We studied the genomic status of BCL6 in 23 cases of nodular lymphocyte predominance Hodgkin lymphoma (NLPHL) and 40 cases of classical Hodgkin lymphoma (cHL), using dual-color interphase fluorescence in situ hybridization (FISH). The BCL6rearrangement was identified in 48% of NLPHL cases and was not detected in cHL cases. As a confirmation, sequential or simultaneous immunohistochemistry (IHC) and FISH using CD20 or BCL6 antibodies and BCL6 DNA probes was performed in 8 NLPHL cases. The BCL6-associated translocations, t(3;22)(q27;q11), t(3;7)(q27;p12), and the most probable t(3;9)(q27;p13), were identified in 3 cases. A consistent expression of BCL6 protein in popcorn cells with the highest number of intensely stained cells in cases with a genomic BCL6rearrangement was shown by IHC. These findings support the hypothesis of a germinal center B cell–derived origin of NLPHL, indicate a significant role of BCL6 in the pathogenesis of NLPHL, and provide further evidence of the genetic diversity underlying the pathogenesis of NLPHL and cHL.
Introduction
On the basis of morphologic, immunohistochemical, and clinical features, 2 distinct biologic entities of Hodgkin lymphoma (HL) have been recognized: nodular lymphocyte predominance HL (NLPHL) and classical HL (cHL).1,2 Several lines of evidence indicate that the neoplastic cells of HL originate from a germinal center (GC) B cell that has been selected and stimulated by antigen.3-8 Accordingly, immunohistochemical studies showed that neoplastic cells of NLPHL (popcorn cells) were consistently associated with a BCL6+/CD138− phenotype typical of GC cells. The phenotype of Reed-Sternberg (RS) cells of cHL (mostly BCL6−/CD138+, rarely BCL6+/CD138−) suggested, however, that cHL is a heterogeneous entity comprising tumors of GC and post-GC B-cell origin.9-11
The BCL6 protein regarded as a GC marker12-14 is a multifunctional regulator of lymphocyte differentiation and immune response (for reviews, see Staudt et al15 and Ye16). The gene encoding BCL6 located at 3q27 is a frequent target of promiscuous chromosomal translocations found in approximately 1 of 6 malignant lymphomas, mainly of a diffuse large B-cell type.15,17-19 At the molecular level, allBCL6-associated translocations lead to up-regulation ofBCL6 transcription by a mechanism of promoter exchange,20 21 probably preventing down-regulation of its transcription that occurs upon plasmacytic differentiation.
Molecular mechanisms and genes that play key roles in the pathogenetic pathways underlying the neoplastic process in HL remain largely unknown. Following up on our recent finding ofBCL6 rearrangement in 2 NLPHL cases,22 we studied the genomic status of BCL6 in a series of cases diagnosed as NLPHL and cHL, using dual-color interphase fluorescence in situ hybridization (FISH).
Study design
Patient material
Twenty-three NLPHL and 40 cHL cases were retrieved from the files of the Department of Pathology, Catholic University of Leuven, Leuven, Belgium (12 NLPHL and 40 cHL cases), and the Department of Pathology, Stichting PAMM (Laboratorium voor Pathologische Anatomie en Medische Microbiologie), Eindhoven, the Netherlands (11 NLPHL cases). In 12 NLPHL cases (nos. 1-12) and all cHL cases, cytogenetic specimens stored at −20°C were available for FISH analysis. In the 11 NLPHL cases (nos. 13-23) not previously subjected to cytogenetic analysis, the interphase FISH was performed either on tissue imprints and/or single cells isolated from frozen tissue sections or on frozen tissue sections. In 2 cases (nos. 15 and 17), consecutive biopsies obtained at time of relapse were additionally examined (a indicates original samples; b, relapsed samples).
G-banding analysis
Cytogenetic analysis of 52 HL cases was performed according to standard methods. Chromosomal aberrations are presented in accordance with the International System for Human Cytogenetic Nomenclature.23
FISH analysis
FISH analysis of the BCL6 gene was performed with the Locus Specific Identifiers (LSI) BCL6 Dual Color, Break Apart Rearrangement Probe (Vysis, Bergisch-Gladbach, Germany) according to the manufacturer's instructions. The FISH data were collected on a Leica DMRB fluorescence microscope (Leica, Wetzlar, Germany) equipped with a cooled black-and-white charged-couple-device camera (Photometrics, Tuscon, AZ) run by Quips SmartCapture FISH Imaging Software (Vysis).
Immunohistochemistry (IHC)
Immunostaining for BCL6 was performed on formalin-fixed, paraffin-embedded sections with the UltraVision detection system (Lab Vision, Fremont, CA) and the BCL6 antibody (Ab-1, mouse monoclonal antibody Clone P1F6, Neomarkers, Fremont, CA; and PG-Bp6, DAKO, Glostrub, Denmark).
Results of the interphase FISH analysis were validated by sequential IHC and FISH analysis or combined immunophenotyping and interphase cytogenetics (FICTION) analysis applied on frozen tissue sections (10 μm) from, respectively, 4 (nos. 1, 3-5) and 4 (nos. 13-16) NLPHL cases. In the first approach, CD20 (L26; DAKO) immunostained frozen tissue sections were reviewed and popcorn cells were localized and recorded. Subsequently, these sections were subjected to BCL6-FISH and the previously recorded cells were evaluated. FICTION with CD20 and LSI BCL6 was performed according to the previously described protocol.24
Results and discussion
Initially, metaphase and interphase cells from 2 previously reported NLPHL cases22 with structural aberrations of 3q27 identified as the t(3;22)(q27;q11) (case 1) and the complex der 3 t(3;?;1) (3pter→3q27::?::1q21→ter) (case 2) were reanalyzed with LSI BCL6 (Table1). This double-color FISH assay applied in case 1 confirmed a rearrangement of BCL6(BCL6-R; split of green and orange signals) due to the t(3;22)(q27;q11), which is a known immunoglobulin λ (Ig λ)–mediated BCL6 variant translocation.25The hybridization pattern in case 2 (2 green signals on the der 3 and an orange signal on the del 7(p12)), shown in Figure1D pointed to BCL6-R due to the cryptic t(3;7)(q27;p12) associated with an additional duplication of the rearranged 3′end of BCL6 and a 1q translocation. It is noteworthy that an analogous t(3;7) resulting in the Ikaros-BCL6 rearrangement was recently described in diffuse large B-cell lymphoma (DLBCL).26 27
Cytogenetic and FISH findings in 11 NLPHL cases with the 3q27/BCL6 rearrangement
| Case (LN sample) . | Status . | Cytogenetics/3q27 abnormalities . | Interphase FISH patterns found with LSI BCL6 (no. of nuclei) . | Interpretations of FISH results . | Confirmation of BCL6-R . | |
|---|---|---|---|---|---|---|
| Ploidy* . | Status of 3q27/BCL6 . | |||||
| 1 | D | t(3;22)(q27;q11) | 1F1G1O (10) | 2n | 1xchr3, t(3;22)(q27;q11) | IHC/FISH |
| (BCL6/BCL6) | ||||||
| 2(a) | D | der(3)t(3;?;1)(q27;?;q21) | ND | ND | ||
| 2(b) | R | der(3)t(3;?;1)(q27;?;q21) | 2F4G2O (6) | 4n† | 2xchr.3, 2xder(3)t(3;7;3;1)(3pter_q27::7p12→7pter:: 3q27::7p12::1q21→1qter), 2xder(7)t(3;7)(q27;p12)† | ND |
| 3 | D | NK | 1F1G1O (3) | 2n | 1xchr3, t(3;?)(q27;?) | IHC/FISH |
| 2F2G2O (3) | 4n | 2xchr3, 2x t(3;?)(q27;?)† | (CD20/BCL6) | |||
| (BCL6/BCL6) | ||||||
| 4 | D | NK | 1F1G1O (18) | 2n | 1xchr.3, t(3;?)(q27;?) | IHC/FISH |
| 2F1G1O (3) | 3n | 2xchr.3, t(3;?)(q27;?) | (CD20/BCL6) | |||
| 2F2G2O (1) | 4n | 2xchr.3, 2x t(3;?)(q27;?) | ||||
| 5F1G1O (1) | 6n | 5xch.3, t(3;?)(q27;?) | ||||
| 6F2G2O (2) | 8n | 6xch.3, 2x t(3;?)(q27;?) | ||||
| 5 | D | NM | 1F1G1O (4) | 2n | 1xchr.3, t(3;?)(q27;?) | IHC/FISH |
| 2F1G1O (1) | 3n | 2xchr.3, t(3;?)(q27;?) | (CD20/BCL6) | |||
| 2F2G2O (6) | 4n | 2xchr.3, 2x t(3;?)(q27;?) | ||||
| 6 | D | NM | 2F1O (6) | 2n | 2xchr.3, der(?)t(3;?)(q27;?)† | ND |
| 13 | D | ND | 1F2G2O (1) | 3n | 1xchr.3, 2xt(3;?)(q27;?) | FICTION |
| 2F1G1O (4) | 3n | 2xchr.3, t(3;?)(q27;?) | (CD20 +BCL6) | |||
| 3F1G1O (2) | 4n | 3xchr.3, t(3;?)(q27;?) | ||||
| 14 | D | ND | 1F2G1O (6) | 3n | 1xchr.3, t(3;?)(q27;?), der(3) t(3;?)(q27;?) | FICTION |
| 2F4G2O (1) | 6n | 2xchr.3, 2x t(3;?)(q27;?), 2xder(3) | (CD20 + BCL6) | |||
| t(3;?)(q27;?) | ||||||
| 15(a) | D | ND | 2F1G1O (10) | 3n | 2xchr.3, t(3;?)(q27;?) | FICTION |
| (CD20 +BCL6) | ||||||
| 15(b) | R | ND | 2F1G (2) | 3n | 2xchr.3, der(3) t(3;?)(q27;?) | FICTION |
| 3F1G (6) | 4n | 3xchr.3, der(3) t(3;?)(q27;?) | (CD20 + BCL6) | |||
| 16 | D | ND | 1F1G1O (12) | 2n | 1xchr.3, t(3;?)(q27;?) | FICTION |
| 1F2G2O (6) | 3n | 1xchr.3, 2x t(3;?)(q27;?) | (CD20 + BCL6) | |||
| 2F1G1O (2) | 3n | 2xchr.3, t(3;?)(q27;?) | ||||
| 3F1G1O (1) | 4n | 3xchr.3, t(3;?)(q27;?) | ||||
| 6F2G2O (2) | 8n | 6xchr.3, 2xt(3;?)(q27;?) | ||||
| 17(a) | D | ND | 2F1G (7) | 3n | 2xchr.3, der(3)t(3;?)(q27;?) | ND |
| 17(b) | R | ND | 2F1G (8) | 3n | 2xchr.3, der(3)t(3;?)(q27;?) | ND |
| 3F1G (3) | 4n | 3xchr.3, der(3)t(3;?)(q27;?) | ||||
| Case (LN sample) . | Status . | Cytogenetics/3q27 abnormalities . | Interphase FISH patterns found with LSI BCL6 (no. of nuclei) . | Interpretations of FISH results . | Confirmation of BCL6-R . | |
|---|---|---|---|---|---|---|
| Ploidy* . | Status of 3q27/BCL6 . | |||||
| 1 | D | t(3;22)(q27;q11) | 1F1G1O (10) | 2n | 1xchr3, t(3;22)(q27;q11) | IHC/FISH |
| (BCL6/BCL6) | ||||||
| 2(a) | D | der(3)t(3;?;1)(q27;?;q21) | ND | ND | ||
| 2(b) | R | der(3)t(3;?;1)(q27;?;q21) | 2F4G2O (6) | 4n† | 2xchr.3, 2xder(3)t(3;7;3;1)(3pter_q27::7p12→7pter:: 3q27::7p12::1q21→1qter), 2xder(7)t(3;7)(q27;p12)† | ND |
| 3 | D | NK | 1F1G1O (3) | 2n | 1xchr3, t(3;?)(q27;?) | IHC/FISH |
| 2F2G2O (3) | 4n | 2xchr3, 2x t(3;?)(q27;?)† | (CD20/BCL6) | |||
| (BCL6/BCL6) | ||||||
| 4 | D | NK | 1F1G1O (18) | 2n | 1xchr.3, t(3;?)(q27;?) | IHC/FISH |
| 2F1G1O (3) | 3n | 2xchr.3, t(3;?)(q27;?) | (CD20/BCL6) | |||
| 2F2G2O (1) | 4n | 2xchr.3, 2x t(3;?)(q27;?) | ||||
| 5F1G1O (1) | 6n | 5xch.3, t(3;?)(q27;?) | ||||
| 6F2G2O (2) | 8n | 6xch.3, 2x t(3;?)(q27;?) | ||||
| 5 | D | NM | 1F1G1O (4) | 2n | 1xchr.3, t(3;?)(q27;?) | IHC/FISH |
| 2F1G1O (1) | 3n | 2xchr.3, t(3;?)(q27;?) | (CD20/BCL6) | |||
| 2F2G2O (6) | 4n | 2xchr.3, 2x t(3;?)(q27;?) | ||||
| 6 | D | NM | 2F1O (6) | 2n | 2xchr.3, der(?)t(3;?)(q27;?)† | ND |
| 13 | D | ND | 1F2G2O (1) | 3n | 1xchr.3, 2xt(3;?)(q27;?) | FICTION |
| 2F1G1O (4) | 3n | 2xchr.3, t(3;?)(q27;?) | (CD20 +BCL6) | |||
| 3F1G1O (2) | 4n | 3xchr.3, t(3;?)(q27;?) | ||||
| 14 | D | ND | 1F2G1O (6) | 3n | 1xchr.3, t(3;?)(q27;?), der(3) t(3;?)(q27;?) | FICTION |
| 2F4G2O (1) | 6n | 2xchr.3, 2x t(3;?)(q27;?), 2xder(3) | (CD20 + BCL6) | |||
| t(3;?)(q27;?) | ||||||
| 15(a) | D | ND | 2F1G1O (10) | 3n | 2xchr.3, t(3;?)(q27;?) | FICTION |
| (CD20 +BCL6) | ||||||
| 15(b) | R | ND | 2F1G (2) | 3n | 2xchr.3, der(3) t(3;?)(q27;?) | FICTION |
| 3F1G (6) | 4n | 3xchr.3, der(3) t(3;?)(q27;?) | (CD20 + BCL6) | |||
| 16 | D | ND | 1F1G1O (12) | 2n | 1xchr.3, t(3;?)(q27;?) | FICTION |
| 1F2G2O (6) | 3n | 1xchr.3, 2x t(3;?)(q27;?) | (CD20 + BCL6) | |||
| 2F1G1O (2) | 3n | 2xchr.3, t(3;?)(q27;?) | ||||
| 3F1G1O (1) | 4n | 3xchr.3, t(3;?)(q27;?) | ||||
| 6F2G2O (2) | 8n | 6xchr.3, 2xt(3;?)(q27;?) | ||||
| 17(a) | D | ND | 2F1G (7) | 3n | 2xchr.3, der(3)t(3;?)(q27;?) | ND |
| 17(b) | R | ND | 2F1G (8) | 3n | 2xchr.3, der(3)t(3;?)(q27;?) | ND |
| 3F1G (3) | 4n | 3xchr.3, der(3)t(3;?)(q27;?) | ||||
LN indicates lymph node; D, at diagnosis; R, at relapse; NK, normal karyotype; NM, no mitosis; ND, not done; F, fused signal; G, green signal; and O, orange signal.
2n indicates diploid; 3n, triploid; 4n, tetraploid; 8n, octaploid.
Interpretation based on further FISH studies (I.W. et al, in preparation).
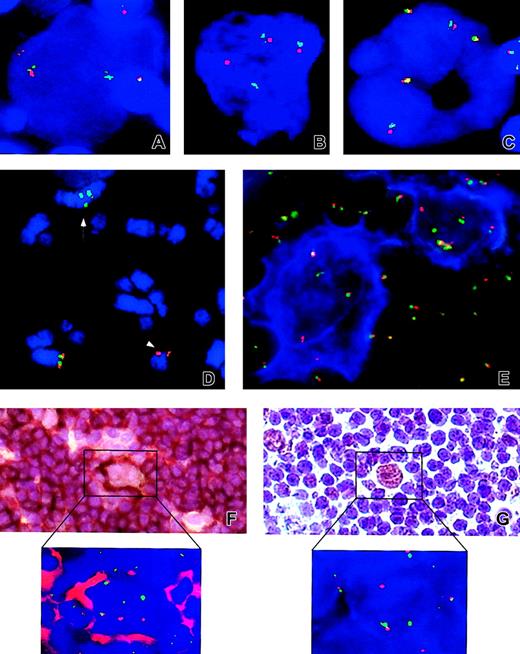
Examples of FISH and IHC results.
Analyzed cases: NLPHL cases no. 4 (A), no. 13 (B,E), no. 2 (D), no. 3 (F and inset), and no. 1 (G and inset) and classical HL case no. 24 (C). Applied probes: LSI BCL6 (A-E, F inset, G inset). (D) Arrow and arrowhead indicate the der(3) t(3;7;3;1) and the der(7)t(3;7) chromosomes, respectively. (E) An example of FICTION with CD20 (blue) and LSI BCL6 (F-G and insets of both panels). Examples of sequential IHC and FISH with CD20 and LSIBCL6, and anti-BCL6 and LSI BCL6, respectively. Note the split of LSI BCL6 signals in atypical cells from NLPHL cases (A-E,F inset, and G inset) and presence of only fused signals in the cHL case (C). Original magnifications × 630 (A-C,E), × 400 (F-G), and × 1000 (F-G insets).
Examples of FISH and IHC results.
Analyzed cases: NLPHL cases no. 4 (A), no. 13 (B,E), no. 2 (D), no. 3 (F and inset), and no. 1 (G and inset) and classical HL case no. 24 (C). Applied probes: LSI BCL6 (A-E, F inset, G inset). (D) Arrow and arrowhead indicate the der(3) t(3;7;3;1) and the der(7)t(3;7) chromosomes, respectively. (E) An example of FICTION with CD20 (blue) and LSI BCL6 (F-G and insets of both panels). Examples of sequential IHC and FISH with CD20 and LSIBCL6, and anti-BCL6 and LSI BCL6, respectively. Note the split of LSI BCL6 signals in atypical cells from NLPHL cases (A-E,F inset, and G inset) and presence of only fused signals in the cHL case (C). Original magnifications × 630 (A-C,E), × 400 (F-G), and × 1000 (F-G insets).
In the remaining 21 NLPHL cases and 40 cHL cases without available chromosome spreads, LSI BCL6 FISH studies were performed on an interphase cell level. Only large atypical nuclei were evaluated. The number of atypical cells analyzed in each specimen ranged from 6 to 35, with an average of 11 and 16 in NLPHL and cHL samples, respectively.
FISH patterns indicative of the BCL6 rearrangement were found in 9 NLPHL cases, which together with the 2 previously presented cases indicates an occurrence of BCL6-R in 48% (11 of 23) of NLPHL cases. Results of interphase FISH showed that atypical cells identified in cases with BCL6-R displayed different configurations of fused (germ line BCL6) as well as split green and orange signals (rearranged BCL6; Table 1). Balanced green and orange signals indicative of a reciprocalBCL6 translocation were found in 7 samples (cases 1, 3-5, 13, 15a, and 16; Figure 1A-B,E,F inset,G inset). In 6 samples (cases 2, 6, 14, 15b, 17a, and 17b), we observed either loss of the orange or green signal, suggesting a nonreciprocal translocation, or presence of an extra green signal, likely due to a duplication of the der 3 or the 3′ end of BCL6, as detected in case 2 (Figure 1D). Most analyzed cases showed a cell-to-cell variation of the number of signals, most probably reflecting different ploidy levels (ranging from 2n to 8n). Of interest, 2 patients (cases 3 and 6) later developed aBCL6-associated DLBCL. In case 3, a t(3;9)(q27;p13) was identified on metaphases of DLBCL and was retrospectively shown by FISH to be present in NLPHL (I.W. et al, in preparation).
Large atypical cells from the remaining 12 NLPHL cases showed the presence of numerous fused FISH signals indicative of a BCL6germ line configuration (data not shown). The number of signals per cell varied and ranged from 2 to 12, but in 6 and 2 cases predominant cell clones with 3 and 4 fusion signals, respectively, were evident.
The validity of the applied approach (FISH analysis of selected large atypical nuclei) was confirmed by performing either a sequential IHC and FISH with CD20 and LSI BCL6 (on samples 1 and 3-5) or FICTION (on samples 13, 14, 15a, 15b, and 16) on frozen tissue sections from 9 BCL6-R NLPHL cases, respectively. In each of these cases, 4 to 25 informative popcorn cells selected by morphologic criteria were evaluated, and all of them showed fused as well as split green and orange signals (examples are shown in Figure 1E-F,F inset). The reliability of our results is also supported by a finding of a similar incidence of BCL6-R in NLPHL cases coming from 2 different centers (Leuven, 6 of 12; Eindhoven, 5 of 11).
To check whether genomic rearrangements of BCL6 were reflected at the protein level, we analyzed 11 available NLPHL cases, using immunostaining with anti-BCL6 antibody. Assessment of BCL6 immunostaining was carried out blindly in 100 popcorn cells per case without knowledge of the BCL6 gene status. Popcorn cells showed expression of BCL6 in all cases, but the number of positive cells as well as the intensity of immunostaining varied. The percentages of BCL6+ cells ranged from 40% to 97% and were assigned to the following categories: less than 50%, 50% to 75%, and more than 75%. These categories comprised 2, 2, and 7 cases, respectively (Table 2). Of note, the highest percentages of BCL6-positive popcorn cells (ranging from 87% to 97%) and the strongest BCL6 immunostaining were observed in 5 NLPHL cases with a rearrangement of BCL6. These findings are illustrated by a sequential anti-BCL6/BCL6–FISH analysis additionally performed in cases 1 (Figure 1G,G inset) and 3. Taken together, our observations confirmed previous findings of a common expression of BCL6 in popcorn cells9 10 and, in addition, showed that at least in some NLPHL cases a strong expression of BCL6 may result from a genomic rearrangement of the BCL6 gene affected by chromosomal translocations.
Results of the BCL6-immunostaining performed in 11 NLPHL cases
| BCL6+ popcorn cells, % . | No. of cases . | Intensity of immunostaining . | FISH status of BCL6 . |
|---|---|---|---|
| Less than 50 | 2 | Moderate | Germ line |
| 50-75 | 2 | Moderate | Germ line |
| More than 75 | 2 | Moderate to strong | Germ line |
| More than 75 | 5 | Strong | Rearranged |
| BCL6+ popcorn cells, % . | No. of cases . | Intensity of immunostaining . | FISH status of BCL6 . |
|---|---|---|---|
| Less than 50 | 2 | Moderate | Germ line |
| 50-75 | 2 | Moderate | Germ line |
| More than 75 | 2 | Moderate to strong | Germ line |
| More than 75 | 5 | Strong | Rearranged |
Comparison of BCL6 alterations in NLPHL with those reported for DLBCL showed striking similarities. Both lymphoma types showed a similar frequency of BCL6 rearrangements (more than 40% of cases), occurrence of the same type of translocations (eg, t(3;22), t(3;7), t(3;9)) involving either Ig or non-Ig loci, and a common expression of BCL6 protein. A relationship between the 2 lymphomas was illustrated in 2 patients (nos. 3 and 6), who presented consecutively NLPHL and DLBCL. These findings further support a GC B cell–derived origin of popcorn cells and indicate that genomic rearrangements ofBCL6 may play an important role in the initiation of NLPHL, just as they do in DLBCL.
The finding of frequent BCL6 rearrangement in NLPHL remains in contrast with results obtained in cHL. This latter group comprised 23 cases of nodular sclerosis (NS) HL and 17 cases of mixed-cellularity (MC) HL. Atypical cells from all 40 cases analyzed by interphase FISH showed the presence of numerous fused (but not split) LSI BCL6 signals, pointing to the germ line BCL6gene (an example is shown in Figure 1C). The difference in the frequency of BCL6 breaks in NLPHL versus cHL is highly significant (P < .001 by Fisher exact test). As in NLPHL, the number of signals per cell in cHL cases varied and ranged from 2 to 16, most likely reflecting the genomic instability of HL-related neoplastic cells, previously reported by other groups.24 28-31
In summary, we show here the frequent occurrence of BCL6rearrangement in NLPHL, which was detected by FISH in 11 (48%) of 23 analyzed cases, but not in NS and MC subtypes of cHL (0 of 40 cases;P < .001, Fisher exact test). These findings remain in agreement with immunohistochemical data showing consistent expression of BCL6 in NLPHL and lack of or low-level expression of BCL6 in RS cells9 10 and reinforce the concept of distinct, entity-related mechanisms of pathogenesis in these tumors.
We thank Ursula Pluys and Rens van Hezik for skillful technical assistance, Emina Torlakovic for double-checking the BCL6 immunostaining, and Rita Logist for editorial help. These research results are from the Interuniversity Poles of Attraction program initiated by the Belgian Prime Minister's Office, Science Policy Programming. All scientific responsibility is assumed by the authors.
Prepublished online as Blood First Edition Paper, September 12, 2002; DOI 10.1182/blood-2002-05-1592.
Supported by grant G025298 from the Fund for Scientific Research (FWO), Flanders, Belgium, and by the Interdisiziplinäres Zentrum für Klinische Krebsforschung (IZKF), Kiel, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anne Hagemeijer, Center for Human Genetics, University of Leuven, Campus Gasthuisberg O&N6, Herestraat 49, B-3000 Leuven, Belgium; e-mail:anne.hagemeijer@med.kuleuven.ac.be.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal