Abstract
Signal transducer and activator of transcription (STAT) proteins are a 7-member family of cytoplasmic transcription factors that contribute to signal transduction by cytokines, hormones, and growth factors. STAT proteins control fundamental cellular processes, including survival, proliferation, and differentiation. Given the critical roles of STAT proteins, it was hypothesized that inappropriate or aberrant activation of STATs might contribute to cellular transformation and, in particular, leukemogenesis. Constitutive activation of mutated STAT3 has in fact been demonstrated to result in transformation. STAT activation has been extensively studied in leukemias, and mechanisms of STAT activation and the potential role of STAT signaling in leukemogenesis are the focus of this review. A better understanding of mechanisms of dysregulation of STAT signaling pathways may serve as a basis for designing novel therapeutic strategies that target these pathways in leukemia cells.
Introduction
Signal transducer and activator of transcription (STAT) proteins are a family of latent cytoplasmic transcription factors involved in cytokine, hormone, and growth factor signal transduction.1-7 STAT proteins mediate broadly diverse biologic processes, including cell growth, differentiation, apoptosis, fetal development, transformation, inflammation, and immune response. The intent of this review is to provide a brief synopsis of the role of STAT activation in signal transduction, the structure of STAT proteins, mechanisms of aberrant signal transduction, and the role of STAT proteins in normal and malignant hematopoiesis. The review focuses in particular on the role of STAT activation in leukemogenesis.
STATs in signal transduction
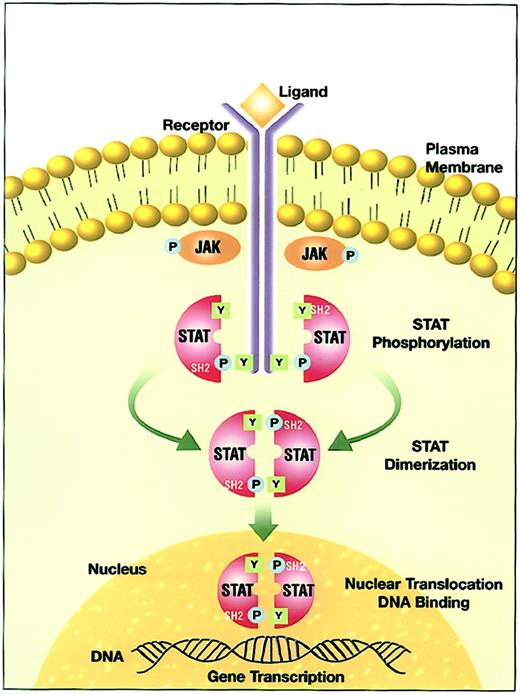
The interaction of a cytokine with its ligand-binding receptor α subunit is the first step in the formation of a signaling-competent receptor complex. This process involves the oligomerization of the ligand-bound subunit with either another subunit or a separate, signal-transducing β subunit.8 9 This oligomerization initiates the process of signal transduction by activation of the receptor-associated Janus family tyrosine kinases (JAKs) through cross-phosphorylation (Figure 1). Immediate targets of the activated JAKs are the cytoplasmic portions of the receptors and receptor-associated proteins. The tyrosine phosphorylated sites become docking elements for Src homology 2 (SH2)– and phosphotyrosyl-binding domain-containing proteins present in the membrane or the cytoplasmic compartment. Prominent among these are the STATs. Receptor-recruited STATs are phosphorylated on a single tyrosine residue in the carboxy terminal portion. The modified STATs are released from the cytoplasmic region of the receptor subunits to form homodimers or heterodimers through reciprocal interaction between the phosphotyrosine of one STAT and the SH2 domain of another. Following dimerization, STATs rapidly translocate to the nucleus and interact with specific regulatory elements to induce target gene transcription.
JAK-STAT signal transduction pathway.
Ligand-induced receptor oligomerization activates JAKs that subsequently phosphorylate tyrosine residues on the cytoplasmic portion of the receptor. The quiescent STAT monomers are then recruited to the activated receptor complex via the interaction of the SH2 domains with phosphotyrosine docking sites. STATs are phosphorylated by the JAKs on a conserved tyrosine residue in the c-terminal domain to form STAT homodimers or heterodimers. STATs dissociate from the receptor after the dimerization and translocate into the nucleus. In the nucleus, STATs bind to specific response elements and induce gene transcription.
JAK-STAT signal transduction pathway.
Ligand-induced receptor oligomerization activates JAKs that subsequently phosphorylate tyrosine residues on the cytoplasmic portion of the receptor. The quiescent STAT monomers are then recruited to the activated receptor complex via the interaction of the SH2 domains with phosphotyrosine docking sites. STATs are phosphorylated by the JAKs on a conserved tyrosine residue in the c-terminal domain to form STAT homodimers or heterodimers. STATs dissociate from the receptor after the dimerization and translocate into the nucleus. In the nucleus, STATs bind to specific response elements and induce gene transcription.
STAT family members and chromosomal localization
STAT proteins were originally discovered in interferon (IFN)–regulated gene transcription in the early 1990s.10-12 Since then, a number of cytokines have been recognized to activate various STAT proteins (Table1). Seven members of the STAT family of transcription factors have been identified in mammalian cells: STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6.
Phenotypes of STAT knock-out mice
| STAT . | Cytokine affected . | Knockout phenotype . |
|---|---|---|
| STAT1 | IFN-α/β, γ | Defective IFN-dependent immune responses |
| High susceptibility to bacterial/viral infections13 14 | ||
| STAT2 | IFN-α/β | Defective type I IFN-dependent immune responses15 |
| STAT3 | IL-2, IL-6, IL-7, IL-9, IL-10, IL-11, IL-15, IL-21, EGF, OSM, G-CSF*, TPO, LIF, GH | Early fetal death16 17 Impaired T-cell proliferation in response to IL-618 and IL-219† |
| Impaired IL-10–mediated anti-inflammatory responses20‡ | ||
| Defective wound healing in skin211-153 | ||
| Delayed involution of mammary gland after weaning221-155 | ||
| STAT4 | IL-12 | Impaired Th1 cell development23 24 |
| STAT5a and/or STAT5b | IL-2*, IL-3, IL-5, IL-7, IL-9, IL-15, G-CSF, GM-CSF, EPO, TPO, GH, PRL | Loss of mammary gland development and lactogenesis251-154 Loss of sexually dimorphic growth in males261-154 |
| Defective granulocyte proliferation in response to GM-CSF27# | ||
| Impaired cell growth in response to IL-228 291-160 | ||
| Defective natural killer (NK) cell development291-160 | ||
| Infertility in females301-164 | ||
| Fetal anemia311-164 | ||
| Reduced number of NK cells and impaired IL-2–induced T-cell proliferation321-164 | ||
| STAT6 | IL-4, IL-13 | Impaired Th2 differentiation, defective IgE class switch33-36 |
| STAT . | Cytokine affected . | Knockout phenotype . |
|---|---|---|
| STAT1 | IFN-α/β, γ | Defective IFN-dependent immune responses |
| High susceptibility to bacterial/viral infections13 14 | ||
| STAT2 | IFN-α/β | Defective type I IFN-dependent immune responses15 |
| STAT3 | IL-2, IL-6, IL-7, IL-9, IL-10, IL-11, IL-15, IL-21, EGF, OSM, G-CSF*, TPO, LIF, GH | Early fetal death16 17 Impaired T-cell proliferation in response to IL-618 and IL-219† |
| Impaired IL-10–mediated anti-inflammatory responses20‡ | ||
| Defective wound healing in skin211-153 | ||
| Delayed involution of mammary gland after weaning221-155 | ||
| STAT4 | IL-12 | Impaired Th1 cell development23 24 |
| STAT5a and/or STAT5b | IL-2*, IL-3, IL-5, IL-7, IL-9, IL-15, G-CSF, GM-CSF, EPO, TPO, GH, PRL | Loss of mammary gland development and lactogenesis251-154 Loss of sexually dimorphic growth in males261-154 |
| Defective granulocyte proliferation in response to GM-CSF27# | ||
| Impaired cell growth in response to IL-228 291-160 | ||
| Defective natural killer (NK) cell development291-160 | ||
| Infertility in females301-164 | ||
| Fetal anemia311-164 | ||
| Reduced number of NK cells and impaired IL-2–induced T-cell proliferation321-164 | ||
| STAT6 | IL-4, IL-13 | Impaired Th2 differentiation, defective IgE class switch33-36 |
IFN indicates interferon; IL, interleukin; OSM, oncostatin M; G, granulocyte; GM, granulocyte/macrophage; CSF, colony-stimulating factor; TPO, thrombopoietin; LIF, leukemia inhibitory factor; GH, growth hormone; and PRL, prolactin.
More prominent.
T-cell–selective STAT3 knockout mice.
Macrophage-selective STAT3 knockout mice.
Keratinocyte-selective STAT3 knockout mice.
Mammary gland epithelium-selective STAT3 knockout mice.
STAT5a only.
#STAT5b only.
More prominent in STAT5b.
STAT5a/STAT5b double knock out.
Convincing evidence from genetic mapping studies indicates a common ancestral origin that gave rise to 3 chromosomal clusters of STAT genes through a series of duplication processes (Table2).37
STAT chromosomal localization
| . | Chromosomal localization* . | |
|---|---|---|
| . | Murine . | Human . |
| STAT1 | 1 | 2q32.2 |
| STAT2 | 10 | 12q13.3 |
| STAT3 | 11 | 17q21.2 |
| STAT4 | 1 | 2q32.2 |
| STAT5a | 11 | 17q21.2 |
| STAT5b | 11 | 17q21.2 |
| STAT6 | 10 | 12q13.3 |
| . | Chromosomal localization* . | |
|---|---|---|
| . | Murine . | Human . |
| STAT1 | 1 | 2q32.2 |
| STAT2 | 10 | 12q13.3 |
| STAT3 | 11 | 17q21.2 |
| STAT4 | 1 | 2q32.2 |
| STAT5a | 11 | 17q21.2 |
| STAT5b | 11 | 17q21.2 |
| STAT6 | 10 | 12q13.3 |
The exact chromosomal localizations of the STAT genes in humans were identified in the sequencing of the human genome and can be found on the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/genome/guide/).
Structure of STATs
Previous characterization of the crystal structure of STAT molecules allowed a better understanding of the distinct functional domains within the STAT proteins.38 39 Several domains are conserved in all STAT family members (Figure 2; Table3).
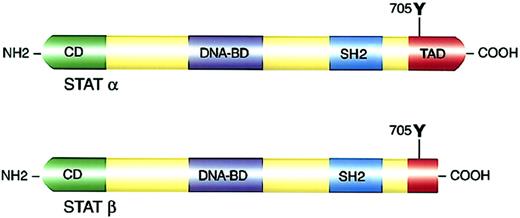
Structure and functional domains of STAT molecules.
(Top panel) Full-length STATα. (Bottom panel) c-Terminal transactivation domain truncation resulting in STATβ isoforms. NH2 indicates amino terminal; COOH, carboxyl terminal; CD, cooperative domain; DNA-BD, DNA binding domain; and TAD, transactivation domain.
Structure and functional domains of STAT molecules.
(Top panel) Full-length STATα. (Bottom panel) c-Terminal transactivation domain truncation resulting in STATβ isoforms. NH2 indicates amino terminal; COOH, carboxyl terminal; CD, cooperative domain; DNA-BD, DNA binding domain; and TAD, transactivation domain.
STATs structure
| Domain . | Role . | Reference . |
|---|---|---|
| Oligomerization domain | Interacts with other proteins and mediates oligomerization of STAT dimers to form tetramers. | 40,41 |
| DNA binding domain | Defines the DNA-binding specificity and mediates distinct signals for specific ligands. | 42-44 |
| SH2 domain | Mediates specific interactions between STAT-receptor, STAT-JAK, and STAT-STAT. | 45-49 |
| Carboxyl-terminal domain | Transcriptional activation domain that is thought to regulate the transcriptional activity of STATs and provide functional specificity. | 50-53 |
| Tyrosine residue | Phosphorylation site in the c-terminal domain approximately 700 residues from the N-terminus that regulates the DNA-binding activity of all STATs. On phosphorylation, mediates STAT dimerization by binding to the SH2 domain of the reciprocal STAT molecule. | 10,12 |
| Serine residue3-150 | A second phosphorylation site in the c-terminal domain.3-151 | 54,55 |
| Domain . | Role . | Reference . |
|---|---|---|
| Oligomerization domain | Interacts with other proteins and mediates oligomerization of STAT dimers to form tetramers. | 40,41 |
| DNA binding domain | Defines the DNA-binding specificity and mediates distinct signals for specific ligands. | 42-44 |
| SH2 domain | Mediates specific interactions between STAT-receptor, STAT-JAK, and STAT-STAT. | 45-49 |
| Carboxyl-terminal domain | Transcriptional activation domain that is thought to regulate the transcriptional activity of STATs and provide functional specificity. | 50-53 |
| Tyrosine residue | Phosphorylation site in the c-terminal domain approximately 700 residues from the N-terminus that regulates the DNA-binding activity of all STATs. On phosphorylation, mediates STAT dimerization by binding to the SH2 domain of the reciprocal STAT molecule. | 10,12 |
| Serine residue3-150 | A second phosphorylation site in the c-terminal domain.3-151 | 54,55 |
All except STAT2 and STAT6.
STAT isoforms
STAT isoforms lacking regions of the c-terminal domain have a competitive dominant-negative (DN) effect on gene induction mediated by the STAT pathway, counteracting the effects of the full-length isoform STATα.60-69 A representative example of the different STAT3 isoforms is described in Table 4. The transcriptional activities of the different isoforms are distinct, suggesting that the balance of these isoforms controls gene activation, leading to distinct biologic responses.
STAT3 isoforms
| . | Description . | Molecular weight (kDa) . |
|---|---|---|
| STAT3α | Full length | 92 |
| STAT3β | Missing c-terminal transactivation domain; functionally distinct, either dominant negative or altered binding | 83 |
| STAT3γ | Lacking tyrosine residue; can still be recruited to tyrosine-phosphorylated receptor proteins by the binding function of the remaining SH2 domain but signaling by the receptor complex terminates | 72 |
| STAT3δ | Unknown | 64 |
| . | Description . | Molecular weight (kDa) . |
|---|---|---|
| STAT3α | Full length | 92 |
| STAT3β | Missing c-terminal transactivation domain; functionally distinct, either dominant negative or altered binding | 83 |
| STAT3γ | Lacking tyrosine residue; can still be recruited to tyrosine-phosphorylated receptor proteins by the binding function of the remaining SH2 domain but signaling by the receptor complex terminates | 72 |
| STAT3δ | Unknown | 64 |
c-Terminally truncated STAT isoforms can be generated by 2 different mechanisms. The first mechanism is alternative mRNA splicing.10,60-68 Splicing joins the coding sequences (exons) by removing the intervening noncoding sequences (introns) from primary transcripts. Alternative splicing generates an enormous repertoire of functional diversity by producing multiple RNAs and proteins from a single gene. In the case of STAT3β, alternative splicing results in truncation of 55 amino acids from the c-terminal of STAT3α and gain of a unique 7-amino acid sequence.60-63STAT3β lacks the Ser727 phosphorylation site, which is proposed to enhance STAT3α DNA binding. The second mechanism that produces c-terminally truncated STAT isoforms is proteolytic processing.69-73 The transcriptional activation domain is truncated to form STAT3β without any amino acid gain. Notably, the proteolytic activity has only been identified in myeloid cell lines and not in cells of lymphoid lineage.
Knock-out mouse models
Studies of targeted deletion of STAT genes in mice have provided insight into the roles of STAT proteins in response to various cytokines in vital biologic functions (Table1).13-36,74-76 Models demonstrated the relevance of the STAT isoforms as mediators of cytokine receptor-specific signaling reactions, eg, STAT1 for IFN-γ,13,14 STAT2 for the immune response associated with type I IFNs,15 STAT3 for early development,16,17 STAT4 for IL-12 action,23,24 STAT5 for mammary gland function,25,26 and STAT6 for IL-4–dependent response.34-36 The details of knock-out mouse model studies are beyond the scope of this review. Of importance is that targeted deletion of STAT did not result in cellular transformation or development of leukemia in any of the knock-out models.76
STATs in myeloid differentiation
The G-CSF and IL-6 family of cytokines, which share gp130 as a common signal transducing subunit, and the GM-CSF and IL-3 family of cytokines are the main cytokines involved in myeloid differentiation. Other hematopoietic growth factors are also implicated to a lesser degree. STAT3 and STAT5 are the major STAT family members governing signal transduction in growth factor–regulated control of myelopoiesis.77,78 Studies with STAT null-mutant mice showed no role for STAT1, STAT4, or STAT6.74
The critical role of STAT3 in myeloid differentiation has been demonstrated with the use of DN mutants.79-81 STAT3 activation by the gp130 family of cytokines in M1 murine myeloid leukemia cells is associated with growth arrest and morphologic differentiation, and blocking IL-6– and LIF-induced activation of STAT3 in DN STAT3 mutants defective in either the tyrosine phosphorylation site (Y705F; STAT3F) or the DNA binding site (EE434-435AA; STAT3D) results in maturation arrest.80-82These data suggest that STAT3 is necessary in gp130-mediated differentiation of myeloid lineage cells. In contrast, the amount of STAT3 protein decreases during differentiation of embryonic stem (ES) cells,83 and STAT3F84 or specific STAT3 antisense oligodeoxynucleotides (ODNs)85 promote differentiation and block self-renewal of ES cells. These contradictory data suggest that cytokines transmit specific signals to direct lineage commitment of pluripotent hematopoietic stem cells and that specific target genes are stimulated in different cells.
G-CSF–induced myeloid differentiation has been demonstrated to be mediated by STAT3 activation.79,86,87 DN STAT3 mutants, STAT3F and STAT3D, prevented G-CSF–induced granulocytic differentiation in murine myeloid LGM-1 cells, but cell proliferation was not impaired.79 These data suggest that STAT3 activation is crucial for G-CSF–induced differentiation but not growth. In a similar fashion, the introduction of DN STAT3 constructs, STAT3F and STAT3D, into mouse myeloid cell lines suppressed G-CSF–induced neutrophilic differentiation,87,88 most probably because of STAT3-mediated up-regulation of the cyclin-dependent kinase (cdk) inhibitor p27 (Kip1).88Other evidence of STAT3 involvement in G-CSF–induced myeloid differentiation is the identification of a novel STAT3 recruitment motif within the G-CSF receptor (G-CSFR).89 The cytoplasmic phosphotyrosine residues Y704 and Y744 of G-CSFR were demonstrated to mediate G-CSF–induced differentiation of M1 leukemia cells.90 These residues have been reported to function as docking sites for STAT3α and STAT3β. Phosphorylated Y704 and Y744 each recruit STAT3 directly, resulting in STAT3 activation.89 Together, these data support a critical role for STAT3 in G-CSF–induced terminal myeloid differentiation.
STAT5 has also been implicated in myeloid differentiation induced by IL-3, G-CSF, and GM-CSF.91-93 The detection of STAT5 mRNA by polymerase chain reaction (PCR) was suggested to represent an early marker of differentiation in ES cells.83 In addition, STAT5 activation has been shown to be necessary for G-CSF–induced myeloid differentiation.92 Ilaria et al92 generated both an NH2-terminal mutant STAT5a/WKR (W255KR→AAA) and c-terminally truncated STAT5a/<53C, lacking the last 53 amino acids of murine STAT5a, which is similar to a naturally occurring isoform of rat STAT5b.94 These DN STAT5 proteins inhibited G-CSF–induced neutrophilic maturation of murine myeloid 32D cells. However, in IL-3–dependent cell lines, the expression of c-terminally truncated DN STAT5 was shown to inhibit growth, suggesting that STAT5 is needed for proliferation.66,92 Likewise, IL-3–induced STAT5-dependent proliferation was suppressed by DN STAT5a mutants without inducing apoptosis.92 In summary, it seems that STAT5 has distinct roles in IL-3–dependent and IL-3–independent pathways.
The antiapoptotic activity of STAT5 was shown to be necessary during the terminal stages of myeloid differentiation.93 For example, primary chicken myeloblasts expressing DN STAT5 were not capable of generating mature neutrophils because of apoptosis, which was reversed by Bcl-2.93 Similarly, bone marrow myeloid cells from STAT5a/STAT5b–knock-out mice showed a differentiation defect and underwent apoptosis during GM-CSF–dependent maturation in vitro. The antiapoptotic protein Bcl-xL was induced in response to GM-CSF and IL-3 through a STAT5-dependent pathway, indicating that antiapoptotic effects of STAT5 are due to induction of the Bcl-x gene.93 These data suggest that STAT5 is required for granulocytic differentiation and has a permissive role in promoting survival and proliferation of differentiating myeloid progenitor cells.
The transcriptional activation domain of STAT proteins provides functional specificity, including commitment to myeloid differentiation.50-53 Therefore, the distinct transactivating capabilities of the α and β isoforms of STAT proteins are biologically significant and appear to affect the cell's fate. STAT3β is activated by signals leading to differentiation, whereas STAT3α is activated by signals leading to proliferation and transformation.61,87 For example, on G-CSF stimulation, STAT3β form is activated in normal human CD34+ bone marrow cells and in leukemic HL-60 cells capable of differentiation in response to G-CSF, whereas full-length STAT3α was the predominant form activated in acute myeloid leukemia (AML) cell lines that proliferate in response to G-CSF, but were unable to differentiate.61 Similarly, functionally distinct isoforms of STAT5 generated by proteolytic activity mediate different effects in mature and immature myeloid cells.67,69 Full-length STAT5a/STAT5b forms are activated in response to IL-3 in mature cells, whereas the c-terminally truncated β isoforms are prevalent in immature murine myeloid cell lines.67,69,95,96 The proteolytic activity that cleaves STAT5α to the STAT5β isoform was identified in murine cell lines representing immature stages in myeloid differentiation, suggesting that this activity may be involved in lineage-specific STAT5 signaling.69 A switch toward the full-length STAT5 isoforms was observed during myeloid differentiation of the murine hematopoietic cell line FdCP1 in response to GM-CSF, indicating the loss of STAT5 proteolytic activity.96Moreover, insertion of a noncleavable STAT5b mutant resulted in impaired differentiation in response to GM-CSF.96Conversely, another study showed enhanced expression of truncated STAT5 during the differentiation process, and IL-5 and GM-CSF were shown to activate the full-length STAT5 in immature human myeloid cells and the truncated STAT5 in mature cells.97 The reasons for these opposing findings are unclear. In summary, the differential activity of STAT isoforms may contribute to defining distinct biologic responses elicited by STAT-mediated gene induction.
STATs and oncogenesis
Dysregulation of STAT signaling pathways, particularly STAT3 and STAT5, has been demonstrated to contribute to malignant cellular transformation.98 99 STAT proteins are postulated to play important roles in oncogenesis by 2 distinct mechanisms: constitutive activity of the full-length molecule and expression of a c-terminally mutated one.
Constitutive activation of STAT1, STAT3, and STAT5 has been demonstrated to be associated with malignant transformation induced by various oncoproteins.59,99-102 Full-length STAT3 is constitutively activated in NIH3T3 fibroblast cell lines transformed by the oncogenic v-Src tyrosine kinase, and the level of constitutive STAT3 activity correlates directly with oncogenic transformation by Src.59,100-102 The transforming ability was suppressed by DN STAT3 mutants, including recombinantly generated STAT3F, STAT3D, STAT3S, and c-terminally truncated splice variant STAT3β isoform.59,102 Similarly, JAK1 and Src have been demonstrated to work together to activate STAT3 in transformed NIH3T3 cells,99 suggesting a model in which STAT3 is recruited to Src by JAK1, before phosphorylation and activation directly by Src.
Constitutively activated STAT may exert its transforming activity through the induction of an antiapoptotic pathway. Inhibition of the STAT3 pathway has been shown to induce apoptosis in breast cancer cell lines.103 Two members of the antiapoptotic Bcl-2 family, Bcl-xL and Mcl-1, were shown to be up-regulated in multiple myeloma cells in which constitutive STAT3 activity was induced by IL-6.104-107 In head and neck cancers, constitutive STAT3 activity with up-regulated epidermal growth factor receptor (EGFR) signaling plays an important role in malignant proliferation through a Bcl-x–induced antiapoptotic mechanism.108-110 These data suggest that constitutive STAT protein activity may induce antiapoptotic pathways in various malignancies. The candidate target genes regulated by the STAT pathways, such as c-myc,cyclin D1, and Bcl-x, appear to contribute to oncogenesis by inducing cell proliferation and survival through the control of cell cycle progression and/or prevention of apoptosis.
A genetically mutated STAT3 (STAT3-C) with the substitution of 2 cysteine residues within the c-terminal loop of the SH2 domain has been demonstrated to possess intrinsic oncogenic potential in the absence of tyrosine phosphorylation and to act as a transforming agent.111 This molecule is constitutively active, forms homodimers spontaneously, independently of tyrosine phosphorylation, migrates to the nucleus, binds to DNA, and induces transcription. At the molecular level, this mutant molecule up-regulates the expression of cyclin D1, Bcl-x, and c-myc. Transfection of STAT3-C into rodent fibroblasts also induces the formation of transformed colonies in soft agar and produces tumors in nude mice. These data suggest that altering the c-terminal domain of STAT3 induces constitutive activation. This observation provides further evidence that STAT3 activation may be oncogenic by itself and is not just a consequence of tyrosine phosphorylation.
Interaction of the STAT pathway with other signaling pathway(s) from the hematopoietic growth factor receptor, eg, the mitogen-activated protein (MAP) kinase pathway, may also play a role in oncogenic transformation.112,113 Activation of the MAP kinase has been demonstrated in AML and in multiple myeloma,112,113and some direct “cross-talk” may exist between these pathways.114 115
STATs in solid tumors
Constitutive activation of STAT proteins has been reported in a number of malignant cell lines and human cancers.98Although this review concentrates on leukemia, the large body of information on the role of STAT proteins in solid tumors will be briefly summarized.
The role of STAT molecules in breast cancer has been extensively studied.116 Constitutive activation of STAT3 and/or STAT1 has been detected in breast carcinoma cell lines103,117-119 and human breast carcinoma nuclear extracts,103,120 but not in cell lines derived from nonmalignant mammary gland epithelium103,118,119 or in cells from healthy human breast tissue.120 STAT3 activity correlates with elevated EGFR and Src expression and with their respective tyrosine kinase activities.98 Abrogation of constitutive STAT3 activity by DN STAT3 mutants induces apoptosis and growth arrest in breast cancer cell lines,103 suggesting a pivotal role for constitutively active STAT3 in breast cancer development, possibly via an aberrant EGFR pathway and/or Src kinase activity. Similarly, human head and neck squamous cell carcinomas (HNSCCs) also display constitutive STAT3 activity, which mediates activation of EGFR tyrosine kinase induced by transforming growth factor α (TGF-α).108-110,121 Down-regulation of STAT3 with DN variants or antisense plasmid gene therapy blocks malignant proliferation, decreases Bcl-xL expression, and induces apoptosis in HNSCCs.108-110 Constitutive activity of STAT5 has also been reported in HNSCCs, with predominance of STAT5b.122 Additionally, TGF-α–stimulated Erb-B-1/-2 complex was shown to activate STAT3 in non–small cell lung cancer.123
Src kinase–mediated activation of STAT3 has been shown to be essential in prostate and ovarian carcinomas.117 Interestingly, enhanced expression of breast cancer susceptibility gene 1(BRCA1) in prostate cancer cell lines was shown to induce constitutive tyrosine and serine phosphorylation of STAT3 and upstream activation of STAT3, JAK1, and JAK2.124 Additionally, autocrine stimulation by IL-6 induces prostate cancer cell growth accompanied by activation of STAT3.125 Likewise, IL-6 treatment of colorectal carcinoma cells induces activation of STAT1 and, to a lesser extent, STAT3.126 Finally, murine B16 melanoma cells display constitutive STAT3 activity with an unknown activating cytokine, and DN STAT3β inhibits activation of STAT3 and suppresses cell growth.127 The presence of STAT3β had no effect on normal fibroblasts, suggesting that only malignant cells are dependent on STAT3 activity for survival. Moreover, in vivo inhibition of activated STAT3 by DN STAT3β caused regression of tumor growth in a syngeneic mouse melanoma model system, providing the proof of principle that STAT molecules might be an appropriate target for anticancer therapy.127
STATs in nonleukemic hematopoietic malignancies
IL-6 signaling mediated by STAT3 transcriptional activity is the major pathway involved in growth and differentiation of B cells into malignant plasma cells.112,128 Indeed, STAT3 is constitutively active in human bone marrow mononuclear cells from patients with multiple myeloma and in the IL-6–dependent human myeloma cell line U266, which expresses high levels of the antiapoptotic protein Bcl-xL.104,105 IL-6–dependent constitutive STAT3 activity signaling confers resistance to apoptosis in U266 cells.104 Inhibition of STAT3 signaling by DN STAT3 or by AG490, an inhibitor of the JAK2 kinase, has been shown to block Bcl-xL expression, with subsequent induction of apoptosis.104 The expression of Mcl-1, another antiapoptotic protein, has been shown to be up-regulated by IL-6 in human myeloma cells through the STAT3 pathway.106Furthermore, the presence of IFN-α, like IL-6, was shown to enhance survival of human myeloma cells through STAT3-mediated up-regulation of the Mcl-1 protein.107 Finally, constitutive activity of STAT3 in murine plasmacytomas and hybridomas in the absence of exogenous growth factors was shown to be associated with the acquisition of an IL-6–independent phenotype.129 These data suggest a fundamental role for STAT3 in oncogenesis in plasma cell myeloma.
Constitutive activity of STAT3 and STAT5, but not STAT1, was demonstrated in the mouse T-cell lymphoma cell line, LSTRA, with overexpression of the Lck protein, a Src family tyrosine kinase.130 In addition, constitutive activity of STAT1 and STAT3 was reported to be related to the presence of Epstein-Barr virus (EBV) DNA in Cherry lymphoblastoid cells (LCL) and Burkitt lymphoma cells131; this activity was associated with IL-10 and Bcl-2 protein expression. Cells with no EBV or IL-10 expression did not have constitutive STAT activity.131Consistent with these results, endogenous IL-10 was shown to induce STAT3 activation in an acquired immunodeficiency syndrome (AIDS)–related Burkitt lymphoma cell line, 2F7, leading to the overexpression of the antiapoptotic protein Bcl-2.132Treatment with anti-CD20 monoclonal antibody, Rituximab, decreased transcription and production of IL-10, which disrupted IL-10 autocrine/paracrine loops, with consequent down-regulation of STAT3 binding activity and, in turn, decreased Bcl-2 expression.132 The significance of STAT3 activation in the apoptotic pathway has been further demonstrated in T-cell large granular lymphocyte (LGL) leukemia associated with antiapoptotic Mcl-1 overexpression.133 Inhibition of STAT3 signaling causes apoptosis of leukemic LGLs and reduced Mcl-1 expression. These results demonstrate that activated STAT3 has an antiapoptotic effect in tumor cells.
STAT3 and/or STAT5 are constitutively activated in human T-cell lymphotrophic virus type I (HTLV-I)–related adult T-cell leukemia/lymphoma134 and HTLV-I–transformed T cells in association with the acquisition of IL-2–independent growth.135,136 Constitutive STAT3 phosphorylation on Tyr705 was detected in self-renewing CD5+ murine B-1 lymphocytes.137 Similar to primary B-1 cells, nuclear extracts of CD5+ B-cell lymphoma cells have been shown to contain a constitutively active STAT3 that is phosphorylated on Tyr705 and Ser727.137 Suppression of STAT3 expression in these cells was associated with a block in the G1 phase of the cell cycle, indicating a role for STAT3 in growth and immunoglobulin production of B-cell lymphoma through control of cell cycle progression. STAT3 and STAT5 were also shown to be constitutively activated in cutaneous lymphomas, including cutaneous anaplastic large T-cell lymphoma,138 Sézary syndrome,138,139and mycosis fungoides.140,141 The abrogation of STAT3 signaling resulted in a decrease in antiapoptotic Bcl-2 protein and an increase in proapoptotic Bax protein, with subsequent induction of apoptosis in mycosis fungoides cells.141
Recently, constitutively activated STAT3 was identified in Hodgkin disease (HD) cell lines.142 Additionally, constitutive phosphorylation of STAT3 and STAT6 was demonstrated in Reed-Sternberg cells from patients with HD.143 STAT6 activation was due in part to IL-13 signaling in these cells, and abrogation of IL-13 signaling resulted in the inhibition of STAT6 phosphorylation and cellular proliferation, suggesting a vital role for STAT6-mediated IL-13 signaling in the development of HD.
STATs in leukemias
The role of aberrant STAT signaling and constitutive STAT activation in leukemias has been a recent focus of intensive research. A growing body of evidence indicates a fundamental causative role for dysregulated STAT signaling mechanisms in both acute and chronic leukemias.
The initial hypothesis implicating STAT activation in leukemogenesis stemmed from studies of the fruit flyDrosophila.144-146 The hopscotch (hop) locus encodes a Drosophila JAK homolog.147 A singleDrosophila STAT gene, called D-STAT,STAT92E, or Marelle, has been identified, which functions in the embryonic development of theDrosophila.148-150 In Drosophila melanogaster, the dominant temperature-sensitive gain-of-function hop JAK kinase mutations hopTum-1 and hopT42increase tyrosine kinase activity and cause clonal proliferation of plasmatocytes, similar to the clonal proliferation of leukemia cells.151-154 These mutations hyperphosphorylate and hyperactivate D-STAT when overexpressed in Drosophila melanogaster cells and lead to a leukemia-like phenotype. However, introducing a lack-of-function D-STAT mutation into these cells did not totally reverse the overproliferation and leukemia-like abnormalities,154 suggesting that hyperactivation of STAT alone is most probably not sufficient for proliferation and survival.
Acute leukemias
Leukemia cells and normal hematopoietic progenitors proliferate in the bone marrow stroma. Long-term bone marrow culture provides a means to examine the interplay between hematopoietic cells and bone marrow stroma.155 The adherent layer in such a system (equivalent to stromal cells) modulates hematopoiesis in vitro. Several groups, including ours, have shown that cultured stroma from a subset of patients with AML produces multiple cytokines.156 157 What needs to be determined is why leukemic cells have a different response to growth factors than normal hematopoietic cells residing in the same microenvironment. Reasons may include differences in the expression of receptor subunits, signal communicating proteins, or downstream target genes.
AML is characterized by maturation arrest of a malignant clone of myeloid cells. Growth factors and growth factor signaling pathways are likely to determine the proliferation and differentiation state of the leukemic blasts in vivo.158 Receptors for growth factors that signal through STAT proteins are present on AML blasts and most of them are known to have intact function.158 Because multipotential nonleukemic hematopoietic cells undergo differentiation, whereas leukemic cells maintain proliferation rather than differentiation, in response to growth factors, aberration of signaling pathways is suggested to contribute to leukemogenesis.
Constitutive activation of STATs has been demonstrated in leukemia cell lines159-163 and blasts from 22% to 100% of patients with AML by various groups.73,113,131,163-167Gouileux-Gruart et al164 found constitutive activation of STAT3 in peripheral blood (PB) cells from 5 patients with AML; constitutive activity of STAT5 was also present in 2 of the 5 patients, and STAT1 was activated in 1 patient. The same group also reported a study of 14 patients with AML; 10 patients (71%) exhibited constitutive STAT1 and STAT3 activity, and 1 patient had STAT5 activity in addition to STAT1 and STAT3.131 In another study, constitutive STAT1 activity was associated with IL-3–independent proliferation in 10 of 20 patients (50%) with AML.165 In a recent study, 18 of 26 (69%) patients with AML exhibited constitutive STAT5 activity.166 Interestingly, this activity was associated with Flt3 phosphorylation in 70% of the cases.166 Hayakawa et al113 found constitutive STAT3 activity in 17 of 23 (74%) and STAT5 activity in 40 of 50 (80%) bone marrow samples from patients with AML. Approximately half of the samples tested revealed activation of the MAP kinase pathway; however MAP kinase activation did not correlate with constitutive STAT3/STAT5 phosphorylation.
In an analysis of 36 pretreatment bone marrow samples from newly diagnosed adult patients with AML, we detected constitutive activation of STAT3 and STAT5 in 10 (28%) and 8 (22%) samples, respectively.73 Activation of both STAT3 and STAT5 was seen in 4 patients (11%). There was no STAT6 activation. In another study, we showed that constitutive STAT3 activity correlated with unfavorable treatment outcome.167 Disease-free survival was significantly shorter in patients with, as compared to without, constitutive STAT3 activity. This was the first demonstration of clinical significance of STAT proteins in any malignancy. It is yet unclear whether this adverse treatment outcome is associated with the presence of constitutive STAT activity itself or with a process that leads to constitutive STAT activity.
Production of hematopoietic cytokines by leukemic blasts, with autocrine/paracrine stimulation of the JAK/STAT pathway, might be a possible mechanism for constitutive STAT activity in AML in some cases.168 IL-6 secretion from the leukemic blasts has been shown to cause constitutive STAT3 activity,168 as was also seen in multiple myeloma.104 However, IL-6 has antiproliferative effects in AML.169 Therefore, the role of IL-6–induced STAT3 activation in leukemogenesis remains controversial.
Aberrant STAT activation may be associated with leukemic transformation by various oncoproteins.170 Several phosphotyrosine kinases, including Bcr-Abl and TEL-JAK2, have been shown to activate the STAT pathway in leukemias, without the need for receptor activation.171-174 However, involvement of these kinases in AML is rare. Aberrant regulation of apoptotic pathways may be another cause of leukemogenesis. Members of the antiapoptotic Bcl-2 family are up-regulated in various malignancies, including AML.105-107,110,175,176 However, there are no data suggesting a relationship between these antiapoptotic molecules and STAT activity in AML. Direct cross-talk between the STAT and MAP kinase pathways in AML has also been suggested to play a role in leukemogenesis.113 114
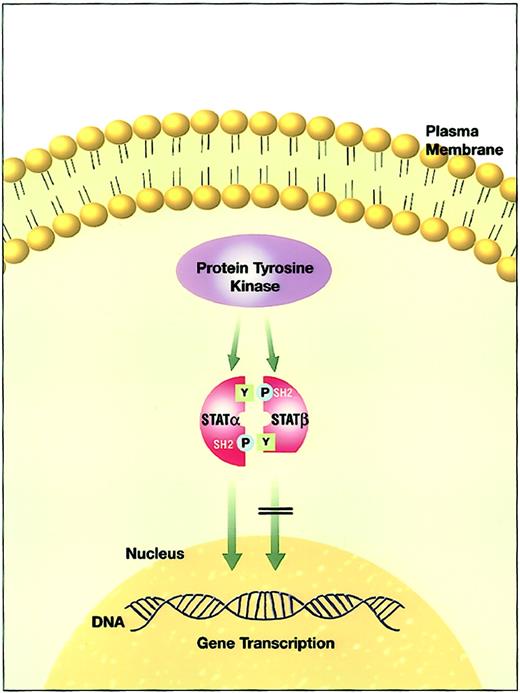
c-Terminally truncated STATβ isoforms have been found in the bone marrow of patients with AML and in several AML cell lines.61,72,73,162,163,167,177 Demonstration of constitutive STAT3 activation in genetically engineered c-terminal STAT3 mutants with resultant neoplastic transformation was the first suggestion that the c-terminal transactivation domain of STAT molecules might play a causative role in oncogenesis.111 In this context, STAT3β isoforms were proposed to play a role in leukemic transformation (Figure 3). We recently demonstrated that truncated STAT proteins are prevalent at relapse of AML and may be involved in disease progression.177Furthermore, the expression of truncated STAT3β isoform in leukemic cells with constitutive STAT3 activity identifies a group of patients with shorter disease-free survival and overall survival.167 It is unclear whether the presence of the STAT3β isoform functions simply to block STAT3 function or has a distinct transcriptional function. Finally, we showed that c-terminally truncated STAT3β isoforms in human AML blasts are generated by a novel serine-dependent proteolytic activity that is different from the activity in murine myeloid cell lines.72 This activity was capable of cleaving both STAT3 and STAT5, but not STAT6, into β isoforms in both the cytoplasm and the nucleus. The cleaved β isoforms retained their DNA binding activity. However, it is still not clear whether one protease or a family of proteolytic enzymes with different substrate specificities is responsible for the production of the STATβ isoforms. Novel therapies targeting the proteolytic activity might hold promise for the treatment of AML and are discussed in “STAT targeting.”
The effect of aberrant truncated STATβ isoform versus normal full-length STATα activity.
The effect of aberrant truncated STATβ isoform versus normal full-length STATα activity.
Constitutive STAT5 activity was recently shown to be associated with spontaneous Flt3 phosphorylation in the majority of AML cases,166 mostly because of mutations in the receptors. Moreover, constitutive STAT5 activity was associated with a low degree of apoptosis without modulation of Bcl-xL levels. Flt3 was suggested to contribute to the inhibition of apoptosis in AML blasts through STAT5 activation. These results are in accordance with previous studies demonstrating a causal relationship between Flt3 mutations and STAT5 activation, with resultant leukemogenesis.178 179
Recent demonstration of a possible role of constitutive STAT3 activity in up-regulation of vascular endothelial growth factor (VEGF) expression and tumor angiogenesis is particularly intriguing,180 especially in view of the evidence of increased angiogenesis in the bone marrow of patients with acute and chronic leukemias181-183 and the prognostic significance of elevated VEGF levels in these patients.184 185
STAT proteins are suggested to be involved in the pathogenesis of acute promyelocytic leukemia (APL).186-188 APL is the M3 subtype of AML in the French-American-British (FAB) classification.189 It is characterized by the reciprocal translocation t(15;17).190 The fusion of the promyelocytic leukemia (PML) gene on chromosome 15q22 with the retinoic acid receptor α (RARα) gene on chromosome 17q21 generates the PML-RARα oncogene. All-trans-retinoic acid (ATRA) directly targets the PML-RARα fusion protein and induces differentiation of leukemic blasts. Reciprocal translocation with 3 other partner genes(PLZF, NPM, and NuMA) also causes APL.190STAT5b was identified as a new gene fused to RARα in APL.186-188 STAT5b-RARα fusion protein results from an interstitial deletion within chromosome 17. Most recently, STAT5b-RARα was shown to block myeloid differentiation through its interaction with a corepressor complex with histone deacetylase activity.187,188 The coiled-coil domain of STAT5b was essential for dimerization of STAT5b-RARα fusion protein and inhibition of normal transcriptional activity via recruitment of the corepressor SMRT (silencing mediator for retinoid and thyroid hormone receptors). Furthermore, STAT5b-RARα and other APL fusion proteins augment STAT3 transcriptional activity.188 However, STAT1 and STAT2 were suggested to play key roles in ATRA-induced proliferation arrest and granulocytic differentiation. ATRA induces expression of the IFN-stimulated transcription factors STAT1, STAT2, and IFN-regulatory factor-1 (IRF-1) during myeloid differentiation.191-197 Additionally, DN STAT1 Y701F suppresses ATRA-induced morphologic differentiation.197 These results indicate that APL might result from aberrant regulation of the STAT3/STAT5 signal transduction pathway and that STAT1/STAT2 activation may be one of the mechanisms of ATRA-induced differentiation.
Studies of the role of STAT proteins in acute lymphoblastic leukemia (ALL) are much less extensive. Constitutive STAT1 activity was demonstrated in PB samples of 1 of 3 patients with ALL, whereas constitutive STAT5 activity was found in all 3 patients.164 Weber-Nordt et al reported constitutive STAT1 activity in only 1 of 24 patients with ALL and STAT5 activity in 15 patients (63%).131 None of the patient samples in either study exhibited constitutive STAT3 activity. The patterns of STAT activation are different in samples from ALL and AML patients; STAT3 activity is most prevalent in AML samples, whereas STAT5 activity is more common in ALL samples.
On the other hand, the t(9;12)(p24;p13) in patients with T-cell ALL, pre-B-cell ALL, and atypical chronic myeloid leukemia (CML) was found to generate the chimeric protein TEL-JAK2, with constitutive tyrosine kinase activity.173,198 This translocation results in fusion of the 3′ functional JH1 kinase domain of JAK2 to the 5′ pointed domain of translocated ets leukemia (TEL), a member of the ETS transcription factor family.173 The TEL-JAK2 fusion protein induced cytokine-independent proliferation in the IL-3–dependent Ba/F3 pre–B-cell line, associated with constitutive activity of STAT1, STAT3, and STAT5.173,174,199,200Constitutive STAT5 activity was also observed in Ba/F3 cell lines transfected with either TEL-Abl or TEL-PDGFβR.174 In addition to these findings in hematopoietic cell lines, TEL-JAK2 transgenic mice were shown to develop T-cell leukemia in association with constitutive STAT5 and STAT1 activity.201 Finally, activation of STAT5 was demonstrated to be essential for induction of myeloproliferative and lymphoproliferative disease by TEL-JAK2 in a murine bone marrow transplant model.202 Mice that received transplants of cells expressing a constitutively active mutant of STAT5a developed a fatal myeloproliferative disease. Furthermore, reconstitution with bone marrow derived from STAT5a/b-deficient mice expressing TEL-JAK2 failed to induce disease.202 SOCS-1, a member of the suppressor of cytokine signaling (SOCS) family of endogenous inhibitors of JAKs and STATs, has been demonstrated to inhibit TEL-JAK2–mediated transformation of Ba/F3 cells with impaired phosphorylation of STAT5.203 Similarly, constitutive expression of a DN form of STAT5a in Ba/F3 cells was shown to interfere with IL-3–independent cellular proliferation mediated by TEL-JAK2.174 These data indicate a cardinal role for STAT5 activity in TEL-JAK2–induced growth factor–independent hematopoietic transformation.
Chronic leukemias
CML is a clonal myeloid disorder characterized by the presence of the Philadelphia (Ph) chromosome, the product of a reciprocal translocation between the chromosomes 9 and 22, t(9;22)(q34;q11).204 This translocation generates theBcr-Abl gene, resulting from the juxtaposition of the c-abl tyrosine kinase locus on chromosome 9 with the breakpoint cluster region (bcr) on chromosome 22. Two different fusion proteins, p190 (190 kDa) and p210 (210 kDa), are produced, depending on the breakpoint site on the bcr gene. The p210 is responsible for CML, whereas p190 results almost exclusively in adult ALL (approximately 30% of patients) and, rarely, AML. The Bcr-Abl chimeric protein is a constitutively activated tyrosine kinase that causes growth factor–independent proliferation and transformation of hematopoietic cells. The JAK/STAT pathway is constitutively activated as a result of this chimeric oncoprotein.
Initial studies of constitutive STAT5 and STAT6 activity in Abelson murine leukemia virus–transformed pre-B cells suggested that activation of the JAK/STAT pathway is involved in oncogenic transformation induced by Abl oncogenes.205,206Pre–B-cell lines transformed with the temperature-sensitive mutant ofv-Abl exhibit DNA-binding activities similar to those of STAT5 and STAT6 at permissive temperatures.205 A shift to nonpermissive temperatures caused inactivation of v-Abl tyrosine kinase, resulting in abrogation of this STAT activation. Fibroblasts transformed by this oncoprotein, v-Abl, also exhibit constitutive STAT activities. However, STAT1 and STAT6, but not STAT5, are constitutively activated in fibroblasts, suggesting that the pattern of STAT activity is cell-lineage dependent.206
The observation that the v-Abl oncogene causes downstream constitutive STAT activity led to further studies in Ph-positive CML cell lines and patient samples.171,172,207-212Constitutive STAT5 activity was demonstrated in Bcr-Abl–positive CML and ALL cell lines, PB samples of patients with CML, and hematopoietic cell lines transfected in vitro with Bcr-Abl, leading to malignant transformation. Carlesso et al171 were the first to demonstrate constitutive STAT1 and STAT5 activity in human Ph chromosome–positive CML cell lines. No constitutive STAT activity was detected in any of the Bcr-Abl–negative cell lines. IL-3–dependent cell lines transfected with p210 Bcr-Abl displayed constitutive STAT1 and STAT5 activity, with resultant cytokine independence. Moreover, this STAT activation by Bcr-Abl was direct, without involvement of JAK kinases.171 Ilaria and Van Etten172 confirmed that Bcr-Abl directly activates specific STAT proteins with a low level of JAK tyrosine phosphorylation. Further, DN JAK mutants failed to block Bcr-Abl–induced STAT5 activation. STAT5 and, to a lesser extent, STAT1 and STAT3 were constitutively activated in p210- and p190-transformed Ba/F3 cells, rendering those cells IL-3 independent.172 Additionally, p190 induced strong STAT6 activity, in contrast to the p210 isoform. This finding has further significance because STAT6 is known to be activated by IL-4, a cytokine regulating Th2 T-cell development and function. This was the first demonstration of the effect of p190 on STAT activation. Frank and Varticovski207 subsequently extended these findings, demonstrating that phosphorylation of STAT1 and STAT5 was greater in Ba/F3 cells transfected with the p190 isoform than in cells transformed by p210 Bcr-Abl. It was suggested that the magnitude of the phosphorylation of STAT proteins by the p190 and p210 isoforms may be a determinant in the biologic effects of Bcr-Abl.
The essential role of STAT5 activation in Bcr-Abl–induced cell growth and transformation was further confirmed using DN STAT5 isoforms that inhibit Bcr-Abl–dependent STAT5 phosphorylation, with subsequent inhibition of gene transcription and cell growth.210,211 A direct physical interaction between Bcr-Abl protein and STAT activity was also proposed.207 A phosphorylated tyrosine residue, Tyr177 (Y177), in Bcr-Abl was shown to share homology with the tyrosine phosphorylation site of STAT1 and STAT5. Ba/F3 cells expressing the Y177F mutant had decreased STAT1 and STAT5 activity, suggesting that Bcr-Abl may interact with signaling pathways through this conserved site to confer growth independence.207 Furthermore, Nieborowska-Skorska et al212 showed that STAT5 activation by Bcr-Abl was dependent on the presence of functional SH2 and SH3 domains in the Bcr-Abl protein. Mutations of both the SH2 and SH3 domains completely abolished the ability of Bcr-Abl to activate STAT5.212 213 These studies provide further evidence that cellular transformation by Bcr-Abl requires STAT5 activity.
Studies investigating possible downstream targets of Bcr-Abl and STAT5 activity have concentrated on genes regulating survival. STAT5 has been demonstrated to play an important role in antiapoptotic activity mediated by Bcr-Abl.212,214-217 Abrogation of STAT5 activity by DN STAT5 mutants was shown to impair Bcr-Abl–dependent protection from apoptosis and leukemogenesis. Furthermore, a constitutively active STAT5 mutant restored antiapoptotic, proliferative, and leukemogenic properties in STAT5 activation–deficient Bcr-Abl mutants.212 Interestingly, the expression of Bcr-Abl in IL-3–dependent cell lines resulted in increased expression of the antiapoptotic Bcl-xL protein via STAT5 phosphorylation.214-217 Blockade of the Bcr-Abl kinase activity by the Bcr-Abl–tyrosine kinase inhibitor CGP 57148 (imatinib mesylate, STI-571, Gleevec) in Bcr-Abl–expressing cell lines and CD34+ cells from patients with CML suppressed STAT5 binding to the Bcl-x promoter, down-regulated the expression of Bcl-xL, and induced apoptosis.216Similarly, apoptosis mediated by imatinib mesylate was shown to correlate with inhibition of STAT5 activity and reduction in overexpressed Bcl-xL.217 Imatinib mesylate rendered the Bcr-Abl–expressing cells vulnerable to apoptosis, whereas Bcr-Abl–negative cells were not affected. These data suggest that STAT5 activity plays an important role in Bcr-Abl–induced resistance to apoptosis, with resultant uncontrolled cell proliferation and leukemogenesis.
In contrast to these findings, a study suggested that there is not a definitive requirement for STAT5 in Bcr-Abl–mediated transformation.218 Using mice lethally irradiated and reconstituted with Bcr-Abl–infected bone marrow cells deficient for STAT5a/5b, Sexl et al218 showed that Abl-induced and Bcr-Abl–induced transformation were independent of STAT5. STAT1 and STAT3 were not activated in STAT5a/5b-deficient cell lines. The presence of a redundant pathway to replace STAT5 activity could not be demonstrated.
Chronic myelomonocytic leukemia (CMML) is a clonal myeloproliferative disorder frequently associated with the chromosomal translocation t(5;12)(q33;p13), which results in TEL-PDGFβR tyrosine kinase fusion protein.219 STAT1 was shown to be activated in Ba/F3 cells transformed by TEL-PDGFβR.220 Interestingly, TEL-PDGFβR itself was suggested to be the kinase directly involved in tyrosine phosphorylation of STAT1. Recently, the same group extended their results to demonstrate that transformation by TEL-PDGFβR causes hyperphosphorylation of STAT5 on tyrosine residues.221However, full transformation of IL-3–dependent Ba/F3 cells by TEL-PDGFβR required engagement of a combination of signaling intermediates, phosphatidylinositol 3-kinase (PI3K) and phospholipase C-γ (PLCγ), as well as activation of STAT5, suggesting that constitutive activation of STAT5 by itself may not be sufficient for transformation.
STAT activity has also been investigated in CLL. CLL is characterized by slow proliferation of a malignant clone of differentiated mature B lymphocytes.222 Leukemic B lymphocytes from PB samples of 32 patients with CLL were shown to possess constitutive serine, but not tyrosine, phosphorylation of STAT1 and STAT3, using specific antibodies against the phosphorylated Ser727 residues.223 STAT1 and STAT3 were studied because of their roles in IL-2 and IL-6 signaling pathways important for lymphocyte functions, and other STAT proteins were not studied. The lack of tyrosine phosphorylation was thought to correlate with the slow growth of CLL cells. It was proposed that serine phosphorylation may enhance the transcriptional signal physiologically induced by STAT activation in response to hematopoietic cytokines, leading to gradual accumulation of malignant B lymphocytes.224 225 However, the significance of this finding in CLL pathobiology remains undetermined.
STAT targeting
In light of previous developments suggesting that aberrant STAT signaling contributes to malignant transformation, targeting STAT signaling appears to be an attractive approach to inhibiting leukemogenesis.226 227 A number of strategies are being developed to design specific inhibitors that disrupt STAT signaling.
Targeting of cytokine receptors with monoclonal antibodies or receptor antagonists is one possible strategy. Because the autocrine and paracrine activation of cytokine receptors has been reported to play a role in inappropriate STAT activation leading to oncogenesis, blocking these loops might prove beneficial in the treatment of leukemias. The IL-6 superantagonist Sant7 is known to be a potent inducer of apoptosis in multiple myeloma cell lines.228 Inhibition of IL-6 receptor signaling by Sant7 was shown to block constitutive STAT3 activity and to inhibit cell growth in U266 myeloma cells.105 Moreover, the feasibility of this approach was demonstrated with the successful use of monoclonal antibodies raised against IL-6 in a patient with plasma cell leukemia.229However, this strategy remains to be implemented in leukemias.
Inhibition of specific kinases is another approach to disrupting STAT activation. Because tyrosine kinase phosphorylation is critical for STAT activation, kinase inhibitors have become the focus of intensive investigation. Specific inhibition of JAK2 activity by a tryphostin family tyrosine kinase blocker, AG490, was shown to block the in vitro growth of ALL cells by inducing apoptosis.230AG490 inhibited leukemic cell infiltration in vivo in an ALL mouse model, with no deleterious effects on mouse hematopoiesis.231 In U266 myeloma cells and mycosis fungoides cell lines, constitutive STAT3-DNA binding activity was inhibited by AG490, resulting in growth arrest.105,140Furthermore, AG490 was shown to inhibit STAT3-mediated antiapoptotic Bcl-xL expression and, therefore, promote apoptosis in U266 cells.105 Similarly, AG490 inhibits Mcl-1 and induces apoptosis in LGL leukemia, with a corresponding decrease in STAT3-DNA binding activity.133 AG490 and another tyrosine kinase inhibitor selective for Src, PD180970, have been shown to inhibit constitutive STAT3 activity, resulting in growth suppression and apoptosis in breast cancer cell lines.104 Moreover, the Bcr-Abl–tyrosine kinase inhibitor imatinib mesylate inhibits the growth of cells expressing the Bcr-Abl, TEL-Abl, and TEL-PDGFR fusion proteins,221 all known to transmit signals through the STAT5 pathway.174 As direct corroborative evidence, blockade of Bcr-Abl kinase activity by imatinib mesylate was demonstrated to induce apoptosis of Ph+ cell lines and CD34+ cells from patients with CML by suppressing the STAT5-dependent expression of Bcl-xL antiapoptotic protein,216 as described earlier in “Chronic leukemias.” Finally, because phosphorylation of the Ser727 residue plays an important role in the STAT signaling pathway in addition to tyrosine phosphorylation, inhibition of serine kinase activation might be another rational therapeutic intervention.58 59
Negative regulation of cytokine signaling by inducibly expressed endogenous proteins is being actively explored as another strategy to block the STAT signaling pathway.114,232-236 SOCS family of proteins (SOCS1-SOCS7 and cytokine-inducible SH2-containing [CIS] protein) negatively modulates STAT signaling by directly binding to JAKs to inhibit tyrosine kinase activity.234,235 SOCS proteins also function by competitive blocking of STAT binding to phosphotyrosine binding sites237 and ubiquitin-mediated proteasome-dependent degradation of the STATs.203,238 The protein inhibitors of activated STATs (PIAS) family are specific inhibitors of STAT proteins.114,236,239 PIAS1 and PIAS3 directly interact with STAT1 and STAT3, respectively, to specifically block STAT-mediated gene transcription through the inhibition of STAT-DNA binding activity.114,236 Additionally, a STAT3-interacting protein (StIP1) has been demonstrated to regulate STAT3-mediated cytokine signal transduction.240 StIP1 was postulated to be a scaffold protein that forms a STAT3-StIP1-JAK complex and enhances the functional interaction of JAKs and STAT3-JAK.240 StIP1 preferentially binds to unphosphorylated STAT3, and overexpression of StIP1 mutants blocks IL-6–induced STAT3 activation. Pharmacologically designed small molecule mimics of SOCS or PIAS proteins or StIP1 mutants to block STAT activity may offer benefit in the treatment of leukemias.
Protein tyrosine or serine phosphatases counteract the effects of kinases to dephosphorylate active STAT proteins. The protein tyrosine phosphatases SHP-1 and SHP-2 and serine/threonine phosphatase, PP2A, are known to be involved in the regulation of STAT1, STAT3, and STAT5 signaling.241-244 Specific customized compounds that induce phosphatase activities to down-regulate phosphorylated STATs may have potential as therapeutic agents.
Dimerization of STAT proteins in the cytoplasm by phosphotyrosine-SH2 interaction is a critical step in STAT activation and subsequent gene transcription. Ideal candidates to interfere with dimerization would be SH2-like peptides recognizing phosphotyrosine residues of the STATs or small molecule peptide mimetics with phosphotyrosine residues that specifically bind to SH2 sequence of STATs. Disruption of STAT3 dimerization by the SH2 domain–binding phosphotyrosyl peptide, PY*LKTK, was demonstrated to block STAT3-mediated DNA binding activity, gene regulation, and cell transformation in vitro and in vivo.245 Because STAT proteins are directly and selectively targeted, nonspecific side effects are theoretically expected to be much less than with other strategies that block STAT upstream signaling.
Intracellular depletion of STAT proteins by antisense ODNs represents another effective approach to direct interference with STAT signaling. STAT3 antisense ODNs were shown to specifically decrease STAT3 levels in ES cells, with resulting impaired LIF-dependent inhibition of differentiation.85 Antisense ODNs against STAT1 were very effective in reducing intracellular STAT1 levels in human liver fat-storing cells, with concomitant inhibition of PDGF- and EGF-induced mitogenesis.246 Similarly, targeting of STAT3 using antisense ODNs resulted in the inhibition of EGF-mediated cell growth in human squamous cell carcinoma cell lines.109,110,121This approach has also been proven useful in decreasing STAT3 activation in LGL leukemia cells and B-cell lymphoma cells, with a corresponding decrease in their proliferative capacity.133 137 These observations indicate that intracellular depletion of STATs by antisense ODNs is a promising therapeutic strategy that deserves further study.
Disruption of STAT-DNA binding is another potential strategy for intervention in the STAT signaling pathway. Intracellular delivery of short double-stranded DNA pieces (decoy ODNs) carrying the consensus STAT-binding sequences is being investigated as an approach to manipulating gene expression.247 Decoy ODNs have been shown to prevent binding of STAT proteins to the promoter regions of targeted genes and interfere with cellular mitogenesis and T-cell development.248-250 Modulation of endogenous gene transcription by introduction of excess amounts of decoy ODNs into cells has the potential to disrupt STAT signaling.
Dominant-negative STAT isoforms have been used to inhibit STAT signaling pathways in several studies, with resultant loss of function.59-63,65,69,70,79,81,102,104,105,121,127Defective DN STAT mutants or DN STATs lacking the c-terminal domain retain the ability to be activated and form dimers with endogenous STATs. On the one hand, these molecules fail to transcribe signals and thus suppress STAT functions. The effectiveness of this strategy has been well established in in vitro and in vivo tumor models.59,102,104,105,121,127 On the other hand, on the basis of the observation that the c-terminal transactivation domain of STAT molecules might play a causative role in oncogenesis,103 constitutively active truncated STAT3β isoforms have been suggested to be involved in leukemic transformation.72,73,167,177 A novel serine-dependent proteolytic activity is responsible for the truncation of the STAT3 c-terminal domain in human AML blasts.72 The detailed characterization and cloning of the proteolytic activity, and the use of serine protease inhibitors (serpins)251 or subsequent design of custom-made targeted therapies to interfere with this activity, could have considerable potential in the treatment of leukemias.
Modulation of STAT activity by pharmacologic and biologic agents such as IFN-α and ATRA has been well documented. Although IFN-α induces the STAT signal transcription pathway, as explained earlier,10-12 chronic systemic administration of IFN-α has been reported to cause loss of constitutively active STAT1 and STAT3 DNA-binding abilities in precursor melanoma lesions, with associated STAT3 dephosphorylation.252 Interestingly, the loss of STAT proteins was suggested to be responsible for IFN-α resistance in cutaneous T-cell lymphoma and melanoma.253,254 These paradoxical observations remain to be explained. Additionally, STAT proteins were suggested to be involved in ATRA-induced growth inhibition and myeloid differentiation of APL cells.195,197 In myeloid leukemia cell lines, ATRA was shown to activate STAT1, STAT2, p48, and IRF-1 expression as essential molecules in IFN-α signal transduction.191-195,197Furthermore, ATRA induces IFN-α synthesis and potentiates the antiproliferative properties of IFN-α.196,255 ATRA was shown to restore IFN sensitivity by up-regulating STAT1 expression in IFN-resistant breast cancer cell lines.256 The cross-talk between retinoic acid and IFN signaling suggests a potentially useful synergistic combination in the treatment of leukemias. In addition to biologic agents, the cytotoxic agent fludarabine was shown to cause specific depletion of STAT1 mRNA and protein in lymphocytes and in CLL cells.257 These studies strongly suggest that modulation of STAT activity by biologic or pharmacologic agents represents an effective treatment strategy against leukemias.
Conclusion and future directions
In the past several years, compelling evidence has accumulated emphasizing the role of STAT proteins in leukemogenesis. Constitutive activation of STATs has now been clearly demonstrated in acute and chronic leukemias. c-Terminally truncated STATβ isoforms have also been detected in leukemic blasts from bone marrow of patients with AML. The constitutive activation of STAT3β isoform has been reported to be associated with poor outcome. Given that these molecules transduce a complex array of physiologic signals regulating fundamental cellular functions, including proliferation, differentiation, and programmed cell death, which are obviously perturbed in leukemias, inappropriate STAT signaling is not surprising. However, mechanisms of STAT activation and the significance of STAT activation in leukemic transformation still remain to be determined. Because leukemogenesis is a multistep process, STAT activation is probably not the only contributor. It is yet unclear whether constitutive STAT activity itself is the cause or the result of a transforming process.
The identification of the range of target genes turned on by STATs may provide important insights into the role of STAT signaling pathways in the development of leukemias. Understanding the molecular and biologic mechanisms by which aberrant STAT signaling is involved in cellular transformation is of paramount importance for the development of tailored therapeutic approaches to interrupting STAT signaling in leukemic blasts.
We thank Dr David A. Frank (Dana-Farber Cancer Institute, Boston, MA) for his critical review of the manuscript, and Sherilyn L. Smail and Benjamin D. Richey for preparation of the color figures.
Prepublished online as Blood First Edition Paper, December 12, 2002; DOI 10.1182/blood-2002-04-1204.
Supported partially by grants CA16056 and CA85580 from the National Cancer Institute. M.B. is a recipient of The Cancer and Leukemia Group B Clinical Research Award supported by Ortho Biotech, Inc.
References
Author notes
Meir Wetzler, Leukemia Section, Department of Medicine, Roswell Park Cancer Institute, Elm and Carlton St, Buffalo, NY 14263; e-mail:meir.wetzler@roswellpark.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal