Abstract
We undertook to systematically analyze the entire mitochondrial genome by gene amplification and direct sequencing in 10 patients with myelodysplasia; results were compared with concomitantly studied 8 healthy volunteers as well as mtDNA sequences in a standard database. Nucleotide changes that were present in our healthy controls as well as those in published databases were counted as polymorphisms. Overall, there was no increase in the number of mtDNA genes harboring polymorphisms or “new” mutations between our patients and healthy controls, although there were a few more mtDNA changes resulting in amino acid changes in myelodysplasia (9 in 8 controls versus 16 in 10 patients). Thirty new mutations, all nucleotide substitutions, were found among the 10 patients, distributed throughout the mitochondrial genome; 5 mutations resulted in amino acid changes. None of the mutations in controls produced amino acid changes. We were not able to confirm previously described mutations in sideroblastic anemia or “hot spots” in the cytochrome c oxidase I and II genes. Our data do not support a major role for mitochondrial genomic instability in myelodysplasia, and they fail to reproduce previous reports of significant or widespread mitochondrial mutations in this disease. Modest changes in mutation numbers and mitochondrial microsatellites may be evidence of increased mutagenesis in mtDNA, or, more likely, a reflection of limited clonality among hematopoietic stem cells in this bone marrow failure syndrome.
Introduction
The myelodysplastic syndromes (MDSs) are a heterogenous group of hematologic diseases, characterized by bone marrow failure and an increased risk of malignant transformation.1 Cytogenetic abnormalities are highly prevalent and correlate with prognosis and progression to leukemia.2-4 The more aggressive categories of MDSs, especially disease secondary to exposure to alkylating drugs, topoisomerase inhibitors, and radiation, share risk factors with acute leukemia and show stereotypical chromosome abnormalities.5-7 As in other premalignant conditions, genomic instability has been implicated in myelodysplasia. However, as with acute myeloid leukemia, the basis for the genomic instability required to generate the number of genetic lesions of a fully malignant phenotype has not been explained. In particular, there is little evidence of defects in mismatch repair and spindle checkpoint genes or for other hypothesized mechanisms.1,8 Ras gene mutations are the most frequent molecular abnormalities present in myelodysplasia, followed by p15 gene hypermethylation, FLT3 duplication, and p53 mutation, but these abnormalities are neither uniformly present nor characteristic of the syndromes.9 10
One clue to a possible basis for genomic instability in myelodysplasia may be provided by the constitutional disease called Pearson syndrome, in which sideroblastic anemia accompanies pancreatic abnormalities.11,12 In this inherited disorder, a number of deletions of mtDNA, as well as direct repeats, deletion-dimers, deletion-multimers, or duplications were observed.12Additionally, genetic changes in mtDNA have been hypothesized more widely to play important roles in senescence, malignancy,13 and autoimmune disease.14,15 In comparison to the nuclear genome, mtDNA shows some modification of the universal genetic code,16-18 a paucity of introns, and lack of histone protection. Past evidence has indicated that the mtDNA repair capacity of mitochondria is limited, and the proximity of mtDNA to sites of active oxygen species generation is suggestive that mtDNA might be more susceptible to mutation than nuclear DNA. Although the limited repair capacity hypothesis has been validated experimentally in some experimental systems,19,20 recent data have shown the existence of base excision repair mechanisms in mammalian mtDNA.21 22
The observation of sideroblastic anemia in Pearson syndrome as well as the theoretical concerns described earlier led to speculation that mtDNA abnormalities might be pathophysiologic in human MDS. Gattermann et al20 initially described abnormalities in the cytochrome c oxidase (CO) gene in patients with acquired idiopathic sideroblastic anemia and later in refractory anemia with excess blasts, lacking ringed sideroblasts.23Mutations leading to amino acid changes have been described in a variety of genes and in different MDS subtypes.23 Even more dramatic data supporting a role for mtDNA mutations in MDS have recently been published by Reddy et al24; they reported that mtDNA COI and COII genes contained large numbers of mutations and evidence of a number of single nucleotide substitutions in a large proportion of their patients.
Because mtDNA abnormalities might be reflective of an increased susceptibility to mutagenesis in patients with bone marrow failure, as well as provide a link to the development of nuclear genomic instability, cytogenetic abnormalities, and leukemic transformation, we undertook a systematic determination of mtDNA sequence in patients with MDS. Our results do not support a role for abnormalities in mtDNA in these syndromes, and the minor changes that were found may be explicable as the result of simpler features of the underlying pathophysiology of these diseases or naturally occurring mutations in mtDNA.
Patients, materials, and methods
Patients and healthy control subjects
Patients with MDS and controls are described in Table 1. Blood and bone marrow specimens were collected after informed consent was obtained following protocols approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute. MDS was diagnosed by peripheral blood and bone marrow findings according to criteria of the French-American-British classification system.25
Patients and healthy controls
| No. . | Age, y/sex . | Diagnosis . | Chromosomal analysis . |
|---|---|---|---|
| 1 | 38/F | Healthy control | ND |
| 2 | 37/M | Healthy control | ND |
| 3 | 41/F | Healthy control | ND |
| 4 | 41/M | Healthy control | ND |
| 5 | 42/M | Healthy control | ND |
| 6 | 48/M | Healthy control | ND |
| 7 | 42/M | Healthy control | ND |
| 8 | 78/M | Healthy control | ND |
| 9 | 58/M | RARS | 46XY |
| 10 | 77/M | RAEB | 46XY |
| 11 | 61/M | RA | 46XY, 11q− |
| 12 | 42/F | RA | 46XX, t(1:13) |
| 13 | 56/M | RAEB | 46XY |
| 14 | 62/M | RARS | 46XY |
| 15 | 60/M | RA | 46XY, 20q− |
| 16 | 64/M | RA | 47XY, +8 |
| 17 | 64/M | RAEB | 46XY, 20q− |
| 18 | 65/M | RARS | 46XY |
| No. . | Age, y/sex . | Diagnosis . | Chromosomal analysis . |
|---|---|---|---|
| 1 | 38/F | Healthy control | ND |
| 2 | 37/M | Healthy control | ND |
| 3 | 41/F | Healthy control | ND |
| 4 | 41/M | Healthy control | ND |
| 5 | 42/M | Healthy control | ND |
| 6 | 48/M | Healthy control | ND |
| 7 | 42/M | Healthy control | ND |
| 8 | 78/M | Healthy control | ND |
| 9 | 58/M | RARS | 46XY |
| 10 | 77/M | RAEB | 46XY |
| 11 | 61/M | RA | 46XY, 11q− |
| 12 | 42/F | RA | 46XX, t(1:13) |
| 13 | 56/M | RAEB | 46XY |
| 14 | 62/M | RARS | 46XY |
| 15 | 60/M | RA | 46XY, 20q− |
| 16 | 64/M | RA | 47XY, +8 |
| 17 | 64/M | RAEB | 46XY, 20q− |
| 18 | 65/M | RARS | 46XY |
M indicates male; F, female; ND, not done; RA, refractory anemia; RARS, refractory anemia with ring sideroblast; and RAEB, refractory anemia with excess blast.
mtDNA extraction and amplification
Total DNA extraction.
Mononuclear cells from bone marrow and peripheral blood were separated by density gradient centrifugation and washed twice in phosphate-buffered saline. DNA was extracted using QIAamp DNA blood mini kit (Qiagen, Valencia, CA). Extracted DNA was resuspended in TE buffer (pH 7.5) containing 10 mM Tris (tris(hydroxymethyl) aminomethane) and 1 mM EDTA (ethylenediaminetetraacetic acid).
Oligonucleotide primers.
To directly sequence the entire mtDNA, we used 20 primer pairs based on a modification of a published protocol26 to obtain 20 partially overlapping segments (Table 2). The COI and COII genes were amplified using additional 6 and 3 primer pairs, respectively, for the confirmation of mutations in these regions. The COI gene at mtDNA map position 5904 to 7445 was reamplified to produce 6 completely overlapping polymerase chain reaction (PCR) fragments to examine the reported “hot spots,” using the following primers: F5700, 5′-TAAGCACCCTAATCAACTGGC-3′; R6262, 5′-GCCTCCACTATAGCAGATGCG-3′; F5999, 5′-TCTAAGCCTCCTTATTCGAGC-3′; R6526, 5′-ATAGTGATGCCAGCAGCTAGG-3′; F6242, 5′-CGCATCTGCTATAGTGGAGG-3′; R6526, 5′-ATAGTGATGCCAGCAGCTAGG-3′; F6426, 5′-GCCATAACCCAATACCAAACG-3′; R7030, 5′-TGGGCTACAACGTAGTACGTG-3′; F6744, 5′-GGCTTCCTAGGGTTTATCGTG-3′; R7255, 5′-TTTCATGTGGTGTATGCATCG-3′; F7075, 5′-GAGGCTTCATTCACTGATTTCC-3′; and R7792, 5′-GGGCAGGATAGTTCAGACGG-3′. Similarly, the COII gene at mtDNA map position 7586 to 8269 was reamplified by PCR reaction to produce 3 completely overlapping fragments: F7215, 5′-CGACGTTACTCGGACTACCC-3′; R7792, 5′-GGGCAGGATAGTTCAGACGG-3′; F7645, 5′-TATCACCTTTCATGATCACGC-3′; R8215, 5′-GACGATGGGCATGAAACTG-3′; F7901, 5′-TGAACCTACGAGTACACCGACTAC-3′; R8311, 5′-AAGTTAGCTTTACAGTGGGCTCTAG-3′.
Primer sets used to amplify mtDNA
| Sets . | Position . | Length, bp . | Forward primer . | Reverse primer . |
|---|---|---|---|---|
| 1 | 15-921 | 907 | CACCCTATTAACCACTCACG | ACTTGGGTTAATCGTGTGACC |
| 2 | 873-1769 | 897 | GGTTGGTCAATTTCGTGCCAG | GCCAGGTTTCAATTTCTATCG |
| 3 | 1657-2660 | 1004 | CTTGACCGCTCTGAGCTAAAC | AGAGACAGCTGAACCCTCGTG |
| 4 | 2417-3557 | 1141 | CACTGTCAACCCAACACAGG | AGAAGAGCGATGGTGAGAGC |
| 5 | 3441-4728 | 1288 | ACTACAACCCTTCGCTGACG | TTATGGTTCATTGTCCGGAGAG |
| 6 | 4447-5882 | 1436 | TTGGTTATACCCTTCCCGTAC | GCTGAGTGAAGCATTGGACTG |
| 7 | 5700-6526 | 827 | TAAGCACCCTAATCAACTGGC | ATAGTGATGCCAGCAGCTAGG |
| 8 | 6426-7255 | 830 | GCCATAACCCAATACCAAACG | TTTCATGTGGTGTATGCATCG |
| 9 | 7215-8215 | 1001 | CGACGTTACTCGGACTACCC | GACGATGGGCATGAAACTG |
| 10 | 8164-9059 | 749 | CGGTCAATGCTCTGAAATCTGTG | GTGGCGCTTCCAATTAGGTG |
| 11 | 8903-9848 | 946 | CCCACTTCTTACCACAAGGC | GAAAGTTGAGCCAATAATGACG |
| 12 | 9754-10556 | 803 | AGTCTCCCTTCACCATTTCCG | GGAGGATATGAGGTGTGAGCG |
| 13 | 10386-11600 | 1215 | GGATTAGACTGAACCGAATTGG | CTGTTTGTCGTAGGCAGATGG |
| 14 | 11403-12189 | 787 | GACTCCCTAAAGCCCATGTCG | AAGCCTCTGTTGTCAGATTCAC |
| 15 | 11901-12876 | 976 | TGCTAGTAACCACGTTCTGGTG | GATATCGCCGATACGGTTG |
| 16 | 12793-13611 | 819 | TTGCTCATCAGTTGATGATACG | TCGAGTGCTATAGGCTTGTC |
| 17 | 13518-14388 | 871 | CATCATCGAAACCGCAAAC | TTAGCGATGGAGGTAGGATTCG |
| 18 | 14189-15396 | 1208 | ACAAACAATGGTCAACCAGTAAC | TTATCGGAATGGGAGGTGATTC |
| 19 | 15260-16451 | 1192 | AGTCCCACCCTCACACGATTC | GCGAGGAGAGTAGCACTCTTG |
| 20 | 16097-336 | 809 | TACATTACTGCCAGCCACCATG | TTAAGTGCTGTGGCCAGAAG |
| Sets . | Position . | Length, bp . | Forward primer . | Reverse primer . |
|---|---|---|---|---|
| 1 | 15-921 | 907 | CACCCTATTAACCACTCACG | ACTTGGGTTAATCGTGTGACC |
| 2 | 873-1769 | 897 | GGTTGGTCAATTTCGTGCCAG | GCCAGGTTTCAATTTCTATCG |
| 3 | 1657-2660 | 1004 | CTTGACCGCTCTGAGCTAAAC | AGAGACAGCTGAACCCTCGTG |
| 4 | 2417-3557 | 1141 | CACTGTCAACCCAACACAGG | AGAAGAGCGATGGTGAGAGC |
| 5 | 3441-4728 | 1288 | ACTACAACCCTTCGCTGACG | TTATGGTTCATTGTCCGGAGAG |
| 6 | 4447-5882 | 1436 | TTGGTTATACCCTTCCCGTAC | GCTGAGTGAAGCATTGGACTG |
| 7 | 5700-6526 | 827 | TAAGCACCCTAATCAACTGGC | ATAGTGATGCCAGCAGCTAGG |
| 8 | 6426-7255 | 830 | GCCATAACCCAATACCAAACG | TTTCATGTGGTGTATGCATCG |
| 9 | 7215-8215 | 1001 | CGACGTTACTCGGACTACCC | GACGATGGGCATGAAACTG |
| 10 | 8164-9059 | 749 | CGGTCAATGCTCTGAAATCTGTG | GTGGCGCTTCCAATTAGGTG |
| 11 | 8903-9848 | 946 | CCCACTTCTTACCACAAGGC | GAAAGTTGAGCCAATAATGACG |
| 12 | 9754-10556 | 803 | AGTCTCCCTTCACCATTTCCG | GGAGGATATGAGGTGTGAGCG |
| 13 | 10386-11600 | 1215 | GGATTAGACTGAACCGAATTGG | CTGTTTGTCGTAGGCAGATGG |
| 14 | 11403-12189 | 787 | GACTCCCTAAAGCCCATGTCG | AAGCCTCTGTTGTCAGATTCAC |
| 15 | 11901-12876 | 976 | TGCTAGTAACCACGTTCTGGTG | GATATCGCCGATACGGTTG |
| 16 | 12793-13611 | 819 | TTGCTCATCAGTTGATGATACG | TCGAGTGCTATAGGCTTGTC |
| 17 | 13518-14388 | 871 | CATCATCGAAACCGCAAAC | TTAGCGATGGAGGTAGGATTCG |
| 18 | 14189-15396 | 1208 | ACAAACAATGGTCAACCAGTAAC | TTATCGGAATGGGAGGTGATTC |
| 19 | 15260-16451 | 1192 | AGTCCCACCCTCACACGATTC | GCGAGGAGAGTAGCACTCTTG |
| 20 | 16097-336 | 809 | TACATTACTGCCAGCCACCATG | TTAAGTGCTGTGGCCAGAAG |
PCR conditions.
The PCR mixture consisted of 50 to 100 ng total DNA, 0.8 μM pair of primers, 0.4 mM of each deoxynucleoside triphosphate (dNTP), 2 U Taq DNA polymerase (TaKaRa LA Taq, Shiga, Japan), 5 μL 10 × buffer, and H2O in a total volume of 50 μL, and hybridizations were performed in a DNA thermal cycler (Perkin-Elmer, Foster City, CA) with one cycle of 96°C for 1 minute, followed by 36 cycles of 94°C for 30 seconds, 52°C for 50 seconds, 72°C for 1 minute, and, finally, one cycle of 72°C for 5 minutes followed by cooling to 4°C. A sample of amplified DNA was electrophoresed in 1.5% agarose and stained with ethidium bromide to assess the purity and size of the PCR products.
Sequence analysis
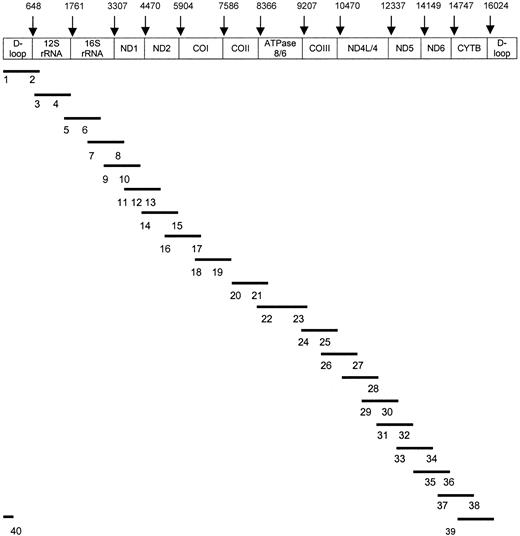
PCR products were purified with the QIA quick PCR purification kit protocol (Qiagen), and cycle sequencing was performed in a volume of 20 μL using 8 μL BigDye Terminator Ready Reaction Kit (Applied Biosystems, Foster City, CA), 10 pmol forward or reverse primer, and 50 to 100 ng PCR product. Using a DNA thermal cycler (Perkin-Elmer), cycle sequence conditions were as follows: 25 cycles of 96°C for 10 seconds, 50°C for 5 seconds, 60°C for 4 minutes, and finally cooling to 4°C. Fluorescent-labeled DNA was purified by DyeEx column (Qiagen) and then sequenced by 310 ABI sequencer/genetic analyzer (Applied Biosystems). Forty oligonucleotide primers derived from Levin et al26 were used in sequencing the entire mtDNA genome (Figure 1).
Sequencing strategy for total mtDNA.
Bars (–) show the 20 amplified mtDNA fragments. Numbers (1-40) indicate sequencing primers corresponding to the PCR products. The 40 sequence primers26 include 1 (F15), 2 (R921), 3 (F873), 4 (F1234), 5 (F1657), 6 (F2105), 7 (F2972), 8 (R3006), 9 (F3441), 10 (F3931), 11 (F4447), 12 (F4976), 13 (F5318), 14 (F5700), 15 (R6526), 16 (F6426), 17 (R7255), 18 (F7215), 19 (F7645), 20 (F8164), 21 (F8539), 22 (F9309), 23 (R9403), 24 (F9754), 25 (F10127), 26 (F11001), 27 (R11166), 28 (R11927), 29 (F11901), 30 (F12357), 31 (F12793), 32 (F13188), 33 (F13518), 34 (R14388), 35 (R14926), 36 (R15396), 37 (F15260), 38 (R16451), 39 (F16097), and 40 (R336). Original primer names are shown in parentheses.
Sequencing strategy for total mtDNA.
Bars (–) show the 20 amplified mtDNA fragments. Numbers (1-40) indicate sequencing primers corresponding to the PCR products. The 40 sequence primers26 include 1 (F15), 2 (R921), 3 (F873), 4 (F1234), 5 (F1657), 6 (F2105), 7 (F2972), 8 (R3006), 9 (F3441), 10 (F3931), 11 (F4447), 12 (F4976), 13 (F5318), 14 (F5700), 15 (R6526), 16 (F6426), 17 (R7255), 18 (F7215), 19 (F7645), 20 (F8164), 21 (F8539), 22 (F9309), 23 (R9403), 24 (F9754), 25 (F10127), 26 (F11001), 27 (R11166), 28 (R11927), 29 (F11901), 30 (F12357), 31 (F12793), 32 (F13188), 33 (F13518), 34 (R14388), 35 (R14926), 36 (R15396), 37 (F15260), 38 (R16451), 39 (F16097), and 40 (R336). Original primer names are shown in parentheses.
Determination of polymorphisms and mutations
Sequence experimentally obtained was compared with the 2001 Revised Cambridge Reference Sequence27,28(http://infinity.gen.emory.edu/mitomap.html) using the Blast2 program29(http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.htlm) and the database search tool, MitoAnalyzer30(http://www.cstl.nist.gov/biotech/strbase/mitoanalyzer.html, 2000) to obtain preliminary evidence for polymorphisms, mutations, and translation of amino acids. All automated results were manually confirmed. Data from patients with MDS also were compared with those obtained from concurrently tested healthy subjects, and we treated nucleotide substitutions found in both patients and healthy controls as polymorphisms. Thus, polymorphic sequence variants were identified from both the reported and unpublished database of polymorphisms in MITOMAP (http://infinity.gen.emory.edu/mitomap.html) and sequence changes observed in healthy individuals. When a mtDNA nucleotide change was only observed in patients with MDS, it was considered a new mutation. New mutations were confirmed by reamplification of the region using a separate cell specimen for DNA extraction.
Mitochondrial genomic instability
We determined the mitochondrial genomic instability using 11 mitochondrial markers: poly (C) tracts (starting bp position at 303, 311, 3566, 12 385, and 16 184); (CA)n microsatellite, starting bp at 514; poly (A) tract starting at 12 418 bp; deletions.31
Statistical analysis
The Mann-Whitney U test was used to evaluate statistical differences in the numbers of affected mtDNA genes, newly detected mtDNA mutations, and amino acid changes between patients and healthy volunteers; P < .05 was considered significant.
Results
mtDNA PCR products
The entire 16 569-bp mitochondrial genome was completely amplified using 20 overlapping 809- to 1288-bp PCR products, yielding single distinct bands for all 20 primer sets. These PCR products were then subjected to direct sequencing.
mtDNA nucleotide changes in healthy controls and patients
Among hematologic healthy control subjects, there were 102 mtDNA sequence variants, which consisted of 75 polymorphisms already listed in a published polymorphism database (http://infinity.gen.emory.edu/mitomap.html) and 27 new sequence variations not previously recorded, including unpublished mtDNA polymorphisms (Table 3). A total of 126 mtDNA nucleotide changes were noted among the 10 patients (Table 4), 30 nucleotide variants (mean = 3.0) were identified as mutations, and 5 mutations were predicted to produce amino acid change (Table 5; Figure2). The 102 nucleotide changes (mean = 12.8) among the 8 healthy individuals were distributed throughout the mitochondrial genome, and none of the new sequence variants led to amino acid changes (Table 5).
Mitochondrial DNA variants in bone marrow specimens of healthy controls
| No. . | Age, y/sex . | Polymorphism . | Mutation . | Affected mtDNA gene . | Amino acid change . |
|---|---|---|---|---|---|
| 1 | 38/F | 8602T>C | ATPase 6 | Phe→Leu | |
| 14212T>C | ND6 | No | |||
| 16326A>G | HV1 | ||||
| 16360C>T | HV1 | ||||
| 2 | 37/M | 4688T>C | ND2 | No | |
| 7022T>C | COI | No | |||
| 8697G>A | ATPase 6 | No | |||
| 9214A>G | COIII | His→Arg | |||
| 10463T>C | tRNA arginine | ||||
| 11812A>G | ND4 | No | |||
| 13368G>A | ND5 | No | |||
| 13965T>C | ND5 | No | |||
| 14233A>G | ND6 | No | |||
| 15607A>G | CYTB | No | |||
| 15927G>A | tRNA threonine | ||||
| 16294C>T | HV1 | ||||
| 16296C>T | HV1 | ||||
| 3 | 41/F | 150C>T | HV2 | ||
| 723A>G | 12S rRNA | ||||
| 1721C>T | 16S rRNA | ||||
| 3197T>C | 16S rRNA | ||||
| 3861A>G | ND1 | No | |||
| 5836A>G | tRNA tyrosine | ||||
| 7768A>G | COII | No | |||
| 10262A>G | ND3 | No | |||
| 13017A>G | ND5 | No | |||
| 13617T>C | ND5 | No | |||
| 13637A>G | ND5 | Glu→Arg | |||
| 14182T>C | ND6 | No | |||
| 16270C>T | HV1 | ||||
| 4 | 41/M | 150C>T | HV2 | ||
| 7669C>T | COII | No | |||
| 5 | 42/M | 1018G>A | 12S rRNA | ||
| 2416T>C | 16S rRNA | ||||
| 2789C>T | 16S rRNA | ||||
| 3594C>T | ND1 | No | |||
| 4104A>G | ND1 | No | |||
| 5022A>C | ND2 | Thr→Pro | |||
| 6663A>G | COI | Ile→Val | |||
| 7175T>C | COI | No | |||
| 7256C>T | COI | No | |||
| 7274C>T | COI | No | |||
| 7771A>G | COII | No | |||
| 8155G>A | COII | No | |||
| 8026G>A | COII | No | |||
| 9221A>G | COIII | No | |||
| 10115T>C | ND3 | No | |||
| 10799C>T | ND4 | No | |||
| 11944T>C | ND4 | No | |||
| 12693A>G | ND5 | No | |||
| 13650C>T | ND5 | No | |||
| 13803A>G | ND5 | No | |||
| 14566A>G | ND6 | No | |||
| 15884G>A | HV1 | ||||
| 16390G>A | NC | ||||
| 6 | 48/M | 215A>G | HV2 | ||
| 520C>CAC | NC | ||||
| 1888G>A | 16S rRNA | ||||
| 2141T>C | 16S rRNA | ||||
| 8697G>A | ATPase 6 | No | |||
| 9117T>C | ATPase 6 | No | |||
| 10463T>C | tRNA arginine | ||||
| 11719G>A | ND4 | No | |||
| 12741C>T | ND5 | No | |||
| 13368G>A | ND5 | No | |||
| 13965T>C | ND5 | No | |||
| 13966A>G | ND5 | Thr→Ala | |||
| 15607A>G | CYTB | No | |||
| 16296C>T | HV1 | ||||
| 16324T>C | HV1 | ||||
| 7 | 42/M | 89T>C | HV2 | ||
| 101G>A | HV2 | ||||
| 106G>C | HV2 | ||||
| 198C>T | HV2 | ||||
| 235A>G | HV2 | ||||
| 309C>T | HV2 | ||||
| 521-522delAC | NC | ||||
| 663A>G | 12S rRNA | ||||
| 1736A>G | 16S rRNA | ||||
| 4248T>C | ND1 | No | |||
| 4824A>G | ND2 | Thr→Ala | |||
| 5460G>A | ND2 | No | |||
| 6734G>A | COI | No | |||
| 6794A>G | COI | No | |||
| 7960A>G | COI | No | |||
| 8027G>A | COII | Ala→Thr | |||
| 12007G>A | ND4 | No | |||
| 16111C>T | HV1 | ||||
| 16290C>T | HV1 | ||||
| 16319G>A | HV1 | ||||
| 16360C>T | HV1 | ||||
| 8 | 78/M | 3197T>C | 16S rRNA | ||
| 5495T>C | ND2 | No | |||
| 11467A>G | ND4 | No | |||
| 12372G>A | ND5 | No | |||
| 13617T>C | ND5 | No | |||
| 15218A>G | CYTB | Thr→Ala | |||
| 16142C>T | HV1 | ||||
| 16210A>G | HV1 | ||||
| 16256C>T | HV1 | ||||
| 16270C>T | HV1 | ||||
| 16399A>G | NC |
| No. . | Age, y/sex . | Polymorphism . | Mutation . | Affected mtDNA gene . | Amino acid change . |
|---|---|---|---|---|---|
| 1 | 38/F | 8602T>C | ATPase 6 | Phe→Leu | |
| 14212T>C | ND6 | No | |||
| 16326A>G | HV1 | ||||
| 16360C>T | HV1 | ||||
| 2 | 37/M | 4688T>C | ND2 | No | |
| 7022T>C | COI | No | |||
| 8697G>A | ATPase 6 | No | |||
| 9214A>G | COIII | His→Arg | |||
| 10463T>C | tRNA arginine | ||||
| 11812A>G | ND4 | No | |||
| 13368G>A | ND5 | No | |||
| 13965T>C | ND5 | No | |||
| 14233A>G | ND6 | No | |||
| 15607A>G | CYTB | No | |||
| 15927G>A | tRNA threonine | ||||
| 16294C>T | HV1 | ||||
| 16296C>T | HV1 | ||||
| 3 | 41/F | 150C>T | HV2 | ||
| 723A>G | 12S rRNA | ||||
| 1721C>T | 16S rRNA | ||||
| 3197T>C | 16S rRNA | ||||
| 3861A>G | ND1 | No | |||
| 5836A>G | tRNA tyrosine | ||||
| 7768A>G | COII | No | |||
| 10262A>G | ND3 | No | |||
| 13017A>G | ND5 | No | |||
| 13617T>C | ND5 | No | |||
| 13637A>G | ND5 | Glu→Arg | |||
| 14182T>C | ND6 | No | |||
| 16270C>T | HV1 | ||||
| 4 | 41/M | 150C>T | HV2 | ||
| 7669C>T | COII | No | |||
| 5 | 42/M | 1018G>A | 12S rRNA | ||
| 2416T>C | 16S rRNA | ||||
| 2789C>T | 16S rRNA | ||||
| 3594C>T | ND1 | No | |||
| 4104A>G | ND1 | No | |||
| 5022A>C | ND2 | Thr→Pro | |||
| 6663A>G | COI | Ile→Val | |||
| 7175T>C | COI | No | |||
| 7256C>T | COI | No | |||
| 7274C>T | COI | No | |||
| 7771A>G | COII | No | |||
| 8155G>A | COII | No | |||
| 8026G>A | COII | No | |||
| 9221A>G | COIII | No | |||
| 10115T>C | ND3 | No | |||
| 10799C>T | ND4 | No | |||
| 11944T>C | ND4 | No | |||
| 12693A>G | ND5 | No | |||
| 13650C>T | ND5 | No | |||
| 13803A>G | ND5 | No | |||
| 14566A>G | ND6 | No | |||
| 15884G>A | HV1 | ||||
| 16390G>A | NC | ||||
| 6 | 48/M | 215A>G | HV2 | ||
| 520C>CAC | NC | ||||
| 1888G>A | 16S rRNA | ||||
| 2141T>C | 16S rRNA | ||||
| 8697G>A | ATPase 6 | No | |||
| 9117T>C | ATPase 6 | No | |||
| 10463T>C | tRNA arginine | ||||
| 11719G>A | ND4 | No | |||
| 12741C>T | ND5 | No | |||
| 13368G>A | ND5 | No | |||
| 13965T>C | ND5 | No | |||
| 13966A>G | ND5 | Thr→Ala | |||
| 15607A>G | CYTB | No | |||
| 16296C>T | HV1 | ||||
| 16324T>C | HV1 | ||||
| 7 | 42/M | 89T>C | HV2 | ||
| 101G>A | HV2 | ||||
| 106G>C | HV2 | ||||
| 198C>T | HV2 | ||||
| 235A>G | HV2 | ||||
| 309C>T | HV2 | ||||
| 521-522delAC | NC | ||||
| 663A>G | 12S rRNA | ||||
| 1736A>G | 16S rRNA | ||||
| 4248T>C | ND1 | No | |||
| 4824A>G | ND2 | Thr→Ala | |||
| 5460G>A | ND2 | No | |||
| 6734G>A | COI | No | |||
| 6794A>G | COI | No | |||
| 7960A>G | COI | No | |||
| 8027G>A | COII | Ala→Thr | |||
| 12007G>A | ND4 | No | |||
| 16111C>T | HV1 | ||||
| 16290C>T | HV1 | ||||
| 16319G>A | HV1 | ||||
| 16360C>T | HV1 | ||||
| 8 | 78/M | 3197T>C | 16S rRNA | ||
| 5495T>C | ND2 | No | |||
| 11467A>G | ND4 | No | |||
| 12372G>A | ND5 | No | |||
| 13617T>C | ND5 | No | |||
| 15218A>G | CYTB | Thr→Ala | |||
| 16142C>T | HV1 | ||||
| 16210A>G | HV1 | ||||
| 16256C>T | HV1 | ||||
| 16270C>T | HV1 | ||||
| 16399A>G | NC |
A indicates adenine; C, cytosine; G, guanine; T, thymine; Ala, alanine; Arg, arginine; Glu, glutamic acid; His, histidine; Ile, isoleucine; Leu, leucine; Phe, phenylalanine; Pro, proline; Thr, threonine; Val, valine; ATPase, ATP synthase; CO, cytochrome c oxidase; CYTB, cytochrome b; HV, hypervariable segment; NC, noncoding region; and ND, NADH dehydrogenase.
Mitochondrial DNA aberrations (nucleotide changes) in bone marrow specimens of patients with MDS
| No. . | Age, y/ sex . | Polymorphism . | Mutation . | Affected mtDNA gene . | Amino acid change . |
|---|---|---|---|---|---|
| 9 | 58/M | 204T>C | HV2 | ||
| 4715A>G | ND2 | No | |||
| 10256T>C | ND3 | No | |||
| 16147C>T | HV1 | ||||
| 10 | 77/M | 182C>T | HV2 | ||
| 2015G>A | 16S rRNA | ||||
| 3096T>C | 16S rRNA | ||||
| 5585G>A | NC | ||||
| 11206C>T | ND4 | No | |||
| 11 | 61/M | 189A>G | HV2 | ||
| 200A>G | HV2 | ||||
| 1822T>C | 16S rRNA | ||||
| 4218T>C | ND1 | No | |||
| 5528T>C | tRNA tryptophan | ||||
| 5601C>T | tRNA alanine | ||||
| 7819C>A | COII | No | |||
| 8410C>T | ATPase 8 | No | |||
| 8527A>G | ATPase 6 | Met→Val | |||
| 8932C>T | ATPase 6 | Pro→Ser | |||
| 9540T>C | COIII | No | |||
| 9950T>C | COIII | No | |||
| 10070C>T | ND3 | No | |||
| 11440G>A | ND4 | No | |||
| 14769A>G | CYTB | Asn→Ser | |||
| 15514T>C | CYTB | No | |||
| 16129G>A | HV1 | ||||
| 16209T>C | HV1 | ||||
| 16292C>T | HV1 | ||||
| 16295C>T | HV1 | ||||
| 16311C>T | HV1 | ||||
| 12 | 42/F | 153A>G | HV2 | ||
| 235A>G | HV2 | ||||
| 663A>G | 12S rRNA | ||||
| 1736A>G | 16S rRNA | ||||
| 4248T>C | ND1 | No | |||
| 4541G>A | ND2 | No | |||
| 4824A>G | ND2 | Thr→Ala | |||
| 7830G>A | COII | Arg→His | |||
| 7867C>T | COII | No | |||
| 8027G>A | COII | Ala→Thr | |||
| 8794C>T | ATPase 6 | His→Tyr | |||
| 12007G>A | ND4 | No | |||
| 16111C>T | HV1 | ||||
| 16209T>C | HV1 | ||||
| 16290C>T | HV1 | ||||
| 16319G>A | HV1 | ||||
| 16362T>C | HV1 | ||||
| 13 | 56/M | 199T>C | HV2 | ||
| 204T>C | HV2 | ||||
| 250T>C | HV2 | ||||
| 568C>CC | NC | ||||
| 574A>C | NC | ||||
| 1719G>A | 16S rRNA | ||||
| 4529A>T | ND2 | No | |||
| 5074T>C | ND2 | Ile→Thr | |||
| 8251G>A | COII | No | |||
| 10034T>C | tRNA glycine | ||||
| 10238T>C | ND3 | No | |||
| 12408T>A | ND5 | No | |||
| 12501G>A | ND5 | No | |||
| 14233A>G | ND6 | No | |||
| 15043G>A | CYTB | No | |||
| 15244A>G | CYTB | No | |||
| 15924A>G | tRNA threonine | ||||
| 16129G>A | HV1 | ||||
| 16148C>T | HV1 | ||||
| 16294C>T | HV1 | ||||
| 16374A>C | HV1 | ||||
| 16391G>A | NC | ||||
| 14 | 62/M | 215A>G | HV2 | ||
| 295A>G | HV2 | ||||
| 319T>C | HV2 | ||||
| 489T>C | 12S rRNA | ||||
| 1095T>C | 12S rRNA | ||||
| 1850T>C | 16S rRNA | ||||
| 7789G>A | COII | No | |||
| 10499A>G | ND4L | No | |||
| 11377G>A | ND4 | No | |||
| 13708G>A | ND5 | Ala→Thr | |||
| 13722A>G | ND5 | No | |||
| 14133A>G | ND5 | No | |||
| 15257G>A | CYTB | Asp→Asn | |||
| 16145G>A | HV1 | ||||
| 16231T>C | HV1 | ||||
| 16261C>T | HV1 | ||||
| 15 | 60/M | 7337G>A | COI | No | |
| 12136T>C | ND4 | No | |||
| 14529C>G | ND6 | Gly→Arg | |||
| 15355G>A | CYTB | No | |||
| 15758A>G | CYTB | Ile→Val | |||
| 16259C>T | HV1 | ||||
| 16311T>C | HV1 | ||||
| 16 | 62/M | 504T>C | NC | ||
| 7337G>A | COI | No | |||
| 8697G>A | ATPase 6 | No | |||
| 9899T>C | COIII | No | |||
| 10463T>C | tRNA arginine | ||||
| 12633C>A | ND5 | No | |||
| 13368G>A | ND5 | No | |||
| 14905G>A | CYTB | No | |||
| 15607A>G | CYTB | No | |||
| 15928G>A | tRNA threonine | ||||
| 16163A>G | HV1 | ||||
| 16178T>C | HV1 | ||||
| 16186C>T | HV1 | ||||
| 16189T>C | HV1 | ||||
| 16256C>T | HV1 | ||||
| 17 | 64/M | 3197T>C | 16S rRNA | ||
| 5153A>G | ND2 | No | |||
| 9477G>A | COIII | Val→Ile | |||
| 9667A>G | COIII | Asp→Ser | |||
| 11467A>G | ND4 | No | |||
| 13617T>C | ND5 | No | |||
| 14793A>G | CYTB | His→Arg | |||
| 15218A>G | CYTB | Thr→Ala | |||
| 16256C>T | HV1 | ||||
| 16270C>T | HV1 | ||||
| 16362T>C | HV1 | ||||
| 16399A>G | NC | ||||
| 16428G>A | NC | ||||
| 18 | 65/M | 4580G>A | ND2 | No | |
| 12408T>C | ND5 | No | |||
| 14260A>G | ND6 | No | |||
| 14767T>C | CYTB | No | |||
| 15904C>T | tRNA threonine | ||||
| 16298T>C | HV1 |
| No. . | Age, y/ sex . | Polymorphism . | Mutation . | Affected mtDNA gene . | Amino acid change . |
|---|---|---|---|---|---|
| 9 | 58/M | 204T>C | HV2 | ||
| 4715A>G | ND2 | No | |||
| 10256T>C | ND3 | No | |||
| 16147C>T | HV1 | ||||
| 10 | 77/M | 182C>T | HV2 | ||
| 2015G>A | 16S rRNA | ||||
| 3096T>C | 16S rRNA | ||||
| 5585G>A | NC | ||||
| 11206C>T | ND4 | No | |||
| 11 | 61/M | 189A>G | HV2 | ||
| 200A>G | HV2 | ||||
| 1822T>C | 16S rRNA | ||||
| 4218T>C | ND1 | No | |||
| 5528T>C | tRNA tryptophan | ||||
| 5601C>T | tRNA alanine | ||||
| 7819C>A | COII | No | |||
| 8410C>T | ATPase 8 | No | |||
| 8527A>G | ATPase 6 | Met→Val | |||
| 8932C>T | ATPase 6 | Pro→Ser | |||
| 9540T>C | COIII | No | |||
| 9950T>C | COIII | No | |||
| 10070C>T | ND3 | No | |||
| 11440G>A | ND4 | No | |||
| 14769A>G | CYTB | Asn→Ser | |||
| 15514T>C | CYTB | No | |||
| 16129G>A | HV1 | ||||
| 16209T>C | HV1 | ||||
| 16292C>T | HV1 | ||||
| 16295C>T | HV1 | ||||
| 16311C>T | HV1 | ||||
| 12 | 42/F | 153A>G | HV2 | ||
| 235A>G | HV2 | ||||
| 663A>G | 12S rRNA | ||||
| 1736A>G | 16S rRNA | ||||
| 4248T>C | ND1 | No | |||
| 4541G>A | ND2 | No | |||
| 4824A>G | ND2 | Thr→Ala | |||
| 7830G>A | COII | Arg→His | |||
| 7867C>T | COII | No | |||
| 8027G>A | COII | Ala→Thr | |||
| 8794C>T | ATPase 6 | His→Tyr | |||
| 12007G>A | ND4 | No | |||
| 16111C>T | HV1 | ||||
| 16209T>C | HV1 | ||||
| 16290C>T | HV1 | ||||
| 16319G>A | HV1 | ||||
| 16362T>C | HV1 | ||||
| 13 | 56/M | 199T>C | HV2 | ||
| 204T>C | HV2 | ||||
| 250T>C | HV2 | ||||
| 568C>CC | NC | ||||
| 574A>C | NC | ||||
| 1719G>A | 16S rRNA | ||||
| 4529A>T | ND2 | No | |||
| 5074T>C | ND2 | Ile→Thr | |||
| 8251G>A | COII | No | |||
| 10034T>C | tRNA glycine | ||||
| 10238T>C | ND3 | No | |||
| 12408T>A | ND5 | No | |||
| 12501G>A | ND5 | No | |||
| 14233A>G | ND6 | No | |||
| 15043G>A | CYTB | No | |||
| 15244A>G | CYTB | No | |||
| 15924A>G | tRNA threonine | ||||
| 16129G>A | HV1 | ||||
| 16148C>T | HV1 | ||||
| 16294C>T | HV1 | ||||
| 16374A>C | HV1 | ||||
| 16391G>A | NC | ||||
| 14 | 62/M | 215A>G | HV2 | ||
| 295A>G | HV2 | ||||
| 319T>C | HV2 | ||||
| 489T>C | 12S rRNA | ||||
| 1095T>C | 12S rRNA | ||||
| 1850T>C | 16S rRNA | ||||
| 7789G>A | COII | No | |||
| 10499A>G | ND4L | No | |||
| 11377G>A | ND4 | No | |||
| 13708G>A | ND5 | Ala→Thr | |||
| 13722A>G | ND5 | No | |||
| 14133A>G | ND5 | No | |||
| 15257G>A | CYTB | Asp→Asn | |||
| 16145G>A | HV1 | ||||
| 16231T>C | HV1 | ||||
| 16261C>T | HV1 | ||||
| 15 | 60/M | 7337G>A | COI | No | |
| 12136T>C | ND4 | No | |||
| 14529C>G | ND6 | Gly→Arg | |||
| 15355G>A | CYTB | No | |||
| 15758A>G | CYTB | Ile→Val | |||
| 16259C>T | HV1 | ||||
| 16311T>C | HV1 | ||||
| 16 | 62/M | 504T>C | NC | ||
| 7337G>A | COI | No | |||
| 8697G>A | ATPase 6 | No | |||
| 9899T>C | COIII | No | |||
| 10463T>C | tRNA arginine | ||||
| 12633C>A | ND5 | No | |||
| 13368G>A | ND5 | No | |||
| 14905G>A | CYTB | No | |||
| 15607A>G | CYTB | No | |||
| 15928G>A | tRNA threonine | ||||
| 16163A>G | HV1 | ||||
| 16178T>C | HV1 | ||||
| 16186C>T | HV1 | ||||
| 16189T>C | HV1 | ||||
| 16256C>T | HV1 | ||||
| 17 | 64/M | 3197T>C | 16S rRNA | ||
| 5153A>G | ND2 | No | |||
| 9477G>A | COIII | Val→Ile | |||
| 9667A>G | COIII | Asp→Ser | |||
| 11467A>G | ND4 | No | |||
| 13617T>C | ND5 | No | |||
| 14793A>G | CYTB | His→Arg | |||
| 15218A>G | CYTB | Thr→Ala | |||
| 16256C>T | HV1 | ||||
| 16270C>T | HV1 | ||||
| 16362T>C | HV1 | ||||
| 16399A>G | NC | ||||
| 16428G>A | NC | ||||
| 18 | 65/M | 4580G>A | ND2 | No | |
| 12408T>C | ND5 | No | |||
| 14260A>G | ND6 | No | |||
| 14767T>C | CYTB | No | |||
| 15904C>T | tRNA threonine | ||||
| 16298T>C | HV1 |
Asn indicates Asparagine; Asp, aspartic acid; Gly, glycine; Met, methionine; Ser, serine; and Tyr, tyrosine.
The number and distribution of newly detected mtDNA mutations in bone marrow of patients with MDS and healthy controls
| mtDNA gene . | Patients with MDS . | Healthy controls . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | No. . | M . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | No. . | M . | |
| NC | 0 | 00 | 00 | 5 | 0.5 | 00 | 0 | 3 | 0.4 | |||||||||||||
| 12S rRNA | 0 | 1 | 0.1 | 0 | 1 | 0.1 | ||||||||||||||||
| 16S rRNA | 00 | 0 | 3 | 0.3 | 0 | 00 | 0 | 4 | 0.5 | |||||||||||||
| ND1 | 0 | 1 | 0.1 | 0 | 1 | 0.1 | ||||||||||||||||
| ND2 | 0 | † | 0 | 3 | 0.3 | 0 | 0 | 2 | 0.3 | |||||||||||||
| tRNA | 0 | 1 | 0.1 | 0 | 1 | 0.2 | ||||||||||||||||
| COI | 0 | 0.0 | 0 | 0 | 0 | 2 | 0.3 | |||||||||||||||
| COII | † | 1 | 0.1 | 0 | 2 | 0.3 | ||||||||||||||||
| COIII | 0 | 0.0 | 0 | 1 | 0.1 | |||||||||||||||||
| ATPase 8 | 0 | 1 | 0.1 | 0 | 0.0 | |||||||||||||||||
| ATPase 6 | † | 1 | 0.1 | 0 | 1 | 0.1 | ||||||||||||||||
| COIII | 0 | 0.0 | 0 | 0.0 | ||||||||||||||||||
| ND3 | 0 | 0 | 2 | 0.2 | 0 | 0.0 | ||||||||||||||||
| ND4L | 0 | 0.0 | 0 | 0.0 | ||||||||||||||||||
| ND4 | 0 | 0 | 0 | 3 | 0.3 | 0 | 1 | 0.1 | ||||||||||||||
| ND5 | 0 | 0 | 0 | 3 | 0.3 | 0 | 0 | 000 | 00 | 7 | 0.9 | |||||||||||
| ND6 | † | 0 | 2 | 0.2 | 0 | 1 | 0.1 | |||||||||||||||
| CYTB | †0 | 0 | 3 | 0.3 | 0 | 0.0 | ||||||||||||||||
| Total | 1 | 4 | 8 | 2 | 4 | 2 | 2 | 3 | 1 | 3 | 30 | 3.0 | 0 | 3 | 6 | 0 | 9 | 4 | 3 | 2 | 27 | 3.4 |
| mtDNA gene . | Patients with MDS . | Healthy controls . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | No. . | M . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | No. . | M . | |
| NC | 0 | 00 | 00 | 5 | 0.5 | 00 | 0 | 3 | 0.4 | |||||||||||||
| 12S rRNA | 0 | 1 | 0.1 | 0 | 1 | 0.1 | ||||||||||||||||
| 16S rRNA | 00 | 0 | 3 | 0.3 | 0 | 00 | 0 | 4 | 0.5 | |||||||||||||
| ND1 | 0 | 1 | 0.1 | 0 | 1 | 0.1 | ||||||||||||||||
| ND2 | 0 | † | 0 | 3 | 0.3 | 0 | 0 | 2 | 0.3 | |||||||||||||
| tRNA | 0 | 1 | 0.1 | 0 | 1 | 0.2 | ||||||||||||||||
| COI | 0 | 0.0 | 0 | 0 | 0 | 2 | 0.3 | |||||||||||||||
| COII | † | 1 | 0.1 | 0 | 2 | 0.3 | ||||||||||||||||
| COIII | 0 | 0.0 | 0 | 1 | 0.1 | |||||||||||||||||
| ATPase 8 | 0 | 1 | 0.1 | 0 | 0.0 | |||||||||||||||||
| ATPase 6 | † | 1 | 0.1 | 0 | 1 | 0.1 | ||||||||||||||||
| COIII | 0 | 0.0 | 0 | 0.0 | ||||||||||||||||||
| ND3 | 0 | 0 | 2 | 0.2 | 0 | 0.0 | ||||||||||||||||
| ND4L | 0 | 0.0 | 0 | 0.0 | ||||||||||||||||||
| ND4 | 0 | 0 | 0 | 3 | 0.3 | 0 | 1 | 0.1 | ||||||||||||||
| ND5 | 0 | 0 | 0 | 3 | 0.3 | 0 | 0 | 000 | 00 | 7 | 0.9 | |||||||||||
| ND6 | † | 0 | 2 | 0.2 | 0 | 1 | 0.1 | |||||||||||||||
| CYTB | †0 | 0 | 3 | 0.3 | 0 | 0.0 | ||||||||||||||||
| Total | 1 | 4 | 8 | 2 | 4 | 2 | 2 | 3 | 1 | 3 | 30 | 3.0 | 0 | 3 | 6 | 0 | 9 | 4 | 3 | 2 | 27 | 3.4 |
NC indicates noncoding region including hypervariable segments; M, mean; †, amino acid change; and 0, no amino acid change.
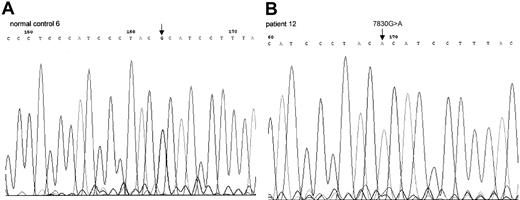
Sequence chromatogram of PCR-amplified mtDNA
COII gene from healthy control 6 (A), patient 12 (B). This 7830G>A mutation led to amino acid change from Arg to His.
Sequence chromatogram of PCR-amplified mtDNA
COII gene from healthy control 6 (A), patient 12 (B). This 7830G>A mutation led to amino acid change from Arg to His.
Hot spot mutations
Statistical comparisons
Compared with the 2001 Revised Cambridge Reference Sequence and with sequences from healthy subjects and patients, we excluded nucleotide changes found in both healthy controls and patients. Although there was a small increase in the number of amino acid changes predicted to result from polymorphisms and mutations in the MDS group (mean = 1.6) compared with healthy individuals (mean = 1.1), this difference was not statistically significant (Table6). There was no difference in the number of affected mtDNA genes in patients with MDS and healthy individuals (Table 5). For mutations alone, only those from patients showed amino acid changes (mean = 0.5) (Table 5).
Amino acid changes among polymorphism and mutation
| mtDNA gene . | Patients with MDS . | Healthy controls . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | No. . | M . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | No. . | M . | |
| ND1 | 0 | 0.0 | 0 | 0 | ||||||||||||||||||
| ND2 | † | † | 2 | 0.2 | † | † | 2 | 0.3 | ||||||||||||||
| COI | 0 | 0.0 | † | 1 | 0.1 | |||||||||||||||||
| COII | †† | 2 | 0.2 | † | 1 | 0.1 | ||||||||||||||||
| ATPase 8 | 0 | 0.0 | 0 | 0 | ||||||||||||||||||
| ATPase 6 | †† | † | 3 | 0.3 | † | 1 | 0.1 | |||||||||||||||
| COIII | †† | 2 | 0.2 | † | 1 | 0.1 | ||||||||||||||||
| ND3 | 0 | 0.3 | 0 | 0 | ||||||||||||||||||
| ND4L | 0 | 0.0 | 0 | 0 | ||||||||||||||||||
| ND4 | 0 | 0.0 | 0 | 0 | ||||||||||||||||||
| ND5 | † | 1 | 0.1 | † | † | 2 | 0.3 | |||||||||||||||
| ND6 | † | 1 | 0.1 | 0 | 0 | |||||||||||||||||
| CYTB | † | † | † | †† | 5 | 0.5 | † | 1 | 0.1 | |||||||||||||
| Total | 0 | 0 | 3 | 4 | 1 | 2 | 2 | 0 | 4 | 0 | 16 | 1.6 | 1 | 1 | 1 | 0 | 2 | 1 | 2 | 1 | 9 | 1.1 |
| mtDNA gene . | Patients with MDS . | Healthy controls . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | No. . | M . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | No. . | M . | |
| ND1 | 0 | 0.0 | 0 | 0 | ||||||||||||||||||
| ND2 | † | † | 2 | 0.2 | † | † | 2 | 0.3 | ||||||||||||||
| COI | 0 | 0.0 | † | 1 | 0.1 | |||||||||||||||||
| COII | †† | 2 | 0.2 | † | 1 | 0.1 | ||||||||||||||||
| ATPase 8 | 0 | 0.0 | 0 | 0 | ||||||||||||||||||
| ATPase 6 | †† | † | 3 | 0.3 | † | 1 | 0.1 | |||||||||||||||
| COIII | †† | 2 | 0.2 | † | 1 | 0.1 | ||||||||||||||||
| ND3 | 0 | 0.3 | 0 | 0 | ||||||||||||||||||
| ND4L | 0 | 0.0 | 0 | 0 | ||||||||||||||||||
| ND4 | 0 | 0.0 | 0 | 0 | ||||||||||||||||||
| ND5 | † | 1 | 0.1 | † | † | 2 | 0.3 | |||||||||||||||
| ND6 | † | 1 | 0.1 | 0 | 0 | |||||||||||||||||
| CYTB | † | † | † | †† | 5 | 0.5 | † | 1 | 0.1 | |||||||||||||
| Total | 0 | 0 | 3 | 4 | 1 | 2 | 2 | 0 | 4 | 0 | 16 | 1.6 | 1 | 1 | 1 | 0 | 2 | 1 | 2 | 1 | 9 | 1.1 |
NC indicates noncoding region including hypervariable segments; M, mean; †, amino acid change; and 0, no amino acid change.
We used homopolymeric tracts and deletions reported for the detection of mitochondrial genomic instability in human tumors. Among the 11 markers, abnormalities were observed in 3 poly (C)s, starting at bp 303, 311, and 16 184, respectively. Although there was a statistical significance difference between MDS group (n = 18, mean = 1.8) and control group (n = 8, mean = 1.0), too few samples were analyzed to exclude the possibility that the C stretch abnormalities had segregated randomly because of their relative frequency in normal populations (see “Discussion”).
Discussion
To date, more than 100 point mutations and more than 200 deletions and rearrangements in mtDNA have been associated with disease, and new mutations are being described every year.15 Ironically, the database of “normal” mtDNA sequences is relatively limited. The classic Cambridge Reference Sequence, based on a consensus analysis of a placenta, the HeLa cell line, and some information from the bovine sequence, has been corrected based on a reanalysis of the original placenta.27 Although other sequences have been reported in the literature and to computerized databases, the origin of the tissues tested has often been from individuals suspected of harboring pathologic mutations or their family members. Even the distinction between polymorphisms, which are common, and new mutations, is poorly demarcated. For these reasons, in the current study we also undertook to determine the sequence of bone marrow mtDNA from a comparable number of age-matched healthy volunteers.
In the current work, we found a large number of polymorphisms as well as apparent new mutations in both patients and controls, with no statistical difference overall between these 2 populations of individuals. For the patients with MDS, there was a modest number of “new” (not present in http://www.gen.emory.edu/mitomap.html) mutations that resulted in predicted amino acid changes in the gene product; among the 10 patients, 5 mutations producing nonsynonymous alterations were found in nicotinamide adenine dinucleotide dehydrogenase (ND)2, COII, adenosine triphosphatase (ATPase) 6, ND6, and cytochrome b(CYTB). No patient showed more than a single such amino acid change, and most of the patients with MDS, therefore, were not distinguishable from healthy individuals. We did not observe small deletions. Large deletions, in particular the common 4.9-kb deletion between the ATPase8 and ND5genes32 33 and other large deletions that characterize Pearson syndrome, or deletions observed in some other constitutional mtDNA diseases would need additional experiments to confirm.
Our results differ and in some respects fail to confirm data from other laboratories that have addressed the role of mtDNA in myelodysplasia. Gattermann et al20 and Gattermann23 used temperature-gradient gel electrophoresis to identify duplexes of amplified mtDNA products and then directly sequenced the implicated region. Looking first at acquired idiopathic sideroblastic anemia, because of its similarity to Pearson syndrome and the obvious mitochondrial involvement, they first reported mutations producing amino acid sequence changes, mainly in the CYTB andCOI genes.20,23 Subsequently, these investigations were expanded to other MDS categories and revealed mutations in CYTB in refractory anemia with excess blasts as well as in early leukemia. In total, new mutations have been reported in 10 sideroblastic anemia cases and 7 other MDSs.23Comparison of these data with our own is not entirely straightforward, partly because the German investigators did not globally query mtDNA in their cases, the number of negative cases is not described, nor were healthy controls included. The putatively pathogenic mutations that they describe are not present in published databases and were not found among our healthy controls. However, in the 3 patients with sideroblastic anemia whom we studied, none showed mutations producing amino acid changes. We cannot exclude that examination of larger numbers or more selected patients with clinical subtypes of sideroblastic anemia might not reveal mutational events, but our data do not support a general abnormality in mtDNA sequence in this syndrome. Reddy et al24 also reported major abnormalities in mtDNA sequence in myelodysplasia, including apparent hot spots: 16 of the 20 MDS cases showed mtDNA mutations, 4 in COI only, 3 in COII only, and 9 in both genes (only portions of these genes were sequenced in this study). The COI changes included deletions, insertions, and substitution mutations;COI mutations were most frequently seen as 7264C>A (25%) and 7289delA (15%), and COII gene mutations were most frequently seen at 7594T>G (30%) and 7595G>C (40%). Even more remarkable, all the substitutional mutations led to amino acid changes or generation of a termination codon. These results are in striking contrast to our own limited number of new mutations, of which none were found at the putative hot spots described by Reddy et al.24 One possible explanation for this discrepancy is misalignment of the sequences obtained from the affected patients with 2001 Revised Cambridge Reference Sequence, which differs from GenBank No. NC 001807 that Reddy et al24 used to design their primers. A 1-bp shift in the sequence comparison using a Blast2 program (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.htlm) would produce such an unexpected result. This plausible explanation would also account for failure to observe the mutations in peripheral blood cells and their higher frequency in more mature marrow cells, for which there is no proposed biologic mechanism.
In addition to the few new mutations, we also noted a modest increase in homopolymeric poly (C) tracts; differences in these tracts have been associated with mitochondrial genomic instability analogous to microsatellites in nuclear genomic instability. Indeed, microsatellite instability has been described in 2 of 26 patients with MDS.34 However, in the mitochondrial forensic community, these regions are referred to as the C-stretch regions; a notable example occurs when a T at 16189 becomes a C, producing a 10C-stretch. Within the population databases, there are a significant number of individuals that have this substitution; for example, the Federal Bureau of Investigation (FBI) database indicates that about 9.8% of Caucasians and 12.6% of African Americans have this C-stretch (http://www.fbi.gov/hq/lab/fsc/backissu/april2002/miller1.htm). Therefore, because our numbers are small, this is probably not a significant difference (T. Parsons, Armed Forces DNA Identification Laboratory [AFDIL], Armed Forces Institute of Pathology [AFIP], Department of Defense [DOD], personal communication, June 2002). While it is tempting to associate both the limited new mutations seen in a few patients and the abnormal homopolymeric tracts with the pathophysiology of MDS, we believe that a more likely and stringent interpretation is that these apparent abnormalities are secondary to the small numbers of stem cell clones operating to support hematopoiesis in this bone marrow failure syndrome. Likely both mtDNA mutations (and also, less frequently, nuclear DNA mutations) and differences in homopolymeric length are prevalent among hematopoietic stem and progenitor cells and are ordinarily of no physiologic consequences. Analysis of total DNA results in an averaging of these abnormalities, a biologic regression to the normal mean. Such abnormalities will be more likely observed when limited numbers of clones are subjected to testing. Tumors, the ultimate clonal population, appear to fix mutations that have arisen randomly.35 For mtDNA, point mutations appear to be abundant in individual cells of normal tissue, which by unconfirmed mechanisms such as the bottleneck theory become homoplasmic for these abnormalities.36 Whether, as is likely, individual clones of cells derived from normal bone marrow progenitors will demonstrate similar phenomena are amenable to experimentation.
We thank Prof Norbert Gattermann (Department of Hematology, University of Dusseldorf, Germany) for helpful discussions and his interest in our work.
This paper is a contribution of the National Institutes of Health (NIH) and National Institute of Standards and Technology (NIST) and is not subject to copyright. Certain commercial equipment, instruments, materials, or companies are identified in this paper to specify the experimental procedure. Such identification does not imply recommendation or endorsement by NIH and NIST, nor does it imply that the materials or equipment identified are the best available for this purpose.
Prepublished online as Blood First Edition Paper, November 21, 2002; DOI 10.1182/blood-2002-06-1825.
M.G.S. and S.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Neal S. Young, Bldg 10, Rm 7C103; NIH; 9000 Rockville Pike, Bethesda, MD 20892-1652; e-mail:youngn@nhlbi.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal