Abstract
The BCL6 gene encodes a POZ/Zinc-finger protein, which acts as a sequence-specific transcriptional repressor. It is expressed in B cells within the germinal centers (GC) and is required for GC formation. In ≈40% of diffuse large cell lymphomas (DLCL) and ≈14% of follicular lymphomas (FL), the BCL6 gene is rearranged by chromosomal translocations, which juxtapose heterologous promoters and 5′ untranslated sequences derived from other chromosomes to the BCL6 coding domain or by mutations in the 5′ regulatory region. To understand the functional consequence of the chromosomal translocations, we have studied the patterns of expression of the promoters found juxtaposed to BCL6 in DLCL and FL during B-lineage differentiation. Distinct heterologous 5′ untranslated regions (IGH, IGL, TTF) were identified fused to the BCL6 coding domain by analysis ofBCL6 cDNAs in two DLCL cases and one mixed follicular lymphoma (MxFL). These three sequences, as well as three other previously identified BCL6 fusion partners (IGHG3, BOB1,H4), were studied for their pattern of expression during B-lineage differentiation by Northern blot analysis of B-cell lines representative of the pre-B, B, immunoblast, and plasma cell stages. In contrast to BCL6, whose transcription is activated only in B cells within the GC, all of the other sequences displayed a broader pattern of expression ranging from constitutive expression throughout B-cell differentiation to persistent expression in immunoblasts and plasma cells. These results indicate that the expression ofBCL6 is deregulated as a consequence of fusion to heterologous promoter regions. The persistent expression of activated BCL6may contribute to lymphomagenesis by blocking B-cell differentiation within the GC.
TRANSLOCATIONS INVOLVING the chromosome band 3q27 are detected in ≈15% of B-cell lymphomas, especially diffuse large cell lymphomas (DLCL).1,2 A gene rearranged at this site, BCL6, has been isolated from 3q27 adjacent to the chromosomal breakpoint.3-9 The BCL6 gene encodes a 95 kD nuclear phosphoprotein with six C-terminal zinc-finger motifs and an N-terminal POZ/ZIN domain homologous to a family of zinc-finger proteins.10,11 In the B-cell differentiation pathway, BCL6 is expressed in mature B cells within germinal centers (GC), but not in immature B-cell precursors or differentiated plasma cells.12,13 The BCL6 protein functions as a DNA-binding transcription repressor involved in the control of germinal center formation and T-cell–dependent antigen response.14 15
Several studies have shown that BCL6 is rearranged in 30% to 40% of DLCL and 6% to 14% of follicular lymphomas (FL).9The translocation breakpoints lie within the 5′ flanking region spanning the promoter and the first noncoding exon or within the first intron. As a result, the 5′ regulatory region containing theBCL6 promoter sequence is either removed or truncated and causes the juxtaposition of BCL6 coding exons (2-10) downstream to the promoter derived from the reciprocal chromosomal partner.9
We and others have shown that translocations involving the chromosomal band 3q27 affect not only the IG gene loci, but also a variety of other loci, such as 1q21, 2q21, 4p11, 5p13, 9p13, 11q23, 12p11, 12q13, and 15q21, a phenomenon termed promiscuous translocation.16 The genes TTF,17BOB1,18 and H419 have recently been isolated and characterized from cases of non-Hodgkin's lymphoma (NHL) carrying t(3;4)(q27;p11), t(3;11)(q27;q23), and t(3;6)(q27;p21) translocations, respectively. Each of the translocations formed a fusion transcript with BCL6 by removing the BCL6 first exon. We have recently shown that in the DLCL cell line, Ly8, theIGHG3-BCL6 chimeric transcript was initiated from theIGHG3 germline transcript promoter (Iγ3), suggesting that the translocation alters BCL6 expression by promoter substitution.20
The analysis of regulation of expression of the promoters of the heterologous genes, which fuse to BCL6 during B-cell differentiation, can provide insights into the functional consequence of translocation. To investigate this, we studied the pattern of expression of a number of BCL6 fusion partner genes by Northern blot hybridization in a panel of cell lines representing various stages of B-lineage differentiation. This analysis showed that, in contrast toBCL6 whose expression is restricted to mature B cells displaying a GC phenotype, all of the fusion partners studied (IGH, IGL, IGHG3, TTF, BOB1, and H4) displayed a different pattern of regulation including a persistence of expression to the plasmacytoid stage. This finding suggests that, driven by the heterologous promoter, BCL6expression is inappropriately continued and may block B-cell differentiation.
MATERIALS AND METHODS
Tumor samples and cell lines.
Lymph node biopsies were obtained from patients undergoing diagnostic evaluation for B-cell lymphoma at the Memorial Sloan-Kettering Cancer Center (MSKCC). Biopsy samples were subjected to histologic, immunophenotypic, cytogenetic, and DNA rearrangement studies as described.21 Three tumors with 3q27 rearrangements were used in this study. Tumor 352 was a DLCL with a t(3;22)(q27;q11), tumor 1020 was a DLCL with a t(3;8;14)(q27;q24;q32) translocation, and tumor 1547 was a MxFL with a t(3;4)(q27;p11) translocation. All three tumors showed BCL6 rearrangements by Southern blot analysis. In addition, the following B-lineage cell lines, which correspond to different stages of B-cell development were also used in this study: 697 (pre-B cell), Ramos, Bjab, (mature B cell) CB33, RD (immunoblast), and U266, Skmm1, JJN3, XG-4, XG-7 and XG-10 (plasma cell).
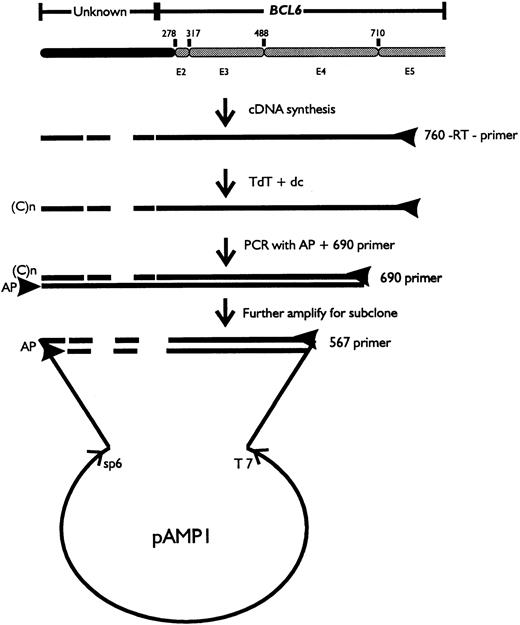
5′ rapid amplification of cDNA ends (RACE) assay and sequencing analysis.
Total RNA was extracted from biopsy samples using RNAgents kit (Promega, Madison, WI). RACE analysis of the 5′ end of BCL6 transcript was performed following the manufacturer's recommendations (GIBCO/BRL, Gaithersburg, MD). Briefly, first strand cDNA was synthesized by reverse transcription using the primer 760 from exon 5: (5′-GTTGAGGAACTCTTCAC-3′), followed by the addition of anchor primer to the 3′ end of the cDNA and polymerase chain reaction (PCR) using the anchor primer and the nest 1 primer 690 from the 3′ end of exon 4: (5′-CAAGTGTCCACAACATGC-3′) (Fig 1). For uracil DNA glycosylase (UDG) cloning, the PCR product was further amplified with the anchor primer and nest 2 primer 567 from the 5′ end of exon 4: (5′-CAUCAUCAUCAUAGGGTTGATCTCAGGATC-3′). The final PCR product was fractionated by agarose gel electrophoresis and the altered transcript was cut out from low-melting gel and purified using the Gene Clean Kit (Bio 101) followed by cloning into vector pAMP1 (GIBCO/BRL). DNA sequencing was performed by the dideoxy chain termination method using the Sequenase sequencing kit (USB, Cleveland, OH) or on the ABI373A DNA sequencer (Perkin Elmer, Applied Biosystem Division, Norwalk, CT).
Northern blot analysis.
A total of 10 μg of total cellular RNA extracted from each cell line was size fractionated on a 1% agarose/formaldehyde gel and transferred to a nylon membrane (Oncor, Gaithersburg, MD). Hybridization was performed in 50% formamide, 3X standard saline citrate (SSC), 10% dextran sulfate, 5X Denhardt's solution, and 0.5% sodium dodecyl sulfate (SDS) at 42°C for 17 hours. The filters were washed in 0.2X SSC, and 0.1% SDS at room temperature for 15 minutes and then at 55°C for another 20 minutes. Probes used in the Northern blot hybridization comprised a 2.3-kb EcoRI cDNA fragment covering exons 3 to 7 of BCL6, a 5.5-kbBamHI/HindIII fragment containing the J region of IGH (provided by J.V. Ravetch, Rockefeller University, New York, NY), an 8-kb BamHI fragment of theIGHG3, which is in the same transcript unit withthe I region of IGHG3 and a probe, pcλ, covering the constant region of the IGL (provided by I.R. Kirsch, National Cancer Institute, Bethesda, MD), a 1.1-kb PCR fragment flanking the first exon and the coding region of TTF(Emb Z35225), a 1.2-kb PCR fragment covering the exons 1 to 5 ofBOB1 (Emb Z49194) and a 1.1-kb PCR fragment flanking the leading sequence and the coding region of H4.
RESULTS
Identification of genes fused to BCL6.
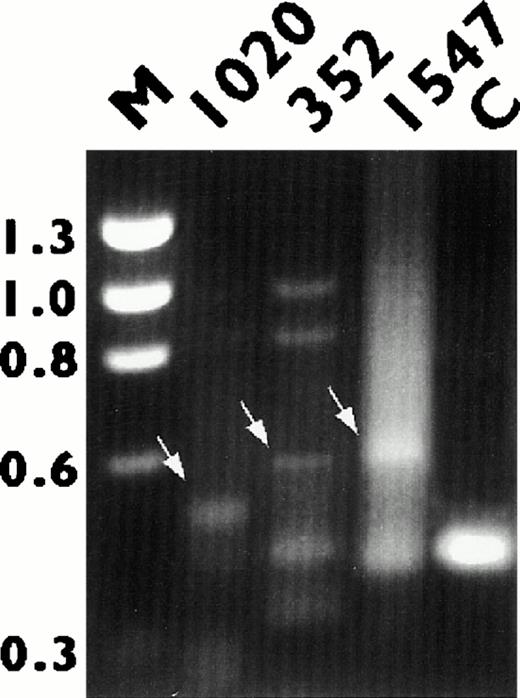
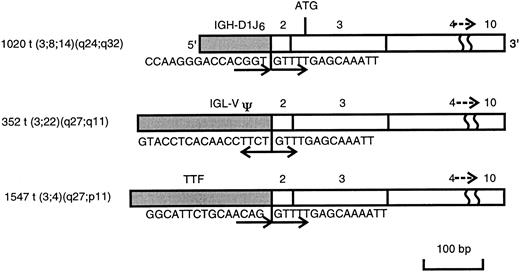
RNAs from three NHL biopsies (352, 1020, and 1547), which carried chromosomal translocations involving 3q27 and which showed BCL6rearrangements by Southern blot analysis, were used in this study. 5′ RACE and electrophoresis analysis showed an ≈400 bp PCR fragment in all cases, which corresponded to the germ line BCL6transcript (Fig 2). Extra PCR bands ranging in size from 500 to 700 kb, possibly representing chimeric or alternatively spliced transcripts were also noted in all three cases (Fig 2). The altered PCR fragments were eluted from the gel, subcloned, and sequenced. Homology search showed that the sequences fused to the 5′ end of BCL6 were identical to the IGH (1020),IGL (352), and TTF (1547) genes, respectively. As shown in Fig 3, 100 bp of sequence derived from the D1 and J6 regions of the IGHgene, 200 bp of the V pseudo region of IGL, and 240 bp of exon 1 of TTF gene were all spliced to the acceptor splice sites of exon 2 of BCL6, leading to complete removal of exon 1 of BCL6, where its promoter is located. The fusion transcripts were in the same transcriptional orientation, except IGL, where the IGLV pseudo region-BCL6 chimeric transcript was in a head-to-head orientation.
Detection of BCL6 fusion transcripts by 5′ RACE analysis. RNA derived from tumors 1020, 352, and 1547 were subjected to reverse transcriptase (RT)-PCR amplification as described in the text. Arrows point to bands representing BCL6 chimeric transcripts. RNA from tumor 1562 (C), which showed no BCL6rearrangement by Southern blot analysis, was used as the negative control. The fragment size of the marker ◊X174 RF DNA/HaeIII is indicated.
Detection of BCL6 fusion transcripts by 5′ RACE analysis. RNA derived from tumors 1020, 352, and 1547 were subjected to reverse transcriptase (RT)-PCR amplification as described in the text. Arrows point to bands representing BCL6 chimeric transcripts. RNA from tumor 1562 (C), which showed no BCL6rearrangement by Southern blot analysis, was used as the negative control. The fragment size of the marker ◊X174 RF DNA/HaeIII is indicated.
Schematic representation of BCL6 fusion transcripts obtained by 5′ RACE analysis on cases 1020, 352, and 1547. The BCL6 transcript is shown in the open box, and the transcript fused to the BCL6 is shown in the closed box. Sequence at the junction of each fusion transcript and its orientation are also shown.
Schematic representation of BCL6 fusion transcripts obtained by 5′ RACE analysis on cases 1020, 352, and 1547. The BCL6 transcript is shown in the open box, and the transcript fused to the BCL6 is shown in the closed box. Sequence at the junction of each fusion transcript and its orientation are also shown.
Expression of BCL6 and its fusion partner genes during the B-cell differentiation.
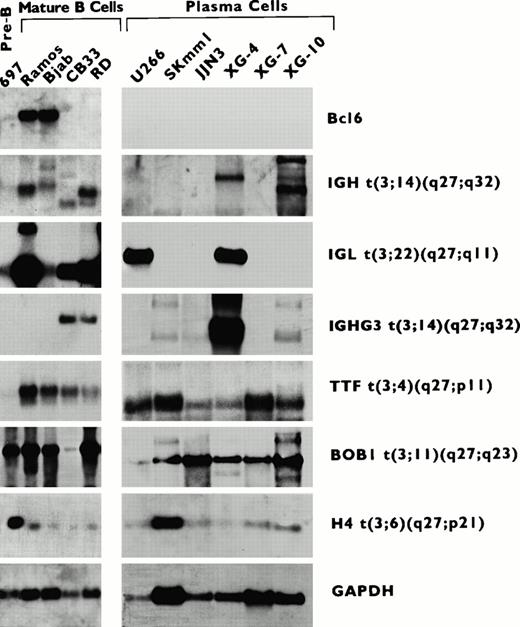
To compare the pattern of expression of BCL6 and its fusion partners during B-cell differentiation, RNAs from a panel of B-cell lines at different stages of differentiation were subjected to sequential Northern blot hybridization with probes derived from theBCL6 fusion partner sequences (IGH, IGL, andTTF) identified in this study. In addition, the expression ofBOB1 and H4 genes and the IGHG3 seqeunce, which have been previously identified to be fused to BCL6 in t(3;11)(q27;q23) and t(3;6) (q27;p21) and t(3;14)(q27;q32) translocations,18-20 also were studied. As shown in Fig 4 and Table1, BCL6 was expressed in cell lines, which corresponded to GC cells (Ramos, Bjab), but not in pre-B cells (697), immunoblasts (CD33, RD), or plasma cells (U266, Skmm1, JJN3, XG-4, XG-7, XG-10). In contrast, all of the BCL6 fusion sequences and genes studied displayed a different pattern of expression. BOB1 andH4 were expressed throughout B-cell differentiation. TTF was also expressed in all stages of B-cell differentiation, except in pre-B cells. IGH and IGL were expressed in pre-B and mature B cells; in addition, they showed persistent expression in both the immunoblatic cell lines. IGHG3 also was expressed in immunoblastic cells, although it was not expressed in pre-B cells and GC B cells. The expression of the IG gene promoters was variable in the plasmacytic cell lines; IGH was expressed in Skmm1, XG-4, and XG-10; IGHG3 was expressed in Skmm1, JJN3, XG-4, and XG-10; while IGL was expressed in U266 and XG-4. These results demonstrate that BCL6 and its fusion partner sequences and genes are regulated differently during B-cell differentiation.
RNA expression of BCL6 and its fusion partner genes in a series of B-cell lines at different stages of differentiation (top panel). Cell lines described in Table1. A mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was used to estimate the amount of blotted RNA. The genes that fused to BCL6 are labeled on the right. The different sizes of IGH, IGHG3, and IGL transcripts represent the products of physiological rearrangements of these genes.
RNA expression of BCL6 and its fusion partner genes in a series of B-cell lines at different stages of differentiation (top panel). Cell lines described in Table1. A mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was used to estimate the amount of blotted RNA. The genes that fused to BCL6 are labeled on the right. The different sizes of IGH, IGHG3, and IGL transcripts represent the products of physiological rearrangements of these genes.
Expression of BCL6 and Genes That Rearrange With It in Chromosomal Translocations in B-Cell Lines
| Cell Line . | Phenotype . | Gene Expression . | ||||||
|---|---|---|---|---|---|---|---|---|
| BCL6 . | IGH . | IGL . | IGHG3 . | TTF . | BOB1 . | H4 . | ||
| 697 | Pre-B (ALL) | − | −/+ | + | − | − | + | + |
| Ramos | B (sBL) | + | + | + | − | + | + | + |
| Bjab | B (eBL) | + | + | + | − | + | + | + |
| CB33(Kat) | Immunoblast (LCL) | − | + | + | + | + | + | + |
| RD(CMV) | Immunoblast (LCL) | − | + | + | + | + | + | + |
| U266 | Plasma cell (MM) | − | − | + | − | + | + | + |
| Skmm1 | Plasma cell (MM) | − | + | − | + | + | + | + |
| JJN3 | Plasma cell (MM) | − | − | − | + | + | + | + |
| XG-4 | Plasma cell (MM) | − | + | + | + | + | + | + |
| XG-7 | Plasma cell (MM) | − | − | − | − | + | + | + |
| XG-10 | Plasma cell (MM) | − | + | − | + | + | + | + |
| Cell Line . | Phenotype . | Gene Expression . | ||||||
|---|---|---|---|---|---|---|---|---|
| BCL6 . | IGH . | IGL . | IGHG3 . | TTF . | BOB1 . | H4 . | ||
| 697 | Pre-B (ALL) | − | −/+ | + | − | − | + | + |
| Ramos | B (sBL) | + | + | + | − | + | + | + |
| Bjab | B (eBL) | + | + | + | − | + | + | + |
| CB33(Kat) | Immunoblast (LCL) | − | + | + | + | + | + | + |
| RD(CMV) | Immunoblast (LCL) | − | + | + | + | + | + | + |
| U266 | Plasma cell (MM) | − | − | + | − | + | + | + |
| Skmm1 | Plasma cell (MM) | − | + | − | + | + | + | + |
| JJN3 | Plasma cell (MM) | − | − | − | + | + | + | + |
| XG-4 | Plasma cell (MM) | − | + | + | + | + | + | + |
| XG-7 | Plasma cell (MM) | − | − | − | − | + | + | + |
| XG-10 | Plasma cell (MM) | − | + | − | + | + | + | + |
Abbreviations: ALL, acute lymphoblastic leukemia; sBL, sporadic-type Burkitt's lymphoma; eBL, endemic-type Burkitt's lymphoma; LCL, EBV-immortalized lymphoblastoid cell line; MM, multiple myeloma.
DISCUSSION
Previous studies of BCL6 expression by immunohistochemical analysis showed it to be highly regulated during B-cell differentiation, being restricted to B cells in the GC, but not in pre-B or mature progenitor cells, including immunoblasts and plasma cells.12 As shown previously and now in this study, chromosomal translocations juxtapose heterologous promoters to theBCL6 coding domain, leading to its deregulated expression, probably shutting off its downregulation during B-cell differentiation. We have identified three heterologous sequences (IGH,IGL, and TTF) fused to BCL6 by translocation. These, as well as three previously identified BCL6 fusion partners (IGHG3, BOB1, and H4), were studied for their patterns of expression during B-cell differentiation by Northern blot analysis. As expected, BCL6 expression was restricted to GC-type B cells. In contrast, all of the BCL6 fusion partner genes displayed a different pattern of expression, including their persistent expression in immunoblasts and some, but not all, plasma cells. The absence of expression of IG genes in some of the plasma cells may be related to their state of differentiation. Although the pattern of expression of BCL6 fusion partners detected here in malignant cell lines may not reflect their physiologic pattern in normal cells, our results clearly show that the heterologous promoters display a regulatory pattern distinct from that of BCL6 in most B cells analyzed.
The results presented here indicate that promoter substitution by chromosome rearrangement leads to constitutive expression ofBCL6, dictated by the expression pattern of the new promoter. Consistent with these findings, recent immunohistochemical analysis of DLCL, Burkitt's lymphoma (BL), and FL biopsy samples also showed high levels of BCL6 protein,12 13 suggesting that its expression is constantly upregulated during B-cell lymphomagenesis. The pattern of expression of most of the promoters juxtaposed to BCL6 studied here, specifically the lack of expression of TTF andIGHG3 in the pre-B–cell line, suggest that expression during late stages of development may be critical for BCL6deregulation. However, additional studies of cells representative of the pre-B–cell stage are necessary to confirm this hypothesis.
A similar change in the pattern of expression during B-cell differentiation following chromosomal translocation was previously reported in the case of the BCL2 gene. This gene is normally expressed during pre-B–cell development, but is downregulated in association with B-cell maturation or to facilitate apoptosis. However, mature B-cell lymphomas with a t(14;18)(q32;q21) translocation express higher levels of BCL2 mRNA than normal mature B cells, indicating that fusion of the BCL2 gene with the IGgene leads to an inappropriately high level of BCL2-IG fusion transcript at the mature B-cell stage leading to abrogation of apoptosis signals and ultimately to lymphomagenesis.22 23
Recent analysis of BCL6 -/- mice showed that BCL6 is required for GC formation and a normal T-cell–dependent antigen response.15 B cells undergo a complex processing within the GC, which includes proliferation, hypermutation of the V region of IGH, and immunoglobulin isotype-switching.24During this process, B cells either differentiate to memory cells or plasma cells, or undergo apoptosis. Normal BCL6 downregulation in the late stage of B-cell differentiation is therefore important for terminal differentiation of B cells. As pointed out above, BCL6is specifically expressed in GC B cells and its expression is required for GC formation.12 15 Such constitutive expression by the activated BCL6 allele may enforce on the cell a GC phenotype, including constitutive proliferation, block of differentiation, or inhibition of apoptosis. The resulting block of normal downregulation of BCL6 may also expose the cells to genetic instability typical of GC leading to lymphoma development.
Supported by Grants No. CA-66999 (to R.S.K.C.) and CA-44029 (to R.D-F.) from the National Institutes of Health/National Cancer Institute, Bethesda, MD.
Address reprint requests to R.S.K. Chaganti, PhD, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10021.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal