent kinase inhibitor 1A (p21, Cip 1) | — | −2.64* | − 2.46* |
| CSF2 (GM-CSF) | M11734 | Colony-stimulating factor 2 (granulocyte-macrophage) | — | −6.96* | −2.14* |
| CTLA-4 | NM_005214 | Cytotoxic T-lymphocyte-associated protein 4 | — | −4.00* | −5.28* |
| CTLA-4 | BG536887 | Cytotoxic T-lymphocyte-associated protein 4 | −2.30* | −4.92* | −6.50* |
| CTLA-4 | U90273 | Cytotoxic T-lymphocyte-associated protein 4 | −2.30* | −4.92* | −7.46* |
| CTLA-4 | AI733018 | Cytotoxic T-lymphocyte-associated protein 4 | −3.25* | −5.28* | −6.06* |
| IFN-γ | M29383 | Interferon, gamma | −2.14* | −10.56* | −9.19* |
| IL-2Rα | NM_000417 | Interleukin 2 receptor, alpha | −2.14* | — | — |
| IL-3 | NM_000588 | Interleukin 3 (colony-stimulating factor, multiple) | — | −12.13* | −5.66* |
| IL-4 | NM_000589 | Interleukin 4 | — | −6.50* | −3.73* |
| IL-4 | NM_000589 | Interleukin 4 | — | −8.00* | −4.00* |
| IL-5 | NM_000879 | Interleukin 5 (colony-stimulating factor, eosinophil) | — | −17.15* | −8.00* |
| IL-13 | NM_002188 | Interleukin 13 | — | −3.73* | −3.48* |
| TNF | NM_000594 | Tumor necrosis factor (TNF superfamily, member 2) | — | −2.46* | — |
| TNFSF5 (CD40L) | NM000074 | Tumor necrosis factor (ligand) superfamily, member 5 (hyper-IgM syndrome) | — | −2.30* | −2.30* |
| MIP-1α | NM_002983 | CCL2 | — | −4.59* | −4.00* |
| CCNA2 | NM_001237 | Cyclin A2 | — | — | 8.57 |
| CCNE2 | AF112857 | Cyclin E2 | — | — | 2.46 |
| CCNE2 | NM_004702 | Cyclin E2 | — | — | 3.25 |
Gene symbol . | Accession no. . | Gene name . | 0 h . | 6 h . | 16 h . |
|---|---|---|---|---|---|
| p21 | NM_000389 | Cyclin-dependent kinase inhibitor 1A (p21, Cip 1) | — | −2.64* | − 2.46* |
| CSF2 (GM-CSF) | M11734 | Colony-stimulating factor 2 (granulocyte-macrophage) | — | −6.96* | −2.14* |
| CTLA-4 | NM_005214 | Cytotoxic T-lymphocyte-associated protein 4 | — | −4.00* | −5.28* |
| CTLA-4 | BG536887 | Cytotoxic T-lymphocyte-associated protein 4 | −2.30* | −4.92* | −6.50* |
| CTLA-4 | U90273 | Cytotoxic T-lymphocyte-associated protein 4 | −2.30* | −4.92* | −7.46* |
| CTLA-4 | AI733018 | Cytotoxic T-lymphocyte-associated protein 4 | −3.25* | −5.28* | −6.06* |
| IFN-γ | M29383 | Interferon, gamma | −2.14* | −10.56* | −9.19* |
| IL-2Rα | NM_000417 | Interleukin 2 receptor, alpha | −2.14* | — | — |
| IL-3 | NM_000588 | Interleukin 3 (colony-stimulating factor, multiple) | — | −12.13* | −5.66* |
| IL-4 | NM_000589 | Interleukin 4 | — | −6.50* | −3.73* |
| IL-4 | NM_000589 | Interleukin 4 | — | −8.00* | −4.00* |
| IL-5 | NM_000879 | Interleukin 5 (colony-stimulating factor, eosinophil) | — | −17.15* | −8.00* |
| IL-13 | NM_002188 | Interleukin 13 | — | −3.73* | −3.48* |
| TNF | NM_000594 | Tumor necrosis factor (TNF superfamily, member 2) | — | −2.46* | — |
| TNFSF5 (CD40L) | NM000074 | Tumor necrosis factor (ligand) superfamily, member 5 (hyper-IgM syndrome) | — | −2.30* | −2.30* |
| MIP-1α | NM_002983 | CCL2 | — | −4.59* | −4.00* |
| CCNA2 | NM_001237 | Cyclin A2 | — | — | 8.57 |
| CCNE2 | AF112857 | Cyclin E2 | — | — | 2.46 |
| CCNE2 | NM_004702 | Cyclin E2 | — | — | 3.25 |
Criteria for inclusion were as follows: at least one present (P) call and FC ≥ 2 and change call ≠ no change (NC). A negative number (*) indicates lower expression in UCB than AB; a positive number, higher expression in UCB than AB; —, the gene did not meet inclusion criteria at that time point; and genes with multiple results indicate multiple probe sets for that gene on the microarray.
NFAT-pathway-associated genes with differential expression in UCB versus AB CD4+ T cells
Gene symbol . | Accession no. . | Gene name . | 0 h . | 6 h . | 16 h . | |
|---|---|---|---|---|---|---|
| NFAT family members | ||||||
| NFAT5 | W61007 | Nuclear factor of activated T cells 5, tonicity responsive | — | −2.14* | — | |
| NFATC1 | U08015 | Nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1 | — | — | 2.00 | |
| NFAT-activating receptors | — | — | — | |||
| FCER1α | BC005912 | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | −2.30* | — | — | |
| Signaling molecules | ||||||
| PAK1 | AU147145 | p21/Cdc42/Rac1-activated kinase 1 | 2.00 | — | — | |
| PAK1 | U51120 | p21/Cdc42/Rac1-activated kinase 1 | 4.92 | — | 3.25 | |
| PAK1 | AU154408 | p21/Cdc42/Rac1-activated kinase 1 | — | 2.14 | — | |
| Vav3 | AF118887 | Vav3 oncogene | — | 3.73 | — | |
| Vav3 | NM_006113 | Vav3 oncogene | — | 2.83 | — | |
| CALM2 | BF591288 | Calmodulin 2 (phosphorylase kinase, delta) | — | −2.00* | — | |
| CAMK4 | AL529104 | Calcium/calmodulin-dependent protein kinase IV | — | — | −2.00* | |
| NFAT/Ca2+-signaling inhibitors | ||||||
| SLA | U44403 | Src-like adaptor | — | −3.25* | −2.64* | |
| SLA | NM_006748 | Src-like adaptor | — | −2.00* | −2.30* | |
| Related transcription factors | ||||||
| C/EBPβ | AL564683 | CCAAT/enhancer binding protein (C/EBP), beta | — | −2.00* | — | |
| JUNB | NM_002229 | JunB proto-oncogene | — | −2.46* | — | |
| NFKBIE (IκBϵ) | BG163267 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | — | −2.00* | — | |
| FOSL1 (Fra-1) | BG251266 | FOS-like antigen 1 | — | — | −2.64* | |
Gene symbol . | Accession no. . | Gene name . | 0 h . | 6 h . | 16 h . | |
|---|---|---|---|---|---|---|
| NFAT family members | ||||||
| NFAT5 | W61007 | Nuclear factor of activated T cells 5, tonicity responsive | — | −2.14* | — | |
| NFATC1 | U08015 | Nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1 | — | — | 2.00 | |
| NFAT-activating receptors | — | — | — | |||
| FCER1α | BC005912 | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | −2.30* | — | — | |
| Signaling molecules | ||||||
| PAK1 | AU147145 | p21/Cdc42/Rac1-activated kinase 1 | 2.00 | — | — | |
| PAK1 | U51120 | p21/Cdc42/Rac1-activated kinase 1 | 4.92 | — | 3.25 | |
| PAK1 | AU154408 | p21/Cdc42/Rac1-activated kinase 1 | — | 2.14 | — | |
| Vav3 | AF118887 | Vav3 oncogene | — | 3.73 | — | |
| Vav3 | NM_006113 | Vav3 oncogene | — | 2.83 | — | |
| CALM2 | BF591288 | Calmodulin 2 (phosphorylase kinase, delta) | — | −2.00* | — | |
| CAMK4 | AL529104 | Calcium/calmodulin-dependent protein kinase IV | — | — | −2.00* | |
| NFAT/Ca2+-signaling inhibitors | ||||||
| SLA | U44403 | Src-like adaptor | — | −3.25* | −2.64* | |
| SLA | NM_006748 | Src-like adaptor | — | −2.00* | −2.30* | |
| Related transcription factors | ||||||
| C/EBPβ | AL564683 | CCAAT/enhancer binding protein (C/EBP), beta | — | −2.00* | — | |
| JUNB | NM_002229 | JunB proto-oncogene | — | −2.46* | — | |
| NFKBIE (IκBϵ) | BG163267 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | — | −2.00* | — | |
| FOSL1 (Fra-1) | BG251266 | FOS-like antigen 1 | — | — | −2.64* | |
Criteria for inclusion were as follows: at least one present (P) call and FC ≥ 2 and change call ≠ no change (NC). A negative number (*) indicates lower expression in UCB than AB; a positive number, higher expression in UCB than AB;—, the gene did not meet inclusion criteria at that time point; and genes with multiple results indicate multiple probe sets for that gene on the microarray.
NFAT-pathway-associated gene expression
Of the NFAT family members, only NFAT5 and NFATc1 showed differential expression comparing UCB and AB. Surprisingly, we did not detect NFATc2 mRNA in either UCB or in adult CD4+ T lymphocytes at any time point in either of the 2 NFATc2 probe sets. Specifically, very low signals were detected by both probe sets and were determined absent in each sample by the MAS 5.0 algorithm (data not shown). This inability to detect NFATc2 could result from the intrinsic low levels of this message and/or possibly reflect a lack of sensitivity of the 2 NFATc2 probe sets. In this regard, in previous reverse transcriptase-polymerase chain reaction (RT-PCR) experiments, which logarithmically amplify the message being evaluated compared with linear amplification of cRNA for hybridization to microarrays, NFATc2 mRNA could not be reproducibly detected at 0 hour but could be detected consistently at 16 hours in adult (data not shown).
NFAT5 (also known as tonicity enhancer binding protein) showed decreased expression in UCB at 6 hours of stimulation. Although originally described as a response to hypertonicity, it has been shown that NFAT5-dependent transcription can be induced by T-cell-receptor (TCR)-dependent signaling events and is necessary for optimal T-cell development.30 Although UCB T lymphocytes up-regulated NFAT5 expression by 6 hours of stimulation, this up-regulation was attenuated when compared with the response seen in AB T lymphocytes. Expression fell to similar levels in both UCB and AB after 16 hours of stimulation. NFATc1 (NFAT2), which has been previously reported to be up-regulated within 3 hours of stimulation,31 was notably higher in UCB than AB at 16 hours. As UCB cells express low constitutive levels of NFATc2, the increase in NFATc1 mRNA at 16 hours of stimulation may indicate a compensatory mechanism.
Interestingly, examination of known NFAT-pathway genes revealed that only p21-activated protein kinase 1 (PAK1) and Vav3 demonstrated higher expression in UCB compared with AB cells. Calmodulin 2 (CALM2) and calmodulin-dependent kinase (CAMKIV) (both calmodulin-related genes) demonstrated lower expression in UCB at 6 and 16 hours, respectively. Src-like adapter protein (SLAP), an NFAT/Ca2+-signaling inhibitor, exhibited lower expression in UCB after stimulation at both time points. Three related transcription factors also showed differential expression with C/EBPβ and JunB showing lower expression in UCB compared with AB at 6 hours, and Fosl1 (Fra-1) demonstrating lower expression by UCB CD4+ T lymphocytes at 16 hours.
Cytokine and cytokine receptor mRNA expression
As cytokines and cytokine receptors play a crucial role in allogeneic inflammatory responses of T lymphocytes, we queried the array data to investigate the expression of these genes in UCB compared with AB at all time points. Fourteen cytokine and 10 cytokine receptor genes showed differential expression between UCB and AB CD4+ T lymphocytes at at least one of the 3 time points (Table 3). Of these, all except IL-16 showed lower mRNA expression in UCB CD4+ T lymphocytes with the majority of the differences seen during stimulation. IL-16 expression, as detected by 2 probe sets, was 3.48-fold and 2.83-fold higher in UCB than AB at 6 hours and 3.03-fold and 2.64-fold higher in UCB than AB at 16 hours.
Differential expression of cytokine and cytokine receptor genes in UCB versus AB CD4+T cells
Gene symbol . | Accession no. . | Gene name . | 0 h . | 6 h . | 16 h . |
|---|---|---|---|---|---|
| Cytokines | |||||
| IL-1β | NM_000576 | Interleukin 1, beta | — | −3.48* | −2.00* |
| IL-1β | M15330 | Interleukin 1, beta | — | −3.03* | |
| IL-3 | NM_000588 | Interleukin 3 (colony-stimulating factor, multiple) | — | −12.13* | −5.66* |
| IL-4 | NM_000589 | Interleukin 4 | — | −6.50* | −3.73* |
| IL-4 | NM_000589 | Interleukin 4 | — | −8.00* | −4.00* |
| IL-5 | NM_000879 | Interleukin 5 (colony-stimulating factor, eosinophil) | — | −17.15* | −8.00* |
| IL-9 | NM_000590 | Interleukin 9 | — | −6.96* | −548.75* |
| IL-10 | NM_000572 | Interleukin 10 | — | −16.00* | −27.86* |
| IL-13 | NM_002188 | Interleukin 13 | — | −3.73* | −3.48* |
| IL-15 | NM_000585 | Interleukin 15 | −2.83* | −3.48* | −2.14* |
| IL-16 | NM_004513 | Interleukin 16 (lymphocyte chemoattractant factor) | — | 3.48 | 3.03 |
| IL-16 | M90391 | Interleukin 16 (lymphocyte chemoattractant factor) | — | 2.83 | 2.64 |
| IL-17 | Z58820 | Interleukin 17 (cytotoxic T-lymphocyte-associated serine esterase 8) | — | −84.45* | −675.59* |
| IL-21 | NM_021803 | Interleukin 21 | — | −7.46* | −8.00* |
| IL-22 | NM_020525 | Interleukin 22 | — | −13.00* | −27.86* |
| IL-22 | AF279437 | Interleukin 22 | — | −39.40* | −512.00* |
| IL-26 | NM_018402 | Interleukin 26 | — | — | −207.94* |
| Cytokine receptors | |||||
| IL-1RN | BE563442 | Interleukin 1 receptor antagonist | — | −2.00* | — |
| IL-1RN | AW083357 | Interleukin 1 receptor antagonist | — | −2.46* | — |
| IL-1RN | AW083357 | Interleukin 1 receptor antagonist | — | −2.30* | — |
| IL-1R1 | NM_000877 | Interleukin 1 receptor, type 1 | — | −2.30* | −2.00* |
| IL-1R2 | NM_004633 | Interleukin 1 receptor, type II | — | −3.48* | — |
| IL-1R2 | U64094 | Interleukin 1 receptor, type II | — | — | −2.64* |
| IL-2Rα | NM_000417 | Interleukin 2 receptor, alpha | −2.14* | — | — |
| IL-2Rβ | NM_000878 | Interleukin 2 receptor, beta | −2.83* | −2.30* | — |
| IL-6R | NM_000565 | Interleukin 6 receptor | — | −2.46* | — |
| IL-6R | S72848 | Interleukin 6 receptor | — | — | −3.25* |
| IL-6R | AV700030 | Interleukin 6 receptor | — | −2.46* | −4.59* |
| IL-7R | NM_002185 | Interleukin 7 receptor | — | — | −2.64* |
| IL-10Rα | NM_001558 | Interleukin 10 receptor, alpha | −2.64* | −4.29* | −2.46* |
| IL-17BR | AF250309 | Interleukin 17B receptor | — | −8.57* | — |
| IL-18R1 | NM_003855 | Interleukin 18 receptor 1 | −2.30* | — | — |
| IL-18RAP | NM_003853 | Interleukin 18 receptor accessory protein | −6.06* | −2.14* | −2.83* |
Gene symbol . | Accession no. . | Gene name . | 0 h . | 6 h . | 16 h . |
|---|---|---|---|---|---|
| Cytokines | |||||
| IL-1β | NM_000576 | Interleukin 1, beta | — | −3.48* | −2.00* |
| IL-1β | M15330 | Interleukin 1, beta | — | −3.03* | |
| IL-3 | NM_000588 | Interleukin 3 (colony-stimulating factor, multiple) | — | −12.13* | −5.66* |
| IL-4 | NM_000589 | Interleukin 4 | — | −6.50* | −3.73* |
| IL-4 | NM_000589 | Interleukin 4 | — | −8.00* | −4.00* |
| IL-5 | NM_000879 | Interleukin 5 (colony-stimulating factor, eosinophil) | — | −17.15* | −8.00* |
| IL-9 | NM_000590 | Interleukin 9 | — | −6.96* | −548.75* |
| IL-10 | NM_000572 | Interleukin 10 | — | −16.00* | −27.86* |
| IL-13 | NM_002188 | Interleukin 13 | — | −3.73* | −3.48* |
| IL-15 | NM_000585 | Interleukin 15 | −2.83* | −3.48* | −2.14* |
| IL-16 | NM_004513 | Interleukin 16 (lymphocyte chemoattractant factor) | — | 3.48 | 3.03 |
| IL-16 | M90391 | Interleukin 16 (lymphocyte chemoattractant factor) | — | 2.83 | 2.64 |
| IL-17 | Z58820 | Interleukin 17 (cytotoxic T-lymphocyte-associated serine esterase 8) | — | −84.45* | −675.59* |
| IL-21 | NM_021803 | Interleukin 21 | — | −7.46* | −8.00* |
| IL-22 | NM_020525 | Interleukin 22 | — | −13.00* | −27.86* |
| IL-22 | AF279437 | Interleukin 22 | — | −39.40* | −512.00* |
| IL-26 | NM_018402 | Interleukin 26 | — | — | −207.94* |
| Cytokine receptors | |||||
| IL-1RN | BE563442 | Interleukin 1 receptor antagonist | — | −2.00* | — |
| IL-1RN | AW083357 | Interleukin 1 receptor antagonist | — | −2.46* | — |
| IL-1RN | AW083357 | Interleukin 1 receptor antagonist | — | −2.30* | — |
| IL-1R1 | NM_000877 | Interleukin 1 receptor, type 1 | — | −2.30* | −2.00* |
| IL-1R2 | NM_004633 | Interleukin 1 receptor, type II | — | −3.48* | — |
| IL-1R2 | U64094 | Interleukin 1 receptor, type II | — | — | −2.64* |
| IL-2Rα | NM_000417 | Interleukin 2 receptor, alpha | −2.14* | — | — |
| IL-2Rβ | NM_000878 | Interleukin 2 receptor, beta | −2.83* | −2.30* | — |
| IL-6R | NM_000565 | Interleukin 6 receptor | — | −2.46* | — |
| IL-6R | S72848 | Interleukin 6 receptor | — | — | −3.25* |
| IL-6R | AV700030 | Interleukin 6 receptor | — | −2.46* | −4.59* |
| IL-7R | NM_002185 | Interleukin 7 receptor | — | — | −2.64* |
| IL-10Rα | NM_001558 | Interleukin 10 receptor, alpha | −2.64* | −4.29* | −2.46* |
| IL-17BR | AF250309 | Interleukin 17B receptor | — | −8.57* | — |
| IL-18R1 | NM_003855 | Interleukin 18 receptor 1 | −2.30* | — | — |
| IL-18RAP | NM_003853 | Interleukin 18 receptor accessory protein | −6.06* | −2.14* | −2.83* |
Criteria for inclusion were as follows: at least one present (P) call and FC ≥ 2 and change call ≠ no change (NC). A negative number (*) indicates lower expression in UCB than AB; a positive number indicates higher expression in UCB than AB;—, the gene did not meet inclusion criteria at that time point; and genes with multiple results indicate multiple probe sets for that gene on the microarray.
Chemokine and chemokine receptor mRNA expression
Chemokines and their receptors have been implicated in the pathogenesis of GVHD32 as well as allograft rejection.33 Additionally, they have been shown to have differential expression in UCB with RANTES, CCR1, CCR2, CCR5, CCR6, and CXCR3, previously described as reduced in UCB.34,35 We therefore queried the array data to determine whether there may be differential expression in additional chemokine and receptor genes between UCB and AB (Table 4). Again, with the exception of CXCL11 (IFN-inducible T-cell α chemoattractant [I-TAC]), UCB demonstrated overall lower chemokine mRNA expression than AB. Differences were seen at all time points with most differences seen during stimulation. In the absence of stimulation, approximately half of the chemokine receptor genes showed lower expression in UCB with the remaining receptors showing lower expression after stimulation. These data suggest possible impact on the immediate response of UCB T lymphocytes to chemokine signals.
Differential expression of chemokine and chemokine receptor genes in UCB versus AB CD4+ T cells
Gene symbol . | Accession no. . | Gene name . | 0 h . | 6 h . | 16 h . |
|---|---|---|---|---|---|
| Chemokine ligands | |||||
| CCL2 (MCP-1) | S69738 | Chemokine (C-C motif) ligand 2 | — | −9.19* | — |
| CCL3 (MIP-1α) | NM_002983 | Chemokine (C-C motif) ligand 3 | — | −4.59* | −4.00* |
| CCL4 (MIP-1β) | NM_002984 | Chemokine (C-C motif) ligand 4 | −4.00* | −8.57* | −8.00* |
| CCL5 (RANTES) | M21121 | Chemokine (C-C motif) ligand 5 | −22.63* | −18.38* | −5.28* |
| CCL5 (RANTES) | NM_002985 | Chemokine (C-C motif) ligand 5 | −13.93* | −9.19* | −6.06* |
| CCL17 (TARC) | NM_002987 | Chemokine (C-C motif) ligand 17 | — | −2.46* | — |
| CCL20 (MIP-3α) | NM_004591 | Chemokine (C-C motif) ligand 20 | — | −3.03* | −8.00* |
| CXCL2 (GRO-β) | M57731 | Chemokine (C-X-C motif) ligand 2 | — | — | −4.00* |
| CXCL5 (ENA-78) | AK026546 | Chemokine (C-X-C motif) ligand 5 | — | — | −3.48* |
| CXCL9 (MIG) | NM002416 | Chemokine (C-X-C motif) ligand 9 | — | −3.48* | |
| CXCL11 (I-TAC) | AF030514 | Chemokine (C-X-C motif) ligand 11 | — | −3.73* | 2.30 |
| CXCL11 (I-TAC) | AF002985 | Chemokine (C-X-C motif) ligand 11 | — | −3.48* | |
| CXCL13 (BLC/BCA-1) | NM_006419 | Chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) | — | — | −19.70* |
| XCL1 | U23772 | Chemokine (C motif) ligand 1 | — | −8.00* | — |
| XCL1 | NM_002995 | Chemokine (C motif) ligand 1 | — | −4.00* | — |
| XCL2 (SCM-1β) | NM_003175 | Chemokine (C motif) ligand 2 | — | −8.00* | — |
| Chemokine receptors | |||||
| CCR1 | AI421071 | Chemokine (C-C motif) receptor 1 | — | — | −6.06* |
| CCR1 | NM_001295 | Chemokine (C-C motif) receptor 1 | — | — | −19.70* |
| CCR2 | NM_000647 | Chemokine (C-C motif) receptor 2 | −6.06* | −7.46* | −9.85* |
| CCR2 | NM_000648 | Chemokine (C-C motif) receptor 2 | −5.28* | −2.64* | −29.86* |
| CCR5 | NM_000579 | Chemokine (C-C motif) receptor 5 | −2.64* | −8.57* | −18.38* |
| CCR6 | NM_004367 | Chemokine (C-C motif) receptor 6 | −10.56* | −8.57* | −8.57* |
| CX3CR1 | U20350 | Chemokine (C-X3-C motif) receptor 1 | −4.00* | — | — |
| CXCR3 | NM_001504 | Chemokine (C-X-C motif) receptor 3 | −2.00* | −2.14* | — |
| CXCR4 | L01639 | Chemokine (C-X-C motif) receptor 4 | — | −2.14* | −2.14* |
| CXCR4 | AF348491 | Chemokine (C-X-C motif) receptor 4 | — | −2.30* | — |
| CXCR6 | NM_006564 | Chemokine (C-X-C motif) receptor 6 | — | −3.03* | −5.66* |
| CXCR6 | U73531 | Chemokine (C-X-C motif) receptor 6 | — | −12.13* | −6.96* |
Gene symbol . | Accession no. . | Gene name . | 0 h . | 6 h . | 16 h . |
|---|---|---|---|---|---|
| Chemokine ligands | |||||
| CCL2 (MCP-1) | S69738 | Chemokine (C-C motif) ligand 2 | — | −9.19* | — |
| CCL3 (MIP-1α) | NM_002983 | Chemokine (C-C motif) ligand 3 | — | −4.59* | −4.00* |
| CCL4 (MIP-1β) | NM_002984 | Chemokine (C-C motif) ligand 4 | −4.00* | −8.57* | −8.00* |
| CCL5 (RANTES) | M21121 | Chemokine (C-C motif) ligand 5 | −22.63* | −18.38* | −5.28* |
| CCL5 (RANTES) | NM_002985 | Chemokine (C-C motif) ligand 5 | −13.93* | −9.19* | −6.06* |
| CCL17 (TARC) | NM_002987 | Chemokine (C-C motif) ligand 17 | — | −2.46* | — |
| CCL20 (MIP-3α) | NM_004591 | Chemokine (C-C motif) ligand 20 | — | −3.03* | −8.00* |
| CXCL2 (GRO-β) | M57731 | Chemokine (C-X-C motif) ligand 2 | — | — | −4.00* |
| CXCL5 (ENA-78) | AK026546 | Chemokine (C-X-C motif) ligand 5 | — | — | −3.48* |
| CXCL9 (MIG) | NM002416 | Chemokine (C-X-C motif) ligand 9 | — | −3.48* | |
| CXCL11 (I-TAC) | AF030514 | Chemokine (C-X-C motif) ligand 11 | — | −3.73* | 2.30 |
| CXCL11 (I-TAC) | AF002985 | Chemokine (C-X-C motif) ligand 11 | — | −3.48* | |
| CXCL13 (BLC/BCA-1) | NM_006419 | Chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) | — | — | −19.70* |
| XCL1 | U23772 | Chemokine (C motif) ligand 1 | — | −8.00* | — |
| XCL1 | NM_002995 | Chemokine (C motif) ligand 1 | — | −4.00* | — |
| XCL2 (SCM-1β) | NM_003175 | Chemokine (C motif) ligand 2 | — | −8.00* | — |
| Chemokine receptors | |||||
| CCR1 | AI421071 | Chemokine (C-C motif) receptor 1 | — | — | −6.06* |
| CCR1 | NM_001295 | Chemokine (C-C motif) receptor 1 | — | — | −19.70* |
| CCR2 | NM_000647 | Chemokine (C-C motif) receptor 2 | −6.06* | −7.46* | −9.85* |
| CCR2 | NM_000648 | Chemokine (C-C motif) receptor 2 | −5.28* | −2.64* | −29.86* |
| CCR5 | NM_000579 | Chemokine (C-C motif) receptor 5 | −2.64* | −8.57* | −18.38* |
| CCR6 | NM_004367 | Chemokine (C-C motif) receptor 6 | −10.56* | −8.57* | −8.57* |
| CX3CR1 | U20350 | Chemokine (C-X3-C motif) receptor 1 | −4.00* | — | — |
| CXCR3 | NM_001504 | Chemokine (C-X-C motif) receptor 3 | −2.00* | −2.14* | — |
| CXCR4 | L01639 | Chemokine (C-X-C motif) receptor 4 | — | −2.14* | −2.14* |
| CXCR4 | AF348491 | Chemokine (C-X-C motif) receptor 4 | — | −2.30* | — |
| CXCR6 | NM_006564 | Chemokine (C-X-C motif) receptor 6 | — | −3.03* | −5.66* |
| CXCR6 | U73531 | Chemokine (C-X-C motif) receptor 6 | — | −12.13* | −6.96* |
Criteria for inclusion were as follows: at least one present (P) call and FC ≥ 2 and change call ≠ no change (NC). A positive number indicates higher expression in UCB than AB; a negative number (*), lower expression in UCB than AB; —, the gene did not meet inclusion criteria at that time point; and genes with multiple results indicate multiple probe sets for that gene on the microarray.
Confirmation of mRNA expression
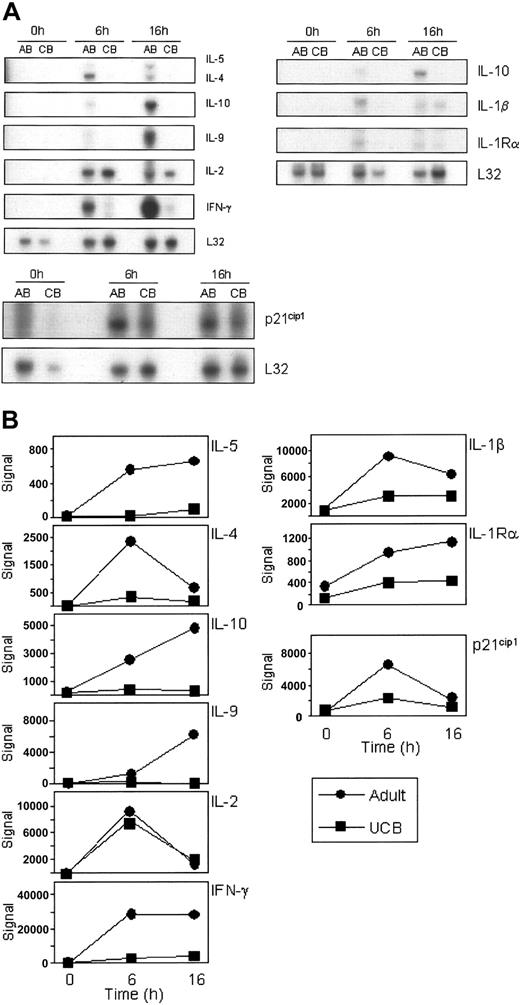
To confirm the data generated from the HG-U133A&B microarrays, RNase protection assays (RPAs) were performed analyzing the same RNA pools that were used for the U133AB gene array analysis. Expression patterns of NFAT-dependent genes including IL-2, IL-4, IFN-γ, and p21 were confirmed by RPA (Figure 2A). Additionally, RPA results for cytokines IL-1β, IL-5, IL-9, IL-10, and receptor IL-1Rα were noted to corroborate the array data. As shown in Figure 2A-B, the experimental results of mRNA expression in the RPAs mirror the expression profiles detected by the microarrays (raw signal) with, for example, IL-4 showing greatest expression at 6 hours on both the microarray and the RPA, while IL-10 expression continues to increase from 0 to 16 hours.
Corroboration of differential mRNA expression of selected probes by multiprobe RNase protection analysis. The same total RNA pools used for the gene array studies were analyzed for mRNA expression by RNase protection arrays (A). Five micrograms of each pool of RNA was used per hybridization to hCK1, hCK2b, and hStress multiprobe templates. Exposure times varied among the gels depending upon the strength of each signal. L32 mRNA is included as a control for input RNA. Representative results from 2 to 3 repeat experiments are shown. (B) Microarray expression data from the HG-U133A&B array corresponding to the above genes were graphed individually, showing the raw signal. ▪ indicates UCB; and •, adult.
Corroboration of differential mRNA expression of selected probes by multiprobe RNase protection analysis. The same total RNA pools used for the gene array studies were analyzed for mRNA expression by RNase protection arrays (A). Five micrograms of each pool of RNA was used per hybridization to hCK1, hCK2b, and hStress multiprobe templates. Exposure times varied among the gels depending upon the strength of each signal. L32 mRNA is included as a control for input RNA. Representative results from 2 to 3 repeat experiments are shown. (B) Microarray expression data from the HG-U133A&B array corresponding to the above genes were graphed individually, showing the raw signal. ▪ indicates UCB; and •, adult.
Analysis of protein expression
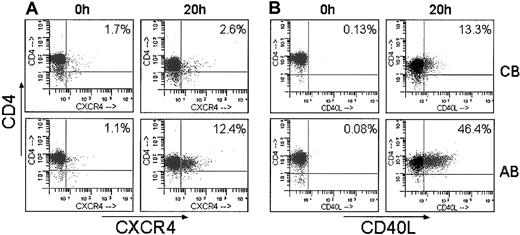
Next, additional confirmatory studies analyzing protein expression by flow cytometry were performed on purified T lymphocytes stimulated for 20 hours. This later time point was chosen to allow for translation of the mRNA into detectable proteins. Correlating with mRNA expression measured by gene array, CXCR4 and CD40L exhibited significantly reduced surface expression in stimulated UCB T lymphocytes. Stimulated cells expressing CXCR4 ranged from 2.1% to 3.4% on UCB compared with 2.5% to 15.7% on AB (n = 4; P < .05); and cells expressing CD40L ranged from 0.74% to 13.3% on UCB compared with 15% to 46.4% on AB (n = 4; P < .05). Representative dot plots are shown in Figure 3. At 20 hours of stimulation, despite the decreased expression detected on the gene array, CTLA-4 protein surface expression did not differ between UCB and AB.
Decreased surface expression of CXCR4 and CD40L in stimulated UCB CD4+ T cells. CD4+ T cells were purified from each UCB and AB sample, stimulated for 20 hours and analyzed by flow cytometry for surface expression of (A) CXCR4 and (B) CD40L. Representative histograms of 4 UCB and 4 adults are shown.
Decreased surface expression of CXCR4 and CD40L in stimulated UCB CD4+ T cells. CD4+ T cells were purified from each UCB and AB sample, stimulated for 20 hours and analyzed by flow cytometry for surface expression of (A) CXCR4 and (B) CD40L. Representative histograms of 4 UCB and 4 adults are shown.
In addition, multiplex cytokine protein measurements were performed using fluorescent microspheres on supernatants obtained from purified T lymphocytes stimulated for 20 hours. NFAT-dependent cytokines including IL-3, IL-4, IL-5, IL-13, GM-CSF, and the chemokine MIP-1α demonstrated strongly reduced protein expression in UCB compared with adult supernatant at 20 hours (Table 5). In addition to NFAT-dependent genes, IL-1β, IL-10, RANTES, MIG, and IL-8 were found to have strongly reduced protein expression in UCB supernatant. Importantly, IL-2 did not demonstrate differential expression by micro-array, RPA, or protein analysis, suggesting NFAT-independent regulation.
Cytokine expression profile of UCB- versus AB-stimulated T-cell supernatants
. | UCB . | AB . |
|---|---|---|
| Cytokines, pg/mL | ||
| IL-2 | 2122 ± 208 | 1815 ± 73 |
| IL-3 | 127 ± 19 | 532 ± 30 |
| IL-4 | 5.7 ± 1 | 125 ± 15 |
| IL-5 | 9 ± 2.8 | 147 ± 3 |
| IL-10 | 232 ± 18 | 20 310 ± 70 |
| IL-13 | <LDL | 410 ± 2 |
| IL-1β | 2 ± 0 | 18.9 ± 2.7 |
| GM-CSF | 168 ± 11 | 1450 ± 186 |
| Chemokines, pg/mL | ||
| RANTES | 118 ± 18 | 452 ± 28 |
| MIG | 17.7 ± 0.9 | 74 ± 32 |
| MIP-1α | 664 ± 77 | 2479 ± 253 |
| IL-8 | 441 ± 17 | 827 ± 65 |
. | UCB . | AB . |
|---|---|---|
| Cytokines, pg/mL | ||
| IL-2 | 2122 ± 208 | 1815 ± 73 |
| IL-3 | 127 ± 19 | 532 ± 30 |
| IL-4 | 5.7 ± 1 | 125 ± 15 |
| IL-5 | 9 ± 2.8 | 147 ± 3 |
| IL-10 | 232 ± 18 | 20 310 ± 70 |
| IL-13 | <LDL | 410 ± 2 |
| IL-1β | 2 ± 0 | 18.9 ± 2.7 |
| GM-CSF | 168 ± 11 | 1450 ± 186 |
| Chemokines, pg/mL | ||
| RANTES | 118 ± 18 | 452 ± 28 |
| MIG | 17.7 ± 0.9 | 74 ± 32 |
| MIP-1α | 664 ± 77 | 2479 ± 253 |
| IL-8 | 441 ± 17 | 827 ± 65 |
Purified CD4+ T cells from one UCB and adult each, were stimulated for 20 hours and supernatants were analyzed in duplicates for cytokine expression using fluorescent microspheres. Numbers are average of duplicate samples (± standard deviation). LDL indicates lower detection level. Representative of 3 performed experiments.
Impaired basal expression of transcription factors
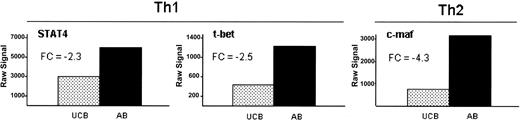
With the observed global decrease in both Th1 and Th2 cytokines and chemokines, we explored the gene expression of related transcription factors prior to stimulation. Th1-related transcription factors STAT436 and T-bet37 and Th2-related transcription factor c-maf38,39 demonstrated reduced gene expression in UCB at 0 hour of 2.3-fold, 2.5-fold, and 4.3-fold, respectively (Figure 4). STAT4 and T-bet remained low at 6 hours of stimulation in UCB, while c-maf exhibited severely reduced expression at both simulation points (data not shown). Interestingly, while NFκB mRNA expression was similar at all time points, there was reduced IκBϵ (NFKBIE) expression in UCB compared with adult at 6 hours of stimulation, suggesting potentially reduced NFκB retention in the cytoplasm at later time points of stimulation (Table 1).
Impaired basal expression of transcription factors associated with Th1 and Th2 phenotype in unstimulated UCB. Th1 (STAT4 and t-bet) and Th2 (c-maf) associated transcription factors are decreased in expression in UCB at 0 hour. All 3 genes met the criteria for inclusion as described in “Materials and methods.” Data are shown here as the raw signal obtained from the Affymetrix HGU133A&B microarray. Fold changes between UCB and AB are indicated (FC).
Impaired basal expression of transcription factors associated with Th1 and Th2 phenotype in unstimulated UCB. Th1 (STAT4 and t-bet) and Th2 (c-maf) associated transcription factors are decreased in expression in UCB at 0 hour. All 3 genes met the criteria for inclusion as described in “Materials and methods.” Data are shown here as the raw signal obtained from the Affymetrix HGU133A&B microarray. Fold changes between UCB and AB are indicated (FC).
Discussion
To gain better insight into differences in global gene expression between UCB and adult T cells, we have performed a microarray-based gene expression analysis of primary purified UCB and adult CD4+ T cells at baseline and after 6 and 16 hours of primary stimulation. Of the approximately 33 000 genes and ESTs available on the array, only 852, 1611, and 1187 met the inclusion criteria (eg, at least one present [P] call and change other than NC and fold change [FC] of at least 2) at 0, 6, and 16 hours, respectively. The largest numbers of differentially expressed genes and ESTs were noted to occur during stimulation. Importantly, at each time point, a larger number of genes exhibited reduced expression in UCB compared with adult than vice versa. This suggests that UCB CD4+ T cells do not up-regulate genes comparable to adult T cells upon stimulation but rather up-regulate a different set of genes as suggested by the patterning on the hierarchical gene tree (Figure 1A).
The focus of this analysis concentrated on NFATc2-dependent genes, as an extension of our previously reported studies outlining deficiencies in UCB NFATc2 protein expression, as well as reduced expression of the NFATc2-dependent genes CTLA-4, IFN-γ, and TNF-α in UCB T cells compared with adult controls.11,23 In this analysis of CD4+ T-cell mRNA expression during primary stimulation, we observed that many NFAT-pathway-related genes and NFATc2-dependent genes exhibited lower expression levels in UCB. In addition to NFAT-regulated cytokine genes, the gene array data revealed a profound overall defect in cytokine expression in UCB T cells, including both Th1 and Th2 cytokines40 as well as chemokines. This may be attributable in part to the reduced expression of Th1- and Th2-associated transcription factors STAT4, T-bet, and c-maf in UCB. Our results thus suggest global immune deficiencies in UCB T cells that are regulated in part at the transcriptional level. Reduced and delayed up-regulation of NFAT-associated genes in UCB may provide the developing neonatal T cells a slower response, thus providing a “window of opportunity” to delineate appropriate level of response to benign environmental antigens versus pathogens. This reduced and slow up-regulation of NFAT-associated genes lends support to immune repertoire development aligned with the Matzinger “Danger Model” that suggests that the immune system does not discriminate between self and nonself but rather between dangerous and harmless entities.41
The dramatically reduced IFN-γ expression (10.6-fold at 6 h; 9.2-fold at 16 h) observed in stimulated UCB T cells did not correlate with residual IFN-γ expression previously reported in NFATc2 gene-deleted mice, suggesting additional deficiencies in UCB.22 The observed reduced mRNA expression of JunB and Fosl1 (Fra-1), both of which partner with NFATc2 to form the AP-1/NFAT transcriptional complex,20 may underlie this severely reduced IFN-γ expression observed upon stimulation of UCB T cells. This deficiency would be expectedly amplified by the observed reduced expression of T-bet in UCB, another transcription factor important for IFN-γ gene expression,42 as no compensation for reduced NFAT through T-bet can occur to up-regulate the IFN-γ gene.
Conflicting data exist regarding expression of IL-2 in UCB T cells.7,10,43,44 IL-2 participates in activation-induced cell death (AICD) of CD4+ T cells and thus contributes to peripheral tolerance through the elimination of selfreactive T cells.45 In our study, we found no difference comparing UCB and adult in expression of IL-2 at any time point by either microarray, RPA, or protein expression analysis. As NFAT complex regulates IL-2 expression, one would expect decreased expression in UCB. The same lack of impaired IL-2 expression was also reported in NFATc2 gene-deleted mice. However, known redundant regulatory mechanisms exist, including other NFAT family members and the NFκB pathway.46,47 NFATc1, a known contributor to IL-2 expression in T cells,46 increases in expression in UCB over AB at 16 hours. Additionally, with reduced expression of NFATc2, transcription of the IL-2 gene may be activated by NFκB.
A possible increase in NFκB transcriptional activity in UCB is suggested by the decreased expression of the NFκB inhibitor IκBϵ (NFKBIE), as well as increased expression of Vav3, a protein that is phosphorylated in response to TCR stimulation and preferentially enhances NFκB-dependent transcription.48 It is interesting to note that Kilpinen et al49 found higher nuclear translocation and activation of NFκB in UCB CD4+ T lymphocytes, suggesting increased activation of NFκB, possibly through defects in inhibitory mechanisms, as suggested by our data. Taken together, our observations of differential expression of NFAT-pathway-associated genes supports previous findings of reduced NFATc2 in UCB while also suggesting possible compensatory pathways for the regulation of IL-2.
While Th1 cytokine deficiency in UCB T cells has been well documented7-12 and has been corroborated by this gene expression data, conflicting data exist regarding Th2 cytokine expression.7,10,13,43,50-52 Several studies have described higher levels of Th2 cytokines in UCB than in adult,50,52 suggesting a Th2 shift that may underlie reduced GVHD observed after UCB transplantation. Our gene array data, including confirmation by RNase protection and cytokine expression analyses, confirms lower Th2 cytokine expression by UCB T cells, including IL-4, IL-5, IL-9, IL-10, and IL-13. Moreover, the reduced mRNA expression of c-maf in UCB, an important transcription factor regulating Th2 cytokine expression, suggests one possible mechanism underlying reduced IL-4 gene expression observed in UCB.38,39
A large number of chemokine and chemokine receptors were also found to have differential expression in UCB and adult T cells. With the exception of CXCL11 (I-TAC) at 16 hours of stimulation, all of the differentially expressed genes were decreased in UCB compared with adult. Several of these chemokines and receptors, including RANTES, CCR1, CCR2, CCR5, CCR6, and CXCR3, have been previously described as reduced in UCB.34,35 Our data corroborate these reports and suggest additional chemokines and chemokine receptors have impaired expression in UCB such as CXCR4, a coreceptor for HIV entry.53 Decreased expression of these chemokines, and particularly their receptors, suggests an additional underlying mechanism for the clinical observations of reduced GVHD allogeneic reactivity observed in UCB transplant recipients, as T-cell migration toward sites of inflammation would be expectedly impaired. This hypothesis is underscored by the report by Sato et al,35 who demonstrated reduced migration of UCB in response to certain inflammatory chemokines. In addition, Fahmy et al33 recently reported the correlation of IFN-γ-inducible protein 10 (IP-10), MIG, I-TAC, RANTES, CXCR3, and CCR5 expression in human cardiac allografts with the presence and grade of acute rejection. Our results show reduced expression of these genes in UCB, with the exception of IP-10, which shows equivalent expression, and I-TAC, which is reduced in UCB at 6 hours but is increased in UCB at 16 hours compared with adult. These observations of reduced chemokine expression and their receptors by UCB CD4+ T cells point to gene regulation underlying neonatal immune development, expectedly limiting T-cell reactivity to low-affinity antigen stimuli.
In addition to the global decreased expression of cytokines, chemokines, receptors, and transcription factors and in concert with the reduced NFATc2 expression, UCB T cells appear to have a profound dysregulation of the Ca2+-signaling cascade. CALM2 and CAMKIV (both calmodulin-related genes) demonstrated lower expression in UCB than AB at 6 and 16 hours, respectively. Surprisingly, the Ca2+ flux inhibitor SLAP was expressed at reduced levels in UCB compared with AB, while IL-16, which has been shown to inhibit Ca2+ influx in stimulated T cells,54 was the only cytokine expressed at higher levels in UCB than AB (∼ 3-fold). A decrease in Ca2+ influx would expectedly result in deficient activation of T lymphocytes through reduced activation of the Ca2+-dependent phosphatase calcineurin and resulting reduced activation of NFAT proteins. One may speculate that increased expression of IL-16 may serve as an active repression mechanism to prevent immediate response of UCB T cells, thus allowing for gradual adaptation to environmental antigens during the ontogeny of the neonatal immune system. Additionally, as the Ca2+ influx may be already inhibited by IL-16, other negative regulatory signals such as SLAP may be less necessary. Further studies to confirm these observations are warranted.
We and others12,13,23 have previously reported increased autoproliferation as well as increased proliferation of UCB cells after primary and secondary stimulation compared with AB cells. Additionally, we have reported increased AICD in UCB T cells after secondary stimulation.12 Recent reports of increased proliferation, apoptosis, dysregulated cell cycling, and overexpression of cyclin genes after stimulation in gene-deleted NFATc2 mice have implicated NFATc2 in cell cycle regulation.29 Our findings of increased cyclin gene expression in stimulated UCB T cells lend further support to this hypothesis. Increased cell turnover may result in the elimination of activated alloreactive donor T cells, thus providing an additional possible underlying etiology in the reduced GVHD observed after UCB transplantation.
Our study provides a comprehensive analysis of the cytokine profile of UCB and AB CD4+ T cells during the early phases of primary stimulation, not only corroborating previous reports but also identifying additional differences in gene regulation underlying reduced UCB T-cell inflammatory responses. Overall, the global reduction of both Th1 and Th2 cytokine gene expression, as well as additional cytokines and receptors, indicates a functional immaturity of UCB T cells, both in the cell-mediated as well as humoral immune response. Taken together, these results not only may explain the reduced GVHD observed after UCB transplantation but also contribute to the elucidation of neonatal tolerance and T-cell repertoire development. The neonatal period of immune tolerance allows for the T cells to learn to distinguish self-antigens from nonself, a process crucial to the prevention of autoimmunity. Our study thus provides a basis for further in-depth analyses of UCB T-cell responses and a rationale for potential targeted GVHD prophylaxis and treatment.
Prepublished online as Blood First Edition Paper, August 28, 2003; DOI 10.1182/blood-2003-05-1732.
Supported by grant no. 6230-98 from The Leukemia and Lymphoma Society of America, the Abraham and Phyllis Katz Foundation and grant RO1-AI47289-01 from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID). M.J.L. is a Leukemia Scholar in Clinical Research.
M.L.V. and M.J.L. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal