Abstract
Genomic aberrations in a series of paired biopsy samples from patients who presented initially with follicle center lymphoma (FCL) and subsequently transformed to diffuse large B-cell lymphoma (DLBCL) were measured by array comparative genomic hybridization (CGH). The consequences of these aberrations on gene expression were determined by comparison with expression analysis on these specimens using cDNA microarrays. A heterogeneous pattern of acquired genomic abnormalities was observed upon transformation, some of which were recurrent in small subsets of patients. Some of the genomic aberration acquired upon transformation, such as gain/amplification of 1q21-q24, 2p16 (REL/BCL11A gene loci), 3q27-q29 (including theBCL6 locus), 7q11.2-q22.1, 12pter-q12, 18q21 (including theBCL2 locus) and Xq, and deletion of 6q22-q24, 13q14-q21 and 17p13 (P53 locus) have been previously implicated in the FCL/DLBCL pathogenesis. In addition, novel genomic imbalances not previously reported in association with FCL transformation, such as overrepresentation of 4p12-pter, 5p12-p15, 6p12.3-p21, 9p23, 9q13-q31, 16q, 17q21, and loss of 1p36.3, 4q21-q23, 5q21-q23, 9q31-qter, 11q24-q25, and 15q23, were identified. We observed a differential expression profile of many genes within regions of gain and deletion upon transformation, including novel target genes associated with FCL transformation. However, other genes did not show deregulated expression despite their location within these areas. In summary, the combination of array CGH and expression analysis provides a more comprehensive picture of the transformation of FCL to DLBCL. This process is associated with the acquisition of a variable spectrum of genomic imbalances affecting recurrent chromosomal areas that harbor overexpressed or underexpressed genes targeted upon transformation.
Introduction
Follicle center lymphoma (FCL) is one of the most common types of lymphoma, comprising about 40% of adult non-Hodgkin lymphomas (NHLs) in Western populations.1 It is characterized by a relatively indolent clinical course and long survival, but it is currently incurable.2 FCL transforms to a more aggressive lymphoma in 25%-60% of patients, an event that represents the outgrowth of a more malignant subclone. Transformation is usually associated with a rapidly progressive clinical course, refractoriness to treatment, and short survival.2,3Several secondary genetic abnormalities have been associated with this histological transformation of FCL, including rearrangements of theMYC gene,4 mutations of the P53tumor suppressor gene,5,6 of the BCL6 gene and of the translocated BCL2 gene,7,8 and inactivation of the P16 and P15 genes by deletion, mutation, and hypermethylation.9 However, these alterations are each observed in only a subset of transformed lymphomas, suggesting that the process of histological transformation can occur by multiple different mechanisms.
In addition to these specific genetic changes, transformation has been associated with cytogenetic alterations such as loss or gain of whole chromosomes, deletions, and amplifications.10-13 However, the data on these chromosomal abnormalities are relatively scarce due to the technical difficulties of obtaining cytogenetic data from paired pretransformation and posttransformation specimens. These difficulties have been partially overcome by the use of comparative genomic hybridization (CGH), which has allowed the description of a high incidence of genomic imbalances in FCL.14-16 There are 3 studies comparing sequential FCL biopsies using conventional CGH, in which DNA is hybridized to normal metaphase chromosomes.17-19 A broad spectrum of alterations were observed accompanying the transformation process and affecting specific chromosomal regions (Table 1). However, the analyses were limited by the low resolution of this form of CGH, which only detects DNA copy number aberrations greater than 10-20 megabases (Mb).14-18 Moreover, with few exceptions,19 the “target” genes within these amplification and deletion sites remain unknown.
Summary of published reports on the genomic characterization of FCL transformation to DLBCL using CGH to chromosomes
| Reference . | n . | Genomic abnormality . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| +2p16 . | +X . | +12q . | +18q21 . | +7 . | +17q . | −4q . | +3q . | −17q . | −13q . | ||
| Hough et al17 | 23 | 4 | 18 | 12 | 1 | 7 | 2 | 4 | 2 | 0 | 2 |
| Goff et al19 | 6 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| Nagy et al29 | 5 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| Present study | 10 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 3 |
| Reference . | n . | Genomic abnormality . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| +2p16 . | +X . | +12q . | +18q21 . | +7 . | +17q . | −4q . | +3q . | −17q . | −13q . | ||
| Hough et al17 | 23 | 4 | 18 | 12 | 1 | 7 | 2 | 4 | 2 | 0 | 2 |
| Goff et al19 | 6 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| Nagy et al29 | 5 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| Present study | 10 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 3 |
The number of cases with the corresponding genomic abnormality is shown.
To begin to address these 2 points, we have performed both genome-wide microarray-based CGH (array CGH) and gene expression analysis20 on a panel of transformed FCLs to determine the genomic deletions and amplifications more precisely and to correlate these genomic alterations with changes in the patterns of gene expression. The array CGH used in this study comprises approximately 2400 BAC and P1 clones for quantitative measurement of DNA copy number changes across the human genome with an average resolution of 1.4 Mb.21 We applied array CGH to paired biopsy specimens of FCL at diagnosis and their clonally related posttransformation diffuse large B-cell lymphoma (DLBCL), as well as to 12 unrelated cell lines known or presumed to be derived from FCL cases. To search for genes deregulated by genomic changes acquired upon transformation, we reanalyzed the recently reported gene expression profiling data of the paired samples.20 We report that FCL transformation is associated with the acquisition of a variable spectrum of genomic imbalances, some of which have not been previously reported in association with FCL transformation. In some cases these genomic changes are associated with changes in expression of genes within the targeted region upon transformation.
Patients, materials, and methods
Tumor specimens
Sequential biopsy specimens from 12 patients with FCL diagnosed and treated at Stanford University Hospital were selected for this study. Overall, 13 biopsy specimens obtained at the time of FCL diagnosis (2 biopsy specimens from case IL111), 2 second biopsy specimens obtained at the time of FCL recurrence, and 10 sequential biopsy specimens obtained at the time of morphologic transformation to DLBCL were evaluated. All lymphoma specimens were rereviewed and classified according to the Revised European-American Lymphoma (REAL) classification.1 The tumors were analyzed by flow cytometry for expression of immunoglobulin (IG) heavy and light chains and B- and T-cell markers. Lymph node histology at the time of transformation was classified as DLBCL in all 10 specimens (T-cell rich, B-cell diffuse large cell lymphoma in specimen IL119B). The clonal relationship between the members of each pair of tumors was confirmed in all cases by examination of IG and/orBCL6 genes and BCL2 gene rearrangements. Standard FCL treatment protocols were used to treat these patients. The patients received between 1 and 6 treatment modalities before the transformation that occurred 2.6 to 9.5 years after FCL diagnosis.
Cell lines
A panel composed of 12 cell lines (Karpas 231, OZ, SUDHL6, BEVA, VAL, Karpas 353, OCI-LY8, PR1, ROS50, DOHH2, RL, Karpas 422) derived from patients with lymphoma, all with t(14;18)(q32;q21), were studied with array CGH. Cell lines were cultured according to standard methods. References for the derivation of the cell lines may be obtained upon request.
Genomic DNA extraction
Genomic DNA was extracted from 5.0 × 106 cells using a commercially available kit as described by the manufacturer (QIAamp Tissue Kit; Qiagen, Valencia, CA). All patient samples were shown to contain between 52% and 91% of tumoral cells by flow cytometry analysis.
Microarray-based CGH
Genome-wide analysis of DNA copy number changes of patient samples and cell lines was performed using array CGH on a microchip with 2460 BAC and P1 clones in triplicate (approximately 7500 elements) in a 12-millimeter square (HumArray 1.14).21 Each clone contains at least one sequence tagged site (STS), allowing linkage to the genome sequence. The array provides an average resolution of 1.4 Mb across the genome. Fabrication and validation of the array, hybridization methods and analytical procedures, and the clone content have been described elsewhere in detail.21Approximately 0.2 μg of test (tumor) and reference genomic DNAs were labeled by random priming using Cy3 and Cy5, respectively. After 48 hours' hybridization, slides were washed and mounted with DAPI (4′,6′-diamidino-2-phenylidole). The images of the arrays were captured using a custom-built CCD camera, and the UCSF SPOT software was used to analyze the images.22 A second program, SPROC, was used to associate clones with each spot and a mapping information file that allows the data to be plotted relative to the position of the BACs on the draft human genome sequence (http://genome.cse.ucsc.edu). A formal data-filtering procedure was then performed, and a SPROC output file consisting of averaged ratios of the triplicate spots for each clone, standard deviations of the replicates, and plotting positions for each clone on the array was obtained. Fluorescent Cy3 (tumor)/Cy5 (normal) log2 ratios were classified as genomic gain (between 0.4 and 0.95), gene amplification (higher than 0.95) or genomic loss (lower than −0.7 for hemizygosity, and lower than −1.5 for homozygous deletion). Increase and decrease of DNA copy number of specific BAC/P1 clones present on the array were confirmed by fluorescence in situ hybridization (FISH) on interphase nuclei from the cell lines, as reported.23 The clones used for FISH (CTC-224F8, RP11-120J1, and RP11-40P17) were obtained from the Children's Hospital Oakland Research Institute, BACPAC Resources (Oakland, CA) or from Research Genetics (Invitrogen, Huntsville, AL). The LSI p16/CEP 9 Dual Color Probe was purchased from Vysis (Downers Grove, IL).
Gene expression analysis
The results of the gene expression changes upon transformation in the analyzed cases have been recently reported.20 To correlate the chromosomal changes to the alterations in gene expression profiles upon transformation, we have searched for changes in the expression of genes located in the regions targeted by chromosomal gains and/or deletions in the corresponding specimens.
Results
Genomic pattern of aberrations in the paired biopsies
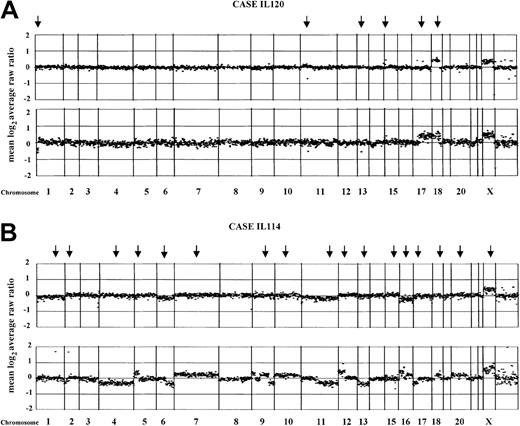
Results of array CGH on 10 paired pretransformation and posttransformation samples (cases from IL105 to IL126) and 2 paired initial and relapse nontransformed samples (cases IL111 and IL112) are summarized in Table 2. Overall, all but 3 of the biopsies showed genomic imbalances. Array CGH detected both small and whole-chromosome areas of gain as well as deletions. It also delineated amplification and deletion borders and distinguished amplification from low-level DNA copy gain (Figure1). Individual examples of array CGH analysis are represented in Figure 2.
Genome-wide analysis of DNA copy-number changes using array CGH in FCL and transformed/relapsed biopsies
| Case no. . | Genomic changes at diagnosis . | Genomic changes at transformed and relapse . |
|---|---|---|
| IL105 | −1p36, +1q21qter,* −6q22qter,* −8p23.1, +8p22,* −9p21 (P16locus), +16p11pter, −17p11pter, +18 | −5q21q23,†+17q11qter† |
| IL114 | Normal | +1q24,†,‡ +2p16 (RELlocus),†,‡ −4,† +5p12p15,† −6q11qter,† +7,† +9p23p24,†+9q13q31,† −9q31qter,† +10,† −11q11qter,† +12pter-q14,† +12q12,†,‡ −13q11qter,† −15q23,† +16, −17p12pter,† +18q11qter,†+20,† +Xq21qter† |
| IL116 | −1p33p36, +1q24qter, +2p16 (REL locus), −4q34qter, −6q11qter, +8q24, +13q21q31, −13q33qter, +17q21q22, +18pter-q21.3,* −18q21.3qter* | +2p16 (REL locus), †,‡ +6p11pter† |
| IL117 | −13q14, +18pter-q21.3 | Same alterations |
| IL119 | Normal | Normal |
| IL120 | −11p14, +15q14, +17p12, +17q21, +18pter-q21.3 | −1p36,† −13q14 (RB locus),†+17q11qter† |
| IL121 | −9p21 (P16 locus), −X* | +18pter-q21.3,† +X† |
| IL123 | −1p36,* +1q21qter,* +2,*+18pter-q21.3 | +21q11qter† |
| IL124 | −6q11qter, −9p11pter, −13q21, −13q31, +17 | +4p11pter,† −4q21qter,† +12 |
| IL126 | −1p34p36,* +1q21qter,* +1q32.1,*,‡ −4, +6p21,*−6q22q24,* +13q32,* −14q12q22,* +15q11q14, −17p11pter,*+17q11-qter,* +18q21.3* | +3q12qter,† −13q21.1† |
| IL111 | −3q28, +8, +18pter-q21.3 | +2p11p25,†,2-153 +3q27q29†,2-153 |
| IL112 | −1p35p36, +8p23.2 | +4p11pter,†,2-153 −8p12p23,†,2-153−9p21.3,†,2-153 +17q21.3†,2-153 |
| Case no. . | Genomic changes at diagnosis . | Genomic changes at transformed and relapse . |
|---|---|---|
| IL105 | −1p36, +1q21qter,* −6q22qter,* −8p23.1, +8p22,* −9p21 (P16locus), +16p11pter, −17p11pter, +18 | −5q21q23,†+17q11qter† |
| IL114 | Normal | +1q24,†,‡ +2p16 (RELlocus),†,‡ −4,† +5p12p15,† −6q11qter,† +7,† +9p23p24,†+9q13q31,† −9q31qter,† +10,† −11q11qter,† +12pter-q14,† +12q12,†,‡ −13q11qter,† −15q23,† +16, −17p12pter,† +18q11qter,†+20,† +Xq21qter† |
| IL116 | −1p33p36, +1q24qter, +2p16 (REL locus), −4q34qter, −6q11qter, +8q24, +13q21q31, −13q33qter, +17q21q22, +18pter-q21.3,* −18q21.3qter* | +2p16 (REL locus), †,‡ +6p11pter† |
| IL117 | −13q14, +18pter-q21.3 | Same alterations |
| IL119 | Normal | Normal |
| IL120 | −11p14, +15q14, +17p12, +17q21, +18pter-q21.3 | −1p36,† −13q14 (RB locus),†+17q11qter† |
| IL121 | −9p21 (P16 locus), −X* | +18pter-q21.3,† +X† |
| IL123 | −1p36,* +1q21qter,* +2,*+18pter-q21.3 | +21q11qter† |
| IL124 | −6q11qter, −9p11pter, −13q21, −13q31, +17 | +4p11pter,† −4q21qter,† +12 |
| IL126 | −1p34p36,* +1q21qter,* +1q32.1,*,‡ −4, +6p21,*−6q22q24,* +13q32,* −14q12q22,* +15q11q14, −17p11pter,*+17q11-qter,* +18q21.3* | +3q12qter,† −13q21.1† |
| IL111 | −3q28, +8, +18pter-q21.3 | +2p11p25,†,2-153 +3q27q29†,2-153 |
| IL112 | −1p35p36, +8p23.2 | +4p11pter,†,2-153 −8p12p23,†,2-153−9p21.3,†,2-153 +17q21.3†,2-153 |
Listed samples nos. IL105 to IL126 were classified as transformed DLBCL. Biopsies nos. IL111 and IL112 were classified as nontransformed relapse.
Genomic abnormalities in the column of genomic changes at diagnosis indicate changes observed in the diagnostic sample that were not seen in the transformation specimen.
Genomic abnormalities in the transformed/relapsed samples column identified in only one of the sequential biopsies, thus representing losses or gains upon transformation or relapse. Only the acquired changes that were not present in the initial biopsies are shown
Genomic abnormalities that represent high-level amplifications.
Abnormalities acquired in nontransformed biopsies (IL111 and IL112).
Normalized copy number variation analysis of paired FCL and DLBCL samples using array CGH.
The figures represent the normalized copy number ratios of genomic DNAs from patients IL120 (A) and IL114 (B). For each figure, the initial FCL biopsies are placed above the posttransformation biopsies (lower panel). Data are plotted as the mean log2 ratio of the triplicate spots for each clone normalized to the genome median log2 ratio. The BAC and P1 clones are ordered by position in the genome, beginning at chromosome 1p and ending at chromosome Xq. Borders between chromosomes are indicated by vertical bars. Arrows indicate genomic changes, which are summarized in Table 1. (A) In both samples from the case IL120, gain of chromosome 18q confirms the clonality of the paired biopsies. In addition, a deletion of approximately 2.3 Mb in 11p14 and a single-copy DNA gain of approximately 1 Mb in 15q14 also were seen. In the transformed biopsy, novel changes included the loss of the terminal portion of chromosome 1p36 (sized approximately 10 Mb), deletion of approximately 1 Mb involving the RB gene locus in 13q14, and gain of the whole arm of chromosome 17. (B) The FCL biopsy from the case IL114 (tumor cell population of 85%) is shown. According to the defined log2 ratios, no changes were observed in the FCL sample. In the transformed biopsy, a number of acquired aberrations are seen, including single-copy DNA alterations of chromosomes 6, 11, and 16, and high-level amplification affecting 1q24 (clone RP11-184N12; log2 ratio + SD, + 1.7 + 0.036), 2p16 (REL/BCL11A gene locus; log2 ratio + SD, + 1.67 + 0.068), and 12q12 (clone RP11-29G23; log2ratio + SD, +0.96 + 0.059).
Normalized copy number variation analysis of paired FCL and DLBCL samples using array CGH.
The figures represent the normalized copy number ratios of genomic DNAs from patients IL120 (A) and IL114 (B). For each figure, the initial FCL biopsies are placed above the posttransformation biopsies (lower panel). Data are plotted as the mean log2 ratio of the triplicate spots for each clone normalized to the genome median log2 ratio. The BAC and P1 clones are ordered by position in the genome, beginning at chromosome 1p and ending at chromosome Xq. Borders between chromosomes are indicated by vertical bars. Arrows indicate genomic changes, which are summarized in Table 1. (A) In both samples from the case IL120, gain of chromosome 18q confirms the clonality of the paired biopsies. In addition, a deletion of approximately 2.3 Mb in 11p14 and a single-copy DNA gain of approximately 1 Mb in 15q14 also were seen. In the transformed biopsy, novel changes included the loss of the terminal portion of chromosome 1p36 (sized approximately 10 Mb), deletion of approximately 1 Mb involving the RB gene locus in 13q14, and gain of the whole arm of chromosome 17. (B) The FCL biopsy from the case IL114 (tumor cell population of 85%) is shown. According to the defined log2 ratios, no changes were observed in the FCL sample. In the transformed biopsy, a number of acquired aberrations are seen, including single-copy DNA alterations of chromosomes 6, 11, and 16, and high-level amplification affecting 1q24 (clone RP11-184N12; log2 ratio + SD, + 1.7 + 0.036), 2p16 (REL/BCL11A gene locus; log2 ratio + SD, + 1.67 + 0.068), and 12q12 (clone RP11-29G23; log2ratio + SD, +0.96 + 0.059).
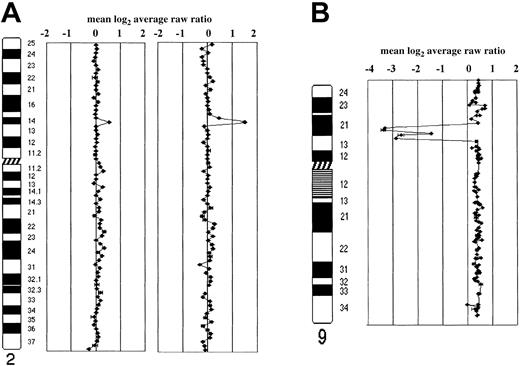
Representative array CGH profiles of transformed FCL and FCL-derived cell lines.
Data are plotted as the mean log2 ratio of the triplicate spots for each clone normalized to the genome median log2ratio. The BAC and P1 clones covering individual chromosomes are shown. (A) Schematic representation of array CGH analysis on chromosome 2 in case IL116. A single-copy gain of a BAC clone in 2p16 containing theREL gene (log2 ratio + SD, 0.52 + 0.046) is shown in the FCL sample (on the left ideogram). In the posttransformed biopsy (on the right ideogram), amplification of the same gene locus (log2 ratio + SD, 1.13 + 0.027) is shown. (B) Homozygous deletion of P16 gene locus in 9p21 in triploid OZ cell line. Because this cell line contains a near-triploid karyotype, DNA copy number baseline is above the ratio 0.4 of the clones into 9p21, locus of the P16 gene, showed a log2 ratio lower than −2, indicating the presence of homozygous deletion sized approximately 5 Mb. FISH using a clone containing the gene confirmed the absence of copies ofP16 in the 3 aberrant chromosomes 9. The homozygous deletion had been reported in diploid OZ cell line using Southern blot analysis.25
Representative array CGH profiles of transformed FCL and FCL-derived cell lines.
Data are plotted as the mean log2 ratio of the triplicate spots for each clone normalized to the genome median log2ratio. The BAC and P1 clones covering individual chromosomes are shown. (A) Schematic representation of array CGH analysis on chromosome 2 in case IL116. A single-copy gain of a BAC clone in 2p16 containing theREL gene (log2 ratio + SD, 0.52 + 0.046) is shown in the FCL sample (on the left ideogram). In the posttransformed biopsy (on the right ideogram), amplification of the same gene locus (log2 ratio + SD, 1.13 + 0.027) is shown. (B) Homozygous deletion of P16 gene locus in 9p21 in triploid OZ cell line. Because this cell line contains a near-triploid karyotype, DNA copy number baseline is above the ratio 0.4 of the clones into 9p21, locus of the P16 gene, showed a log2 ratio lower than −2, indicating the presence of homozygous deletion sized approximately 5 Mb. FISH using a clone containing the gene confirmed the absence of copies ofP16 in the 3 aberrant chromosomes 9. The homozygous deletion had been reported in diploid OZ cell line using Southern blot analysis.25
The pattern of genomic aberrations in the FCL pretransformation biopsies consisted of 54 genomic imbalances (mean per tumor, 4.5; range, 0-12) and included 27 genomic gains (mean, 2.3), 26 losses (mean, 2.2), and one 2-fold amplification affecting clone RP11-249H15 in band 1q32.1 in case IL 126 (including the locus of theIKKE gene). In the transformation/relapse samples, array CGH detected a higher number of chromosomal changes: 77 genomic imbalances (mean per tumor, 6.4; range, 0-20) distributed as 41 gains (mean, 3.4), 32 losses (mean, 2.7), and 4 gene amplification events. The amplifications involved chromosome bands 1q24 (3-fold amplification of clone RP11-184N12 in case IL114, locus of the FcGR2B gene), 2p16 in REL/BCL11A loci (cases IL114 and IL116, showing 3- and 2-fold amplification, respectively), and 12q12 involving clone RP11-29G23 (case IL114, showing 2-fold amplification). The mean number of alterations per tumor was higher in the transformed biopsies than in the relapsed nontransformed specimens (6.7 vs 5).
To search for genomic imbalances appearing de novo in the transformation and relapse phases, array CGH patterns in the paired samples were compared. In all cases there was at least one genomic abnormality present in both paired samples, confirming the clonal relationship between each pair of tumors. The majority of changes accompanying transformation consisted of gains of chromosome material rather than losses (23 vs 12). A heterogeneous pattern of acquired genomic imbalances involving 28 different loci was identified. Some of these changes were observed in 2 or 3 different patients. A number of these genomic imbalances were found exclusively in the transformed DLBCL biopsies, while others were also detected in the initial or relapsed, nontransformed FCL specimens from other patients. The most frequent exclusive aberrations associated with the transformation consisted of gains of chromosomes 12 and Xq (2 cases each). Trisomy of chromosome 7, previously associated with the transformation, was observed in one case.
Molecular dissection of recurrent genomic imbalances in FCL patients and FCL-derived cell lines
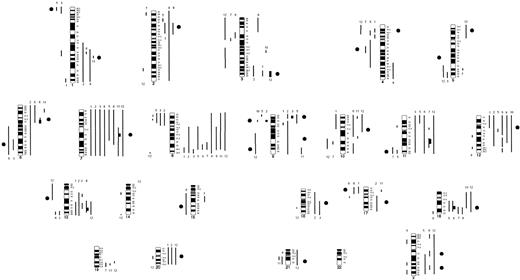
These novel genomic changes suggested a possible association with the transformation of FCL. Because of the small number of patients carrying each particular genomic imbalance, we applied the array CGH to a panel of 12 cell lines with t(14;18)(q32;q21) known or presumed to be derived from FCL cases to assess whether any of the observed changes might be recurrent.24 The mean number of genomic abnormalities present in these cell lines was 11 (range, 2-19) and consisted of 84 genomic gains, 45 losses, and 7 gene amplification sites. The distribution of genomic imbalances is represented in Figure3. Seven amplicons were located at chromosomal bands 3q27-q29, 6p12.3, 7q21.3, 9p23, 11q14-q23, 13q31-q32, and 18q21.3, and the degree of amplification ranged from 2- to 3.5-fold. A region of homozygous loss was identified in the locus ofP16 gene (9p21) in OZ cell line. This cell line showed 3 copies of chromosome 9, and P16 locus was deleted from all the copies (Figure 2B). This homozygous loss already had been reported by Southern blot analysis.25 Most of the genomic imbalances present in the cell lines also were observed in the clinical specimens. An exception was the gain of MYC gene locus in 8q24, which was seen in 10 of 12 cell lines (84%) but in only 2 patients at diagnosis.
Ideogram of gains and losses according to the genomewide array CGH in 12 FCL-derived cell lines.
Lines on the right of each ideogram indicate genomic gains, lines on the left show losses of chromosomal material, and high-level amplifications are symbolized by bold lines. Cell lines are indicated by numbers above each line and correspond to Karpas 231 (no. 1); OZ (no. 2); OCI-LY8 (no. 3); DOHH2 (no. 4); SUDHL6 (no. 5); PR1 (no. 6); Karpas 422 (no. 7); BEVA (no. 8); VAL (no. 9); Karpas 353 (no. 10); ROS50 (no. 11); and RL (no. 12). Black circles localize the regions commonly acquired upon transformation in patients with FCL.
Ideogram of gains and losses according to the genomewide array CGH in 12 FCL-derived cell lines.
Lines on the right of each ideogram indicate genomic gains, lines on the left show losses of chromosomal material, and high-level amplifications are symbolized by bold lines. Cell lines are indicated by numbers above each line and correspond to Karpas 231 (no. 1); OZ (no. 2); OCI-LY8 (no. 3); DOHH2 (no. 4); SUDHL6 (no. 5); PR1 (no. 6); Karpas 422 (no. 7); BEVA (no. 8); VAL (no. 9); Karpas 353 (no. 10); ROS50 (no. 11); and RL (no. 12). Black circles localize the regions commonly acquired upon transformation in patients with FCL.
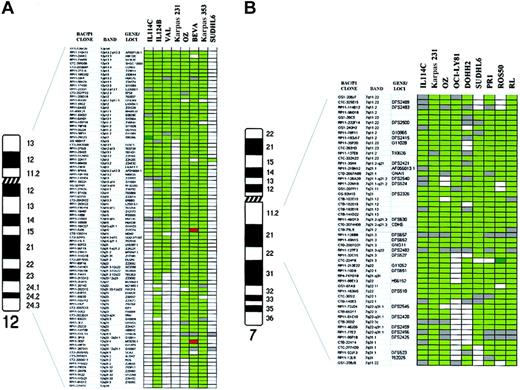
To define more precisely the boundaries of regions of genomic aberration observed in the transformed specimens, we have combined the array CGH data on the patient material and cell lines (Table3). While such a procedure is open to the possibility that some of the aberrations in the cell lines represent adaptation to culture, it may point out genomic regions of highest priority for further investigation for critical genes. Many of the alterations in this combined set of specimens included known loci involved in FCL pathogenesis, such as gain of REL/BCL11A in band 2p16, and gain of 18q21 that was delineated from 18q12 to 18q21.3, including the BCL2 gene locus. In addition, the gain/amplification of 3q was confined to a large area from 3q27 to the telomere in 3q29, including the BCL6 gene locus in all instances. In contrast, gain and amplification of 1q included 2 different peaks at 1q21-q22 in PR-1 cell line (involving the clone RP11-71L20 in the locus of the MCL1 gene) and 1q24 (involving FcGR2B gene locus). One of the abnormalities observed exclusively in the transformation specimens was the gain of chromosome 12, which was also observed in 6 of the cell lines (Figure4A). A low-level DNA copy gain affecting from the 12p telomere to 12q12 was delineated, including amplification of clone RP11-29G23 (12q12) in case IL114. In the SUDHL6 cell line, a gain involving only the clone RP11-32K17 on 12q13 was observed out of the commonly gained interval. Delineation of the genomic gains on chromosome 7 in 8 cell lines and one transformed patient showed a common region of low-level DNA overrepresentation from 7q11.2 to 7q22.1, including a 2-fold amplification peak affecting clone CTC-224F8 (7q21.3) in ROS50 cell line (Figure 4B). FISH confirmed this single amplification. Another aberration detected only in transformed specimens was the gain of chromosome X. All these cases but the PR1 cell line showed gain of the whole long arm of chromosome X. In this cell line, 2 separated regions of gain were delineated in Xq11-q21.3 and Xq25-q26. These genomic gains were flanked by chromosomal deletions, which were confirmed by CGH to chromosomes (data not shown). In addition, novel regions of genomic imbalance not previously associated with FCL transformation were delineated. These included the gain of 9p23-p24, which was seen in one transformed FCL specimen and 3 cell lines, being the smallest interval of overrepresentation defined on 9p23 in OCI-LY8 cell line. FISH analysis using clone RP11-120J1 and the repeated analysis using array CGH study confirmed this single amplification, which was not detected by CGH to chromosomes (data not shown). The gain of 6p12-p21 was also identified in 2 patients (1 at diagnosis and 1 at transformation) and 3 cell lines, including high-level amplification of BAC RP11-40P17 in PR1 cell line at D6S427 locus (6p12.3). This amplification was confirmed by repeating the array CGH study and by using FISH and CGH to chromosomes (data not shown). The gain of 17q, a frequent and common aberration site, was narrowed down to chromosomal band 17q21. Other regions of novel genomic imbalance detected in the transformed samples included overrepresentation of 4p12-pter, 5p12-p15, 9q13-q31 and 16q, and loss of 1p36.3, 4q21-q23, 5q21-q23, 9q31-qter, 11q24-q25, and 15q23.
Delineation of genomic imbalances associated with transformation in follicle center lymphoma by array CGH
| Chromosomal aberrations . | FCL cell lines . | Patient diagnosis . | Patients transformed . | Patients relapsed . | Total no. . | Minimal segment of genomic gain/loss . |
|---|---|---|---|---|---|---|
| −1p36 | 2 | 5 | 1 | 0 | 8 | RP11-51B4 (1p36.32) − RP11-82D16 (1p36.33) |
| +1q24 | 3 | 4 | 13-150 | 0 | 8 | RP11-184N12 (1q24) |
| +2p16 | 3 | 2 | 23-150 | 1 | 8 | GS-274P9 (2p16) |
| +3q27−q29 | 33-150 | 0 | 1 | 1 | 5 | CTD-2091K06 (3q27) − RP11-233N20 (3q29) |
| +4p12−pter | 0 | 0 | 1 | 1 | 2 | RP11-38M16 (4p12) − GS1-118B13 (4pter) |
| −4q21−q22 | 2 | 1 | 2 | 0 | 5 | RP11-115K14 (4q21.1) − RP11-36G19 (4q21.2) |
| +5p12p15 | 1 | 0 | 1 | 0 | 2 | RP11-9G14 (5p12) − RP11-227M19 (5p15.3) |
| −5q21−q23 | 0 | 0 | 1 | 0 | 1 | RP11-203J07 (5q21) − RP11-47L19 (5q23) |
| +6p12−p21 | 33-150 | 1 | 1 | 0 | 5 | RP3-445N2 (6p12.1) − RP11-40p17 (6p12.2 − 6p21.1) |
| −6q22−q24 | 2 | 4 | 1 | 0 | 7 | CTC-268N4 (6q22) − CTD2015J17 (6q24) |
| +7q11−q22 | 83-150 | 0 | 1 | 0 | 9 | RP11-96O16 (7q11.2) − CTC-30512 (7q22) |
| +9p23 | 33-150 | 0 | 1 | 0 | 4 | RP11-117J18 (9p23) |
| +9q13−q31 | 1 | 0 | 1 | 0 | 2 | CTD-2029120 (9q13) − RP11-5K11 (9q31) |
| −9q31−qter | 1 | 0 | 1 | 0 | 2 | RP11-80F13 (9q31) − GS1-135I17 (9qter) |
| +10 | 3 | 0 | 1 | 0 | 4 | GS1-23B11 (10pter) − GS1-261B16 (10qter) |
| −11q24−q25 | 2 | 0 | 1 | 0 | 3 | RP11-145L11 (11q23 − 11q24) − RP1-26N8 (11qter) |
| +12pter−12q12 | 6 | 0 | 2 | 0 | 8 | RP11-110K11 (12p13.3) − RP11-29G23 (12q12) |
| −13q13−q21 | 1 | 2 | 3 | 0 | 6 | RP11-14A4 (13q13); RP11-58D13 (13q14); RP11-234M13 (13q21.1); RP11-205J24 (13q21) |
| −15q23 | 1 | 0 | 1 | 0 | 2 | CTD-2022G15 (15q23) |
| +16 | 2 | 1 | 1 | 0 | 4 | CTD-2371A5 (16pter) − RP11-7D23 (16qter) |
| −17p12−pter | 1 | 2 | 1 | 0 | 4 | RPC-169E11 (17p12) − GS1-68F18 (17pter) |
| +17q21 | 1 | 4 | 2 | 1 | 8 | CTB-305D20 (17q21) |
| +18q21 | 63-150 | 7 | 2 | 0 | 15 | GS-385N22 (18q21.3) − RP11-12J12 (18q22) |
| +20pter−20qter | 3 | 0 | 1 | 0 | 4 | RP11-82O2 (20pter) − RP11-81F12 (20qter) |
| +21q11−qter | 1 | 0 | 1 | 0 | 2 | RP11-96H21 (21q21) − RP11-135B17 (21qter) |
| +Xq11−q21 | 3 | 0 | 1 | 0 | 4 | RP11-177A04 (Xq13) |
| +Xq25−q26 | 3 | 0 | 2 | 0 | 5 | CTD-2015J18 (Xq25) − RP11-97N5 (Xq26) |
| Chromosomal aberrations . | FCL cell lines . | Patient diagnosis . | Patients transformed . | Patients relapsed . | Total no. . | Minimal segment of genomic gain/loss . |
|---|---|---|---|---|---|---|
| −1p36 | 2 | 5 | 1 | 0 | 8 | RP11-51B4 (1p36.32) − RP11-82D16 (1p36.33) |
| +1q24 | 3 | 4 | 13-150 | 0 | 8 | RP11-184N12 (1q24) |
| +2p16 | 3 | 2 | 23-150 | 1 | 8 | GS-274P9 (2p16) |
| +3q27−q29 | 33-150 | 0 | 1 | 1 | 5 | CTD-2091K06 (3q27) − RP11-233N20 (3q29) |
| +4p12−pter | 0 | 0 | 1 | 1 | 2 | RP11-38M16 (4p12) − GS1-118B13 (4pter) |
| −4q21−q22 | 2 | 1 | 2 | 0 | 5 | RP11-115K14 (4q21.1) − RP11-36G19 (4q21.2) |
| +5p12p15 | 1 | 0 | 1 | 0 | 2 | RP11-9G14 (5p12) − RP11-227M19 (5p15.3) |
| −5q21−q23 | 0 | 0 | 1 | 0 | 1 | RP11-203J07 (5q21) − RP11-47L19 (5q23) |
| +6p12−p21 | 33-150 | 1 | 1 | 0 | 5 | RP3-445N2 (6p12.1) − RP11-40p17 (6p12.2 − 6p21.1) |
| −6q22−q24 | 2 | 4 | 1 | 0 | 7 | CTC-268N4 (6q22) − CTD2015J17 (6q24) |
| +7q11−q22 | 83-150 | 0 | 1 | 0 | 9 | RP11-96O16 (7q11.2) − CTC-30512 (7q22) |
| +9p23 | 33-150 | 0 | 1 | 0 | 4 | RP11-117J18 (9p23) |
| +9q13−q31 | 1 | 0 | 1 | 0 | 2 | CTD-2029120 (9q13) − RP11-5K11 (9q31) |
| −9q31−qter | 1 | 0 | 1 | 0 | 2 | RP11-80F13 (9q31) − GS1-135I17 (9qter) |
| +10 | 3 | 0 | 1 | 0 | 4 | GS1-23B11 (10pter) − GS1-261B16 (10qter) |
| −11q24−q25 | 2 | 0 | 1 | 0 | 3 | RP11-145L11 (11q23 − 11q24) − RP1-26N8 (11qter) |
| +12pter−12q12 | 6 | 0 | 2 | 0 | 8 | RP11-110K11 (12p13.3) − RP11-29G23 (12q12) |
| −13q13−q21 | 1 | 2 | 3 | 0 | 6 | RP11-14A4 (13q13); RP11-58D13 (13q14); RP11-234M13 (13q21.1); RP11-205J24 (13q21) |
| −15q23 | 1 | 0 | 1 | 0 | 2 | CTD-2022G15 (15q23) |
| +16 | 2 | 1 | 1 | 0 | 4 | CTD-2371A5 (16pter) − RP11-7D23 (16qter) |
| −17p12−pter | 1 | 2 | 1 | 0 | 4 | RPC-169E11 (17p12) − GS1-68F18 (17pter) |
| +17q21 | 1 | 4 | 2 | 1 | 8 | CTB-305D20 (17q21) |
| +18q21 | 63-150 | 7 | 2 | 0 | 15 | GS-385N22 (18q21.3) − RP11-12J12 (18q22) |
| +20pter−20qter | 3 | 0 | 1 | 0 | 4 | RP11-82O2 (20pter) − RP11-81F12 (20qter) |
| +21q11−qter | 1 | 0 | 1 | 0 | 2 | RP11-96H21 (21q21) − RP11-135B17 (21qter) |
| +Xq11−q21 | 3 | 0 | 1 | 0 | 4 | RP11-177A04 (Xq13) |
| +Xq25−q26 | 3 | 0 | 2 | 0 | 5 | CTD-2015J18 (Xq25) − RP11-97N5 (Xq26) |
Includes one gene amplification event.
Schematic representation of DNA copy-number alterations in patients with transformed FCL and FCL-derived cell lines.
(A) Chromosome 12: array CGH analysis of 2 transformed samples from patients with FCL (IL114C and IL124B) and 6 cell lines (VAL, Karpas 231, OZ, BEVA, Karpas 353, and SUDHL6). Ninety-one BAC and P1-derived clones included in the array are listed in the columns on the left. In dark green, regions of genomic gain; in bright green, regions of genomic amplification; in red, regions of genomic loss; in white, normal log2 ratio; gray squares represent noninformative clones. The log2 ratios for defining genomic imbalances are defined in “Patients, materials, and methods.” The common region of gain was defined between 12p telomere and 12q12, including high-level amplification of clone RP11-29G3 in IL114C (log2 ratio + SD, +0.96 + 0.059). (B) Chromosome 7q: array CGH analysis of 8 cell lines with gain of chromosome 7q (Karpas 231, OZ, OCI-LY8, DOHH2, SUDHL6, PR1, ROS50, and RL) and one transformed patient (IL 114C). Using CGH to chromosomes, the common region of gain in all these samples allowed the delineation of a common region of gain to 7q11-q31 (data not shown). This region is represented here using array CGH. Fifty-two clones covering 7q11-q31 are shown. The different colors represent the genomic aberrations as defined in Figure4A. The common region of gain was delineated between 7q11.2 and 7q22.1. One clone (CTC-224F8 in 7q21.3) showed high-level amplification in ROS50 (log2 ratio + SD, +1.13 + 0.022).
Schematic representation of DNA copy-number alterations in patients with transformed FCL and FCL-derived cell lines.
(A) Chromosome 12: array CGH analysis of 2 transformed samples from patients with FCL (IL114C and IL124B) and 6 cell lines (VAL, Karpas 231, OZ, BEVA, Karpas 353, and SUDHL6). Ninety-one BAC and P1-derived clones included in the array are listed in the columns on the left. In dark green, regions of genomic gain; in bright green, regions of genomic amplification; in red, regions of genomic loss; in white, normal log2 ratio; gray squares represent noninformative clones. The log2 ratios for defining genomic imbalances are defined in “Patients, materials, and methods.” The common region of gain was defined between 12p telomere and 12q12, including high-level amplification of clone RP11-29G3 in IL114C (log2 ratio + SD, +0.96 + 0.059). (B) Chromosome 7q: array CGH analysis of 8 cell lines with gain of chromosome 7q (Karpas 231, OZ, OCI-LY8, DOHH2, SUDHL6, PR1, ROS50, and RL) and one transformed patient (IL 114C). Using CGH to chromosomes, the common region of gain in all these samples allowed the delineation of a common region of gain to 7q11-q31 (data not shown). This region is represented here using array CGH. Fifty-two clones covering 7q11-q31 are shown. The different colors represent the genomic aberrations as defined in Figure4A. The common region of gain was delineated between 7q11.2 and 7q22.1. One clone (CTC-224F8 in 7q21.3) showed high-level amplification in ROS50 (log2 ratio + SD, +1.13 + 0.022).
Expression analysis of the genes located within the common aberrant areas
Examination of gene expression changes upon FCL transformation to DLBCL by cDNA to microarrays identified 2 groups of patients with distinct transformation-associated gene expression profiles.20 One group demonstrated transformation-associated increases in expression of MYC and its target genes, and a second group demonstrated transformation-associated decreases in the expression of MYCand its target genes. In the current study we (1) searched for theMYC gene amplification as a possible explanation for the observed increase in the MYC gene expression; and (2) identified the genes that are contained in the regions of transformation-associated genomic aberrations and determined whether their expression was altered on transformation.
The MYC gene locus on 8q24 was not elevated in copy number in any of the transformed cases according to the array CGH data. Although MYC gene translocation might explain some of the changes in MYC gene expression, the observation of these changes in up to 50% of transformation cases significantly exceeded the reported incidence of MYC translocations upon transformation.4 Other possible genetic mechanisms such as changes in methylation might underlie the observed expression profiles. We also searched for genes harbored in the aberrant genomic areas that were differentially expressed in the transformed samples. We limited our analysis to the set of genes that exhibited at least 2-fold increase or decrease in expression upon transformation. This analysis revealed more than 300 genes located in areas of genomic gain/amplification or loss accompanying the transformation that were overexpressed or underexpressed, respectively (Table4). A number of these genes (NME1 in 17q21, JTV1 in 7p22, CYP51and CUTL1 in 7q22, MXI1 in 10q25,HLA-DRB1 in 6p21.3, CDK2 and CDK4 in 12q,SLC7A5 in 16q24, and AHCY in 20q11) areMYC target genes whose expression was previously observed to be increased upon transformation.20 Therefore, this observation suggests that at least in a subset of cases exhibiting increased expression of the MYC and its target genes upon transformation, genomic imbalances acquired in the transformation process might contribute to the changes in the expression of these genes. Only 3 of the genes (FLJ20706 in 4q23, D13S106Ein 13q14, and PMAIP1 in 18q21.3) exhibited universal expression changes in all the cases with similar chromosomal aberration. The overall number of observed recurrent chromosomal aberrations was small, and therefore further correlation was not possible. The function of these genes is unknown; however, it should be noticed that PMAIP1 was originally cloned from adult T-cell leukemia cell line,26 and it is expressed at high levels in CLL and in some B-NHL,27 and the gene encoding the hypothetical protein FLJ20706 is expressed at high levels in B-cell germinal center cells. In addition, the transformation-associated chromosomal alterations harbored a great number of additional genes that did not experience changes in expression.
List of genes with deregulated expression located at the transformation-associated chromosomal aberrations
| Case . | Chromosome . | Genes . |
|---|---|---|
| IL105 | −5q21q23 | D5S346, GPX3, ADRA1B |
| +17q11qter | SCYA4, KIAA0356, ITGB3, TK1, H3F3B, RPL27, SMT3H2, ACTG1, KPNA2, NME1, NME2, MSF, KPNA2, GCN5L2, HN1, PSCD1, CD79B, ICAM2, CLTC, RARA, SCYA16, ACLY, IGFBP4, STAT3, TUBG2, GCN5L2, RPL27, TK1, SCYA3L1, NXPH3, EAP30, SPAG9, SFRS1, SUPT4H1, PRKAR1A, SMARCD2, MGC10986, BIRC5, CSNK1D, SFRS2, SMT3H2, ALY | |
| IL114 | +1q24 | RER1, TAGLN2, SELL, CREG, DKFZP564B167, CACYBP |
| +2p16 | PEX13, MDH1, XPO1 | |
| −4 | FLJ20706, 4-150FLJ20280 | |
| +5p12p15 | PDCD6, CCT5, NNT, RAD1, SEMA5A, FLJ20303, TRIP13 | |
| −6q11qter | ME1, TTK, BACH2, DKFZP564B0769, KIAA0776, FOXO3A, NMBR, SMPD2, MAP3K5, VNN2, HIVEP2, KIAA1209, RNASE6PL | |
| +7 | POR, FZD9, YWHAG, OGDH, SLC25A13, HNRPA2B1, MCM7, MDH2, KOC1, POLD2, PSMA2, PURB, AHR, GARS, ACTB, ARL4, DKFZP434J154, NUDT1, RAC1, RPA3, HRI, JTV1, KDELR2, FLJ20397, EIF3S9, RALA, KIAA0442, LOC51142, CCT6A, CRIP1, CYP51, PLOD3, GNB2, PMS2L8, STAG3, VGF, CUTL1, KIAA0632, COL1A2, PSMC2, EBNA-2 co-activator, IMPDH1, DLD, ATP6S14, KIAA1170, AKR1B1, EZH2, RHEB2, FLJ20311 | |
| +9p23p24 | ||
| +9q13q31 | TXN, SEC61B, FPGS, PRKACG, BTEB1, CDC20, CTSL, CKS2, C90RF3, CKS2, SEC61B, FLJ20287, TXN | |
| −9q31qter | ORM1, TNFSF8, HSPA5, ASS, COL5A1 | |
| +10 | CHUK, PP, DDX21, ATP5C1, MXI1, VIM, PRDX3, ACF, MGMT, PGAM1, VDAC2, UROS, ERP70, SSH3BP1, TCF8, CREM, COPEB, GDI2, PFKP, PHYH, LOC51008, RET, HNRPF, MBL2, JDP1, CDC2, ANXA7, ZWINT, HK1, PRG1, VCL, PPIF, ANXA11, DLG5, KIAA0261, MPHOSPH1, PLAU, GBF1, ACTR1A, RGS10, ABLIM, MK167, OAT, ECHS1, ADAM8 | |
| −11q11qter | GSTP1, POU2AF1, MS4A7, HTATIP, EMK1, MS4A2, SYT7, ZFPL1, CD6, RASGRP2, FOLR1, KIAA0102, ACTN3, FLJ20189, SCDGF-B, ATM, CASP1, INPPL1, MLL, UBE4A, CBL | |
| +12pter-q14 | PA2G4, LRMP, REA, TUBA3, K-ALPHA-1, LDHB, GAPD, TPI1, DDX11, CS, FLJ22028, KIAA0528, ARHGD1B, FOXM1, MLF2, C1S, PXR1, GPR92, COPS7A, FKBP4, CD69, ATP5B, CD4, CNTN1, SHMT2, PRKAG1, NR4A1, CDK2, HSU47926, MGC5576, HEM1, HNRPA1, MYL6, ATP5G2, EIF4B, FLJ11773, DDIT3, MARS, HSPD1, HSPC228, SAS, RAP1B, CDK4, NEDD1, NR2C1, FLJ11021, ZNF10 | |
| −13q11qter | GJB2, SACS, YDD19, ALOX5AP, HMG1, CG005, D13S106E,4-150 ELF1, AKAP11, SLC25A15, RFP2, FOXO1A, FLJ21562, FBXL3A, GPR18, EFNB2, FLJ10154, ATP11A | |
| −15q23 | RCN2, FBXO22 | |
| +16 | GLG1, AARS, ALDOA, EIF3S8, RPL13 KIAA0251, KNSL4, STX4A, SULT1A3, ITGAL, MAX, ITGAX, UQCRC2, SULT1A1, PHKG2, SULT1A2, PLK, PPP4C, ARL6IP, MHC2TA, TNFRSF17, GSPT1, HIRIP3, NK4, PKMYT1, POLR3K, PRSS22, RNPS1, SEPX1, STUB1, CCNF, BOM045, HMOX2, HMT-1, PMM2, DREV1, SIAH1, SCYD1, CPSF5, GOT2, KIAA0095, E2F4, ATP6D, PP15, RCD-8, CBFB, FLJ13725, FLJ10305, LOC51659, MAP1A/1BLC3, PCOLN3, SLC7A5, TUBB4, CDT1, SLC7A5 | |
| −17p12pter | SERPINF1, DOC2B | |
| +18q11qter | FLJ21172, PMAIP1,4-150 PIK3C3, MADH2, TNFRSF11A | |
| +20 | ADA, PCNA, KIAA1219, CDC25B, GNAS1, PSMA7, MYBL2, AHCY, PTPRA, INSM1, FKBP1A, SNRPB, MAPRE1, E2F1, GSS, BCL2L1, RALY, MMP9, ITGB4BP, PLTP, TOP1, TNFRSF5, B4GALT5, STK15 | |
| +Xq21qter | TMSB4X, SLC25A5, HPRT1, PIM2, ZNF41, DSIPI, SAT, FLJ20811, UBE2A, HDGF, CXORF1, CLIC2, ARD1, ARHGAP4, CETN2, DXS1357E | |
| IL116 | +2p16 | KIAA1048 |
| +6p11pter | LTB, GABBR1, HLA-A, MDFI, PSMB9, HLA-DRB1, RPA40, GLO1 | |
| IL120 | −1p36 | LAP18, RPL11, PABPC4, AKR7A2, HUMHOXY1, CDC42, SMP1, HP1-BP74, EIF4G3, SDHB, NPPA, FGR, NBL1, ENO1, RER1 |
| −13q14 | LCP1, D13S106E,4-150 TSC22, CHCIL | |
| +17q11qter | ||
| IL121 | +18pter−q21.3 | PMAIP14-150 |
| +X | ZNF41, PIM2, SAT, IL13RA1 | |
| IL123 | +21q11qter | HMG14, ERG, DKFZP434C128 |
| IL124 | +4p11pter | TEC, ACOX3, FLJ20280, HMGE, SH3BP2, WHSC1 |
| −4q21qter | HNRPDL, FLJ20706,4-150 NFKB1, FLJ20647, CASP3, FLJ11200 | |
| +12 | ||
| IL126 | +3q12qter | GYG, PXR2b |
| −13q21.1 |
| Case . | Chromosome . | Genes . |
|---|---|---|
| IL105 | −5q21q23 | D5S346, GPX3, ADRA1B |
| +17q11qter | SCYA4, KIAA0356, ITGB3, TK1, H3F3B, RPL27, SMT3H2, ACTG1, KPNA2, NME1, NME2, MSF, KPNA2, GCN5L2, HN1, PSCD1, CD79B, ICAM2, CLTC, RARA, SCYA16, ACLY, IGFBP4, STAT3, TUBG2, GCN5L2, RPL27, TK1, SCYA3L1, NXPH3, EAP30, SPAG9, SFRS1, SUPT4H1, PRKAR1A, SMARCD2, MGC10986, BIRC5, CSNK1D, SFRS2, SMT3H2, ALY | |
| IL114 | +1q24 | RER1, TAGLN2, SELL, CREG, DKFZP564B167, CACYBP |
| +2p16 | PEX13, MDH1, XPO1 | |
| −4 | FLJ20706, 4-150FLJ20280 | |
| +5p12p15 | PDCD6, CCT5, NNT, RAD1, SEMA5A, FLJ20303, TRIP13 | |
| −6q11qter | ME1, TTK, BACH2, DKFZP564B0769, KIAA0776, FOXO3A, NMBR, SMPD2, MAP3K5, VNN2, HIVEP2, KIAA1209, RNASE6PL | |
| +7 | POR, FZD9, YWHAG, OGDH, SLC25A13, HNRPA2B1, MCM7, MDH2, KOC1, POLD2, PSMA2, PURB, AHR, GARS, ACTB, ARL4, DKFZP434J154, NUDT1, RAC1, RPA3, HRI, JTV1, KDELR2, FLJ20397, EIF3S9, RALA, KIAA0442, LOC51142, CCT6A, CRIP1, CYP51, PLOD3, GNB2, PMS2L8, STAG3, VGF, CUTL1, KIAA0632, COL1A2, PSMC2, EBNA-2 co-activator, IMPDH1, DLD, ATP6S14, KIAA1170, AKR1B1, EZH2, RHEB2, FLJ20311 | |
| +9p23p24 | ||
| +9q13q31 | TXN, SEC61B, FPGS, PRKACG, BTEB1, CDC20, CTSL, CKS2, C90RF3, CKS2, SEC61B, FLJ20287, TXN | |
| −9q31qter | ORM1, TNFSF8, HSPA5, ASS, COL5A1 | |
| +10 | CHUK, PP, DDX21, ATP5C1, MXI1, VIM, PRDX3, ACF, MGMT, PGAM1, VDAC2, UROS, ERP70, SSH3BP1, TCF8, CREM, COPEB, GDI2, PFKP, PHYH, LOC51008, RET, HNRPF, MBL2, JDP1, CDC2, ANXA7, ZWINT, HK1, PRG1, VCL, PPIF, ANXA11, DLG5, KIAA0261, MPHOSPH1, PLAU, GBF1, ACTR1A, RGS10, ABLIM, MK167, OAT, ECHS1, ADAM8 | |
| −11q11qter | GSTP1, POU2AF1, MS4A7, HTATIP, EMK1, MS4A2, SYT7, ZFPL1, CD6, RASGRP2, FOLR1, KIAA0102, ACTN3, FLJ20189, SCDGF-B, ATM, CASP1, INPPL1, MLL, UBE4A, CBL | |
| +12pter-q14 | PA2G4, LRMP, REA, TUBA3, K-ALPHA-1, LDHB, GAPD, TPI1, DDX11, CS, FLJ22028, KIAA0528, ARHGD1B, FOXM1, MLF2, C1S, PXR1, GPR92, COPS7A, FKBP4, CD69, ATP5B, CD4, CNTN1, SHMT2, PRKAG1, NR4A1, CDK2, HSU47926, MGC5576, HEM1, HNRPA1, MYL6, ATP5G2, EIF4B, FLJ11773, DDIT3, MARS, HSPD1, HSPC228, SAS, RAP1B, CDK4, NEDD1, NR2C1, FLJ11021, ZNF10 | |
| −13q11qter | GJB2, SACS, YDD19, ALOX5AP, HMG1, CG005, D13S106E,4-150 ELF1, AKAP11, SLC25A15, RFP2, FOXO1A, FLJ21562, FBXL3A, GPR18, EFNB2, FLJ10154, ATP11A | |
| −15q23 | RCN2, FBXO22 | |
| +16 | GLG1, AARS, ALDOA, EIF3S8, RPL13 KIAA0251, KNSL4, STX4A, SULT1A3, ITGAL, MAX, ITGAX, UQCRC2, SULT1A1, PHKG2, SULT1A2, PLK, PPP4C, ARL6IP, MHC2TA, TNFRSF17, GSPT1, HIRIP3, NK4, PKMYT1, POLR3K, PRSS22, RNPS1, SEPX1, STUB1, CCNF, BOM045, HMOX2, HMT-1, PMM2, DREV1, SIAH1, SCYD1, CPSF5, GOT2, KIAA0095, E2F4, ATP6D, PP15, RCD-8, CBFB, FLJ13725, FLJ10305, LOC51659, MAP1A/1BLC3, PCOLN3, SLC7A5, TUBB4, CDT1, SLC7A5 | |
| −17p12pter | SERPINF1, DOC2B | |
| +18q11qter | FLJ21172, PMAIP1,4-150 PIK3C3, MADH2, TNFRSF11A | |
| +20 | ADA, PCNA, KIAA1219, CDC25B, GNAS1, PSMA7, MYBL2, AHCY, PTPRA, INSM1, FKBP1A, SNRPB, MAPRE1, E2F1, GSS, BCL2L1, RALY, MMP9, ITGB4BP, PLTP, TOP1, TNFRSF5, B4GALT5, STK15 | |
| +Xq21qter | TMSB4X, SLC25A5, HPRT1, PIM2, ZNF41, DSIPI, SAT, FLJ20811, UBE2A, HDGF, CXORF1, CLIC2, ARD1, ARHGAP4, CETN2, DXS1357E | |
| IL116 | +2p16 | KIAA1048 |
| +6p11pter | LTB, GABBR1, HLA-A, MDFI, PSMB9, HLA-DRB1, RPA40, GLO1 | |
| IL120 | −1p36 | LAP18, RPL11, PABPC4, AKR7A2, HUMHOXY1, CDC42, SMP1, HP1-BP74, EIF4G3, SDHB, NPPA, FGR, NBL1, ENO1, RER1 |
| −13q14 | LCP1, D13S106E,4-150 TSC22, CHCIL | |
| +17q11qter | ||
| IL121 | +18pter−q21.3 | PMAIP14-150 |
| +X | ZNF41, PIM2, SAT, IL13RA1 | |
| IL123 | +21q11qter | HMG14, ERG, DKFZP434C128 |
| IL124 | +4p11pter | TEC, ACOX3, FLJ20280, HMGE, SH3BP2, WHSC1 |
| −4q21qter | HNRPDL, FLJ20706,4-150 NFKB1, FLJ20647, CASP3, FLJ11200 | |
| +12 | ||
| IL126 | +3q12qter | GYG, PXR2b |
| −13q21.1 |
Genes showing at least 2-fold difference in expression upon transformation. In cases with genomic gain/amplification, overexpressed genes were considered. In cases with genomic deletion, genes with decreased expression are shown.
Genes whose expression changed in two cases.
Discussion
We report the combined examination of genome and gene expression alterations accompanying the transformation of FCL to DLBCL. For this purpose, we have used the recently developed microarray-based comparative genomic hybridization technique, which assembles approximately 2400 BAC/P1 clones for quantitative measurement of DNA copy number aberrations across the human genome21,28 and previously published expression data.20 We find that the transformation of FCL is characterized by the acquisition of a variable spectrum of genomic imbalances. Some of these genomic imbalances are not transformation specific and also can be observed in nontransformed FCL, while others are acquired only upon transformation. In addition novel regions of genomic imbalance associated with transformation were identified. This genomic heterogeneity confirms previous studies of patients with FCL using CGH to chromosomes.17,19,29Moreover, array CGH allowed the structural analysis and delineation of the areas of imbalance in clinical specimens and in cell lines with higher accuracy and precision than CGH to chromosomes, distinguishing gene amplification loci within areas of gain and delineating the borders of the overrepresented loci. Array CGH also was capable of detecting small regions of genomic loss that are undetectable by CGH to chromosomes, including the homozygous loss of the P16 gene locus in the OZ cell line.25 An additional advantage of array CGH was the use of small amounts of genomic DNA (0.2 μg), which makes it suitable for the analysis of clinical material.
Areas of amplification and deletion identified in human tumors are strong candidates to contain genes important for cancer development and progression; these DNA alterations may influence gene expression. Indeed, it is a common assumption that chromosomal gains and losses exert “dosage effects” on the expression of at least some of the genes within these regions. However, confirmatory evidences for this hypothesis are scarce, especially in the process of FCL transformation to DLBCL.19 Limited numbers of analyzed specimens, especially those harboring recurrent chromosomal aberrations, multiplicity of potentially affected genes in each aberrant chromosomal region and lack of knowledge of all the genes in each chromosomal locus make such analysis difficult and contribute to the lack of knowledge of the pathophysiological consequences of the observed chromosomal alterations. Therefore, only cataloging of the chromosomal changes and the accompanying gene expression alterations will result in data that will address these questions. We investigated the genes that may potentially become deregulated in the areas of gain/amplification and loss by reanalyzing the published20 gene expression profiling. Our study identified a number of genes with deregulated expression that may be targeted by the genomic imbalances. However, many of the genes on the expression array that were located in these chromosomal areas did not show marked alteration in their expression within the measurement precision, and in addition, the majority of the genes that showed increased or decreased expression upon transformation were not located within the genomic regions deleted or amplified upon transformation. The limited number of cases with recurrent chromosomal aberrations prevented statistical analysis of the correlation between gene expression versus genomic changes. However, accumulation of similar data will enable such analysis in the future and will allow identification of which genes if any are deregulated by chromosomal aberrations.
Our analysis identified relevant genes that may be novel targets of chromosomal aberrations in FCL. For example, one of the most frequent imbalances was the gain of chromosome 18q, including in all cases chromosomal band 18q21 and the BCL2 gene locus in 18q21.3.17,19,29 As BCL2 gene expression was not altered upon transformation, other targets such as the genePMAIP1 in band 18q21.3 may be activated by this genomic gain. Also frequent was the gain/amplification of 2p16, locus of REL and BCL11A genes. Both REL andBCL11A genes have been found coamplified in both B-NHL and Hodgkin disease.30-34 Our data, however, did not identify marked changes in the expression of the REL gene upon transformation. Other changes acquired exclusively upon transformation included the gain of a segment in chromosome 12 from 12pter to 12q12. Trisomy of chromosome 12 is frequent in numerous B-cell malignancies, being the common region of amplification delineated to 12q12-q15.31,35,36 Recently, copy number gains at 12q12-q14 were reported exclusively in DLBCL cases transformed from previous FCL.17 The repeated alterations of chromosome 12q in lymphoid malignancies suggest that this is a candidate area for further molecular analysis to localize target genes involved in the pathogenesis of B-cell malignancies and especially in FCL transformation. Our study identified overexpression of many genes along 12q, including CDK2 in 12q13.3 and CDK4 in 12q14.1, which have been previously involved in B-cell maligancies.37,38 In previous studies, trisomy of chromosome 7 has been identified as a marker of progression from indolent to aggressive FCL.11,13 According to our new data, the minimal region of gain relies between 7q11.2 and 7q22.1, including one amplification site on 7q21.3. Potential candidate genes located into this area and found overexpressed are CYP51 andCUTL1 (CCATT displacement protein, cux/CDP). This gene regulates normal B lymphopoiesis, and its alteration is correlated with lymphoid abnormalities in mice.39
In addition, several genomic changes not previously reported in association with FCL transformation were identified, including overrepresentation on 6p12.3-p21. This chromosomal site is frequently targeted by translocation and genomic gain that include rearrangement of dominant oncogenes such as PIM1 and CCDN3 in B-NHL and multiple myeloma.40,41 In classical FCL, one case exhibited high-level amplification of 6p21,15 whereas we identified a more centromeric amplicon on 6p12.3. TheHLA-DRB1 gene, which is highly expressed in normal B cells, was found to be overexpressed in one case. Gain of 9p is common in primary mediastinal B-cell lymphoma and classical Hodgkin lymphoma,32,42,43 and according to our data may also have a role in FCL transformation. Other putative targeted gene in FCL transformation was NEM1, located in a novel region of gain in 17q21.33. This gene is a transcription factor and nucleoside diphosphate kinase that is up-regulated by MYC and has a role in the transcriptional regulation of MYC expression and has been involved in human malignancies.44
Several areas of chromosomal deletion also were identified in association with FCL transformation. The deletion of the long arm of chromosome 11 is a common change in mantle cell lymphoma and chronic lymphocytic leukemia, but in the majority of cases this deletion involves band 11q22 where the ATM gene is frequently the targeted gene.45,46 However, the region of deletion delineated here is far telomeric and may target a novel tumor suppressor gene on 11q24-q25. The deletion of the telomeric portion of 1p36.3 included the site of the tumor-suppressor gene TP73, which may be altered by deletion in B-NHL.47 Other areas of genomic loss described in our study will require further analysis to identify putative inactivated tumor suppressor genes.
In summary, the combination of array CGH and expression analysis provide a more comprehensive picture of the aberrant genetic state of FCL and the additional alterations that occur in the subsequent DLBCL than either set of measurements alone. The primary finding is that a heterogeneous set of genomic changes are associated with transformation. In addition, these genomic abnormalities target a fraction of genes that result in deregulated expression upon transformation. Given this heterogeneity and the potentially complex relationship between DNA copy number aberrations and gene expression levels in cancer, accumulation of larger data sets will be required to reveal the specific genetic events that are responsible for FCL transformation.
R.L. is an American Cancer Society Clinical Research Professor.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-07-2119.
Supported by grants from Spanish Ministry of Health FIS-01/0015 and GV2001-116, UICC-ICRETT 460/2001, Spanish Hematology Association (AEHH), grants CA33399 and CA34233 from the USPHS-NIH, grant R01CA83040, and a grant from Vysis Inc.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jose A. Martinez-Climent, Department of Hematology and Medical Oncology, Hospital Clinico, University of Valencia, Avda, Blasco Ibañez, 17, Valencia, 46010 Spain; e-mail:martinez_jos@gva.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal