Abstract
In the search for better protocols of preemptive therapy of human cytomegalovirus (HCMV) infection in hematopoietic stem cell transplant (HSCT) recipients, we conducted a randomized trial comparing antigenemia with the nucleic acid sequence–based assay (NASBA) for determination of HCMV immediate-early messenger RNA (IEmRNA) as the guiding assay for initiation of pre-emptive antiviral treatment. In the IEmRNA arm, antiviral therapy was started upon IEmRNA positivity confirmed the following day, whereas in the antigenemia arm, therapy was started in the presence of either at least 2 pp65-positive leukocytes/2 × 105 examined or a single positive leukocyte confirmed the following day. In both arms, treatment was stopped upon 2 consecutive negative results. All patients were monitored for 3 months after HSCT. The primary end point of the study was duration of anti-HCMV therapy. On the whole, 80 children (41 in the IEmRNA and 39 in the antigenemia arm), recipients of transplants from either a relative or an unrelated donor, completed the study. No patient developed HCMV disease. In the IEmRNA arm, the incidence of HCMV infection was higher compared to the antigenemia arm (80% vs 51%, respectively, P = .0069), as well as the percentage of treated patients (66% vs 44%, respectively, P = .045). However, the percentage of relapses and treated relapses was comparable in the 2 arms. There was no significant difference in median duration of therapy per patient. Although these data indicate that IEmRNA determination does not offer advantages in terms of treatment duration, it can safely replace antigenemia, while semiautomation is the major advantage of the NASBA procedure.
Introduction
Human cytomegalovirus (HCMV) infection still represents the most common and potentially severe viral complication in patients undergoing allogeneic hematopoietic stem cell transplantation (HSCT). In the 1980s, antiviral treatment was started only upon appearance of clinical symptoms of HCMV infection (deferred or symptomatic therapy), which led to a high incidence of fatalities.1 Subsequently, prevention of HCMV disease has become possible following adoption of 2 major approaches. The first is prophylaxis, that is, administration of anti-HCMV drugs to all transplant recipients for a median time of 3 months or 100 days after transplantation. Major drawbacks of such an approach are drug toxicity, occurrence of late (after discontinuation of prophylaxis) HCMV disease (mainly pneumonia), treatment of a substantial proportion of patients not at risk for disease, and low cost-effectiveness.2,3 The second approach is pre-emptive (or pre-symptomatic) therapy, that is, administration of antiviral drugs upon detection of HCMV in blood by any assay, to treat only patients undergoing viral infection and thus at risk of developing overt disease.2,4,5 Assays widely used for this purpose are antigenemia6-9 and DNAemia,4 with treatment starting either when becoming positive or upon reaching a predetermined viral load.10-13 Major drawbacks of this approach are the need for continuous monitoring of HCMV infection in blood and, sometimes, late appearance of HCMV disease. However, the cost of this approach is lower than that of prophylaxis14 and, to avoid delayed onset of disease, some authors suggest that prophylaxis be started following termination of pre-emptive therapy.4,5 A further benefit from the pre-emptive approach is the low-level expression of viral antigens, leading to immune stimulation at early times, which may be beneficial to subsequent recovery of specific T cells and long-term control.
While pre-emptive therapy for solid-organ transplant recipients is mostly started at predetermined levels of viral load in blood, defined either using antigenemia or DNAemia,13,15 pre-emptive therapy for HSCT recipients is currently started upon first confirmed appearance of virus in blood, whatever assay is used. This different strategy has been determined basically by the higher risk of early HCMV-related interstitial pneumonia for HSCT recipients in the posttransplantation period.16 In this respect, pre-emptive therapy can become more effective by using an ultrasensitive diagnostic tool for monitoring HCMV infection in patients undergoing HSCT. Detection of viral transcripts and, in particular, HCMV immediate-early viral transcripts (IEmRNA) by the recently introduced nucleic acid sequence–based amplification assay (NASBA) is a direct marker of HCMV replication and today represents the most sensitive assay for early detection of HCMV infection.17
In a recent retrospective study of HSCT recipients, we found that antigenemia and DNAemia had lower sensitivity (59% and 68%, respectively) than NASBA for IEmRNA, median time to first HCMV detection being 38 days for IEmRNA by NASBA, 40 days for antigenemia, and 41 days for DNAemia, respectively.18 Since HCMV infection can be diagnosed earlier when using NASBA for IEmRNA, therapeutic intervention in the presence of a lower viral load appears possible. In view of this fact, we decided to conduct a prospective, randomized trial to investigate whether use of IEmRNA in place of antigenemia as a guiding parameter for pre-emptive therapy in HSCT recipients could lead to shorter duration of antiviral treatment.
Patients and methods
Patients
From March 2000 to March 2002 a total of 103 pediatric patients underwent an allogeneic HSCT from either an HLA-identical sibling or an unrelated donor (UD). Inclusion criteria for enrollment of patients were serologic evidence of past HCMV infection in either donor (D) or recipient (R), or both, and written informed consent. Exclusion criteria included pairs with both D and R serologic status negative for HCMV; T-cell depletion of the graft; and patients in critical clinical conditions suggesting a life expectancy shorter than 3 months. Twenty donor/recipient pairs were HCMV seronegative and thus not eligible for randomization, and 3 randomized patients (1 allocated in the IEmRNA arm) died in the first 4 weeks after transplantation due to causes not related to HCMV infection. Thus, 80 patients (41 in the IEmRNA arm and 39 in the antigenemia arm) completed the study. The characteristics of the 80 patients are reported in Table 1.
Characteristics of the 80 patients completing follow-up
. | No. of patients (%) according to randomization arm . | . | |
|---|---|---|---|
| Parameter . | IEmRNA n=41 . | Antigenemia n=39 . | |
| Median age (range) | 8 (1-23) | 8 (1-19) | |
| Sex (M/F) | 23/18 | 27/12 | |
| Diagnosis | |||
| AML | 8 (20) | 5 (13) | |
| ALL | 12 (29) | 13 (33) | |
| CML | 1 (2) | 0 | |
| JMML | 2 (5) | 2 (5) | |
| MDS | 5 (12) | 4 (10) | |
| Thalassemia | 6 (15) | 6 (15) | |
| NHL | 2 (5) | 0 | |
| Fanconi anemia | 1 (2) | 0 | |
| SAA | 2 (5) | 1 (3) | |
| DBA | 0 | 1 (3) | |
| Other* | 2 (5) | 7 (18) | |
| HCMV serology | |||
| R+/D- | 17 (41) | 10 (26) | |
| R-/D+ | 4 (10) | 7 (18) | |
| R+/D+ | 20 (49) | 22 (56) | |
| Donor type | |||
| Sibling | 23 (56) | 22 (56) | |
| UD | 18 (44) | 17 (44) | |
| Source of stem cell graft | |||
| Bone marrow | 33 (80) | 29 (74) | |
| Peripheral blood | 3 (7) | 6 (15) | |
| Cord blood | 5 (12) | 4 (12) | |
| Conditioning regimen | |||
| Chemotherapy based | 28 (68) | 27 (69) | |
| TBI based | 13 (32) | 13 (31) | |
| Grades II-IV GVHD | 16 (39) | 12 (31) | |
. | No. of patients (%) according to randomization arm . | . | |
|---|---|---|---|
| Parameter . | IEmRNA n=41 . | Antigenemia n=39 . | |
| Median age (range) | 8 (1-23) | 8 (1-19) | |
| Sex (M/F) | 23/18 | 27/12 | |
| Diagnosis | |||
| AML | 8 (20) | 5 (13) | |
| ALL | 12 (29) | 13 (33) | |
| CML | 1 (2) | 0 | |
| JMML | 2 (5) | 2 (5) | |
| MDS | 5 (12) | 4 (10) | |
| Thalassemia | 6 (15) | 6 (15) | |
| NHL | 2 (5) | 0 | |
| Fanconi anemia | 1 (2) | 0 | |
| SAA | 2 (5) | 1 (3) | |
| DBA | 0 | 1 (3) | |
| Other* | 2 (5) | 7 (18) | |
| HCMV serology | |||
| R+/D- | 17 (41) | 10 (26) | |
| R-/D+ | 4 (10) | 7 (18) | |
| R+/D+ | 20 (49) | 22 (56) | |
| Donor type | |||
| Sibling | 23 (56) | 22 (56) | |
| UD | 18 (44) | 17 (44) | |
| Source of stem cell graft | |||
| Bone marrow | 33 (80) | 29 (74) | |
| Peripheral blood | 3 (7) | 6 (15) | |
| Cord blood | 5 (12) | 4 (12) | |
| Conditioning regimen | |||
| Chemotherapy based | 28 (68) | 27 (69) | |
| TBI based | 13 (32) | 13 (31) | |
| Grades II-IV GVHD | 16 (39) | 12 (31) | |
AML indicates acute myeloid leukemia; ALL, acute lymphoblastic leukemia; CML, chronic myeloid leukemia; JMML, juvenile myelo-monocytic leukemia; MDS, myelodysplastic syndrome; NHL, non-Hodgkin lymphoma; SAA, severe aplastic anemia; DBA, Diamond-Blackfan anemia; R, recipient; D, donor; UD, unrelated donor; TBI, total body irradiation.
Other includes 1 Hodgkin disease, 1 hemophagocytic lymphohistiocytosis, 1 chronic granulomatous disease, 2 sickle cell anemias, 1 Bernard-Soulier syndrome, 1 rhabdomyosarcoma, 1 Ewing sarcoma, and 1 mucopolysaccharidosis.
Among the 41 HSCT recipients enrolled in the NASBA arm, 18 (44%) received the allograft from a UD, whereas 23 (56%) received transplants from an HLA-identical sibling. Of the 39 patients included in the antigenemia arm, 17 (44%) underwent HSCT using a UD, and 22 (56%) received the graft from a sibling (Table 1).
Study design
A prospective, randomized, open-label trial aimed at determining the feasibility of pre-emptive therapy of HCMV infection in HSCT recipients, based on determination of IEmRNA in blood (IEmRNAemia) in comparison to pp65 antigenemia, was carried out following approval from the Policlinico San Matteo ethics committee. Patients' parents gave their written consent to randomization.
The primary end point of the study was to compare the duration of anti-HCMV treatment between the 2 arms. Secondary end points (useful for providing additional data to our present knowledge of the natural and drug-modified history of HCMV infection in HSCT recipients) were the incidence of HCMV infection as defined by the NASBA assay in the IEmRNA arm and by the antigenemia assay in the antigenemia arm; the number of treated patients; the incidence of relapse of infection; the number of patients treated for HCMV relapse; and, finally, the determination of quantitative HCMV DNAemia to investigate whether cutoff DNAemia levels could be proposed for future strategies of pre-emptive therapy.
Patient enrollment was calculated by a sample size evaluation method based on the Mann-Whitney U test. The minimum number of patients to be randomized was 50 per arm, based on a significance level of .05, a study power of 0.80, and hypothesizing a viral clearance of 9 ± 6 days for the antigenemia arm and 6 ± 5 days for the IEmRNA arm.18 An interim analysis was performed yearly to monitor the results of the trial. At the second year, conditional power analysis based on the duration of antiviral treatment in the 2 arms indicated a low probability (< 30% under the most extreme hypothesis) that the study would detect a significant difference if the target sample size (50 patients per arm) was reached. Patient accrual was then stopped.
HCMV infection was defined as active HCMV replication in blood in the absence of clinical manifestations or organ function abnormalities, whereas HCMV disease required documentation of HCMV infection (either by IEmRNAemia or antigenemia) together with clinical symptoms and/or organ function abnormalities.19
Patients were randomized for monitoring by either IEmRNAemia or antigenemia using a block random design for 2 groups on the basis of type of donor (HLA-identical sibling vs UD). In the IEmRNA arm, patients were treated after 2 consecutive IEmRNA-positive results, while treatment was stopped after 2 consecutive negative results. Relapse episodes were treated accordingly. In the antigenemia arm, patients were treated upon detection of either 2 or more pp65-positive leukocytes (see “Virologic follow-up”) or upon first confirmed positivity, when a single positive cell was detected, while therapy was stopped upon 2 consecutive negative results. Relapse episodes were treated similarly. Patients in the IEmRNA arm also were tested prospectively for antigenemia, as patients in the antigenemia arm were tested for IEmRNA, although these assays did not influence treatment, which was decided only on the basis of the test chosen by randomization. In addition, viremia was determined prospectively and DNAemia retrospectively on all blood samples, and results analyzed retrospectively. Monitoring of HCMV infection started when a patient's leukocyte count was higher than 0.5 × 109/L.
Prophylaxis for graft-versus-host disease (GVHD) consisted of cyclosporine-A (Cs-A) alone for patients receiving the allograft from an HLA-identical sibling, whereas patients who received transplants from a UD were given a short course of methotrexate (15 mg/m2 on day +1 and 10 mg/m2 on days +3, +6, and + 11 after transplantation) and antithymocyte globulin (ATG; 3.75 mg/kg/d from day –4 to day –2) in addition to Cs-A. Patients with acute GVHD were treated with steroids as first-line therapy, whereas patients with steroid-resistant disease were treated with extracorporeal photochemotherapy.20 As required, all patients enrolled in the study were given transfusions of leukocyte-depleted red blood cells, platelets, and frozen plasma from blood donors unscreened for HCMV serology. All children were given acyclovir for prophylaxis of herpes simplex virus reactivation at a dosage of 10 mg/kg 3 times a day, starting from day –1 until day +60.
Intravenous ganciclovir (5 mg/kg 2 times a day) was administered for pre-emptive therapy in both arms. Relapse episodes were treated similarly in both arms. In case of either drug toxicity (ie, profound myelosuppression) or the presence of increasing viremia levels in 2 subsequent controls, ganciclovir was replaced by foscarnet (90 mg/kg/2 times a day).
Virologic follow-up
All patients were monitored for HCMV infection until 3 months after HSCT. If patients still showed virologic signs of active HCMV infection after 3 months, follow-up was continued until disappearance of HCMV from blood. Subsequently, monitoring of HCMV infection was performed using antigenemia in all cases at time of the scheduled controls at 6, 9, and 12 months and in the presence of clinical signs or symptoms suggestive of HCMV disease. During follow-up, EDTA (ethylenediaminetetraacetic acid)–anticoagulated blood samples were collected twice a week (Mondays and Thursdays). The donor/recipient (D/R) serostatus was determined by enzyme-linked immunosorbent assay prior to transplantation, using previously reported methods.21
All patients were monitored for HCMV infection by prospective determination of IEmRNA in the NASBA arm and prospective quantitation of antigenemia in the antigenemia arm. Briefly, the qualitative NASBA assay for HCMV IEmRNA determination was performed on 10 μL whole blood, as previously described,18 by using the NucliSens Basic kit (bioMérieux, Boxtel, the Netherlands). Nucleic acids were isolated from 100 μL whole blood (NucliSens Automated Isolation reagents; bioMérieux) according to the method of Boom et al,22 and 1/10 of extracted nucleic acids was used in each NASBA reaction. IE1mRNA (from nucleotide [nt] 171796 to nt 172084) was amplified with a primer containing a T7 promoter and a reverse primer. Amplification products were detected by electrochemiluminescence with an IEmRNA-specific capture probe coupled to paramagnetic beads and a generic ruthenium-labeled detection probe hybridizing with a noncoupling tail of the reverse primer (primers and specific capture probes, not present in the Basic kit, were provided by the manufacturer). A positive and a negative control were included in each test run.
The antigenemia assay was quantitated under a fluorescence microscope by counting the number of pp65-positive/2 × 105 peripheral blood leukocytes (PBLs) examined on cytospin preparations stained with a pool of 3 pp65-specific monoclonal antibodies according to a previously reported23 and recently standardized24 procedure. Viremia was quantified by inoculating 2 × 105 PBLs into human embryonic lung fibroblast cell cultures by the shell vial technique and then, at 16 to 24 hours after inoculation, by counting the number of fibroblast nuclei positive for the HCMV IE antigen p72.25 DNAemia was quantitated by polymerase chain reaction using external standards26 and an internal amplification control.27 HCMV DNA copy numbers were determined in 1/10 volume of nucleic acid extracts (NucliSens Automated Isolation reagents; bioMérieux). This method allowed reproducible HCMV DNA quantification in the range of 101 to 104 genome equivalents (GEs)/10 μL whole blood.
Statistical analysis
Data were analyzed as of October 15, 2002.
Differences between medians were compared by using the Mann-Whitney U test. Differences in percentages were tested using the Pearson chi-square test, while the Fisher exact test was used to evaluate differences in percentages when the total sample size was fewer than 30. All tests were 2-tailed. Curves of number of HCMV-free patients according to antigenemia or IEmRNA assays in the posttransplantation period, as well as curves of number of treatment-free patients and duration of antiviral treatment in the 2 randomization arms, were determined by the Kaplan-Meier method.28 Differences between curves for the 2 arms were determined by the log-rank test. The probabilities of acute GVHD and of 180-day transplantation-related mortality were calculated and expressed as cumulative incidence, as previously described.29,30 P values lower than .05 were considered statistically significant; P values from .05 to .1 were considered not statistically significant, but shown in detail, while P values greater than .1 were expressed as nonsignificant (NS).
Results
Incidence of HCMV infection
In the IEmRNA arm, 33 (80%) of the 41 HSCT recipients completing the study period developed HCMV infection in the posttransplantation period (Table 2). In the antigenemia arm, HCMV infection was detected in 20 (51%) of the 39 patients completing the study. Thus, the number of patients diagnosed as having active HCMV infection was significantly higher in the IEmRNA arm (P = .006). This difference is clearly evidenced by the Kaplan-Meier curves (Figure 1A) showing a statistically significant higher proportion of HCMV-free patients with time in the antigenemia arm (P = .015). This different percentage of infection was due to the different sensitivity of the 2 guiding assays. In fact, evaluation of patients included in the IEmRNA arm by the antigenemia assay, as well as evaluation of patients included in the antigenemia arm by the NASBA assay, showed that the incidence of HCMV infection was comparable in the 2 arms (Table 2). Similarly, when HCMV DNA was determined retrospectively in blood samples of both arms, the results were comparable (Table 2).
Incidence of HCMV infection in the 80 HSCT recipients of the 2 randomization arms according to assay and type of allograft donor
. | No. of patients (%) in the randomization arm . | . | . | . | |
|---|---|---|---|---|---|
| Parameter . | IEmRNA . | Antigenemia . | P* . | Total no. (%) . | |
| Positivity for HCMV | |||||
| IEmRNAemia | 33† (80.5) | 29 (74.4) | NS | 62 (77.5) | |
| Antigenemia | 26 (63.4) | 20† (51.3) | NS | 46 (57.5) | |
| DNAemia | 24 (58.5) | 24 (61.5) | NS | 48 (60.0) | |
| Type of donor | |||||
| UD | 15 of 18 (83.3) | 6 of 17 (35.3) | .006‡ | 21 of 35 (60.0) | |
| Sibling | 18 of 23 (78.3) | 14 of 22 (63.7) | NS | 32 of 45 (71.1) | |
. | No. of patients (%) in the randomization arm . | . | . | . | |
|---|---|---|---|---|---|
| Parameter . | IEmRNA . | Antigenemia . | P* . | Total no. (%) . | |
| Positivity for HCMV | |||||
| IEmRNAemia | 33† (80.5) | 29 (74.4) | NS | 62 (77.5) | |
| Antigenemia | 26 (63.4) | 20† (51.3) | NS | 46 (57.5) | |
| DNAemia | 24 (58.5) | 24 (61.5) | NS | 48 (60.0) | |
| Type of donor | |||||
| UD | 15 of 18 (83.3) | 6 of 17 (35.3) | .006‡ | 21 of 35 (60.0) | |
| Sibling | 18 of 23 (78.3) | 14 of 22 (63.7) | NS | 32 of 45 (71.1) | |
NS indicates not significant.
Chi-square test.
The guiding assay of the relevant arm. Statistical comparison of the 2 guiding assays showed significant difference (P = .006).
Fisher exact test.
Kaplan-Meier curves illustrating the probability of freedom from HCMV infection in blood (A) and freedom from treatment (B) after transplantation as well as during the duration of the antiviral treatment (C-D) in the 2 randomization arms according to the D/R HCMV serostatus.
Kaplan-Meier curves illustrating the probability of freedom from HCMV infection in blood (A) and freedom from treatment (B) after transplantation as well as during the duration of the antiviral treatment (C-D) in the 2 randomization arms according to the D/R HCMV serostatus.
The greater sensitivity of the NASBA versus the antigenemia assay also was demonstrated by the following findings: (1) NASBA detected active HCMV infection in 62 (77%) of 80 patients completing the trial, whereas antigenemia was able to detect only 46 (57%) of 80 patients with active infection (P = .007) (Table 2); (2) on the whole, as many as 387 (25%) of 1555 blood samples were positive for IEmRNA by NASBA, whereas only 296 (19%) of 1585 were positive for antigenemia (P < .001; Table 3); (3) the median time to diagnose HCMV infection in blood was 30 (range, 9-87) and 35 (range, 13-71) days for the NASBA and antigenemia assays, respectively (P = .014).
Kinetics of HCMV infection and response to treatment in blood of HSCT recipients of the 2 arms according to different assays
. | No. of positive blood samples/total no. tested (%) in the randomization arm . | . | . | . | |
|---|---|---|---|---|---|
| Blood samples tested . | IEmRNA . | Antigenemia . | P . | Total no. (%) . | |
| After transplantation | |||||
| IEmRNA | 238/868 (27.4) | 149/687 (21.7) | .009 | 387/1555 (24.9) | |
| Antigenemia | 187/861 (21.7) | 109/724 (15.1) | .001 | 296/1585 (18.7) | |
| DNAemia | 145/838 (17.3) | 84/687 (12.2) | .006 | 229/1525 (15.0) | |
| Before therapy | |||||
| IEmRNA | 71/442 (16.1) | 66/451 (14.6) | NS | 137/893 (15.3) | |
| Antigenemia | 40/435 (9.2)* | 34/469 (7.2)* | NS | 74/904 (8.2) | |
| DNAemia | 37/430 (8.6)* | 33/452 (7.3)* | NS | 70/882 (7.9) | |
| During therapy | |||||
| IEmRNA | 97/220 (44.1)* | 40/104 (38.5)* | NS | 137/324 (42.3) | |
| Antigenemia | 103/220 (46.8)* | 50/117 (42.7)* | NS | 153/337 (45.4) | |
| DNAemia | 65/211 (30.8) | 27/104 (26.0) | NS | 92/315 (29.2) | |
. | No. of positive blood samples/total no. tested (%) in the randomization arm . | . | . | . | |
|---|---|---|---|---|---|
| Blood samples tested . | IEmRNA . | Antigenemia . | P . | Total no. (%) . | |
| After transplantation | |||||
| IEmRNA | 238/868 (27.4) | 149/687 (21.7) | .009 | 387/1555 (24.9) | |
| Antigenemia | 187/861 (21.7) | 109/724 (15.1) | .001 | 296/1585 (18.7) | |
| DNAemia | 145/838 (17.3) | 84/687 (12.2) | .006 | 229/1525 (15.0) | |
| Before therapy | |||||
| IEmRNA | 71/442 (16.1) | 66/451 (14.6) | NS | 137/893 (15.3) | |
| Antigenemia | 40/435 (9.2)* | 34/469 (7.2)* | NS | 74/904 (8.2) | |
| DNAemia | 37/430 (8.6)* | 33/452 (7.3)* | NS | 70/882 (7.9) | |
| During therapy | |||||
| IEmRNA | 97/220 (44.1)* | 40/104 (38.5)* | NS | 137/324 (42.3) | |
| Antigenemia | 103/220 (46.8)* | 50/117 (42.7)* | NS | 153/337 (45.4) | |
| DNAemia | 65/211 (30.8) | 27/104 (26.0) | NS | 92/315 (29.2) | |
NS indicates not significant.
All statistical comparisons between the 2 guiding assays (performed in vertical within each arm and each group of data) by the chi-square test showed statistically significant differences (P < .05), except for the 4 pairs of comparisons marked with asterisks.
As shown in Table 2, a significantly higher incidence of HCMV infection was detected in recipients of a UD allograft randomized in the IEmRNA arm, whereas no difference was detected between the 2 arms in the percentage of patients experiencing viral infection when the donor was an HLA-identical sibling.
With respect to the D/R serostatus, the incidence of HCMV infection was lower, although not significantly, in the D+/R– pairs of the antigenemia group (1 [16.7%] of 6) as compared to R+ HSCT recipients (19 [58%] of 33), regardless of donor HCMV serology (Table 4). This finding suggests that the infection transmitted from the donor to seronegative recipients was mostly short-lived and, thus, not detected by antigenemia.
HCMV infection and antiviral treatment according to D/R serostatus
. | No. of patients (%) in the randomization arm . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | IEmRNA . | . | . | Antigenemia . | . | . | |||||
| . | Total . | HCMV infected . | Treated . | Total . | HCMV infected . | Treated . | |||||
| HCMV serostatus . | n = 41 . | n = 33 . | n = 27 . | n = 39 . | n = 20 . | n = 14 . | |||||
| D+/R+ | 20 | 15 (75.0) | 11 (55.0) | 22 | 15 (68.2) | 13 (59.1) | |||||
| D-/R+ | 17 | 14 (82.4) | 14 (82.4) | 11 | 4 (36.4) | 4 (36.4) | |||||
| D+/R- | 4 | 4 (100) | 2 (50.0) | 6 | 1 (16.7) | 0 | |||||
| R- vs R+, P* | NA | NS | NS | NA | .092 | .027 | |||||
| D- vs D+, P* | NA | NS | .096 | NA | NS | NS | |||||
. | No. of patients (%) in the randomization arm . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | IEmRNA . | . | . | Antigenemia . | . | . | |||||
| . | Total . | HCMV infected . | Treated . | Total . | HCMV infected . | Treated . | |||||
| HCMV serostatus . | n = 41 . | n = 33 . | n = 27 . | n = 39 . | n = 20 . | n = 14 . | |||||
| D+/R+ | 20 | 15 (75.0) | 11 (55.0) | 22 | 15 (68.2) | 13 (59.1) | |||||
| D-/R+ | 17 | 14 (82.4) | 14 (82.4) | 11 | 4 (36.4) | 4 (36.4) | |||||
| D+/R- | 4 | 4 (100) | 2 (50.0) | 6 | 1 (16.7) | 0 | |||||
| R- vs R+, P* | NA | NS | NS | NA | .092 | .027 | |||||
| D- vs D+, P* | NA | NS | .096 | NA | NS | NS | |||||
NA indicates not applicable; NS, not significant.
Fisher exact test.
Pre-emptive therapy
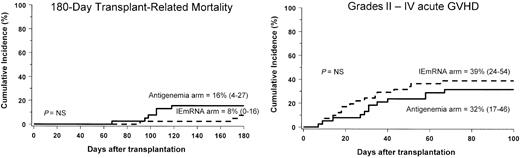
No case of HCMV disease was observed in either arm both during and after the study period. The cumulative incidence of transplantation-related mortality, as well as that of acute GVHD, was not influenced by the randomization arm. In fact, the probability of dying of transplantation-related causes at day +180 was 8% (CI, 0-16) and 16% (CI, 4-27) in the IEmRNA and antigenemia arm, respectively (P = NS, Figure 2A). The cumulative incidence of grades II-IV acute GVHD was 39% (CI, 24-54) and 32% (CI, 17-46) in the IEmRNA and antigenemia arm, respectively (P = NS, Figure 2B).
Cumulative incidence of transplantation-related mortality (left) and acute GVHD (right) in the NASBA (IEmRNA) and antigenemia arms, respectively. P = NS, not significant. Numbers in parentheses indicate 95% confidence interval.
Cumulative incidence of transplantation-related mortality (left) and acute GVHD (right) in the NASBA (IEmRNA) and antigenemia arms, respectively. P = NS, not significant. Numbers in parentheses indicate 95% confidence interval.
Patients were treated according to the criteria reported in “Study design.” Some patients, who had abortive (ie, transient infection, proved by the negative test on a second control after a first positive result) HCMV infection, were not treated. As shown in Table 5, the number of patients treated was significantly higher in the IEmRNA arm (P = .045). This difference also was shown (Figure 1B) by the higher proportion of treatment-free patients in the antigenemia arm with time (P = .06, log-rank test). However, the median duration of the first course of treatment was comparable in the 2 arms. In the IEmRNA arm, there was no difference among the 3 subgroups of patients classified according to the HCMV serostatus when considered separately (Figure 1C). In the antigenemia arm, duration of treatment was longer for the D–/R+ group (Figure 1D). However, this difference was not significant due to the small number of patients studied (P = .089, log-rank test).
Pre-emptive therapy in the 2 randomization arms of the whole transplant patient population, and the 2 subgroups of HSCT recipients from UD and sibling
. | Total patient population in the randomization arm . | . | . | HSCT from UD in the randomization arm . | . | . | HSCT from siblings in the randomization arm . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | IEmRNA . | Antigenemia . | . | IEmRNA . | Antigenemia . | . | IEmRNA . | Antigenemia . | . | |||
| Parameter . | n = 41 . | n = 39 . | P . | n = 18 . | n = 17 . | P . | n = 23 . | n = 22 . | P . | |||
| No. (%) of patients | ||||||||||||
| HCMV-infected (first episode) | 33 (80.5) | 20 (51.3) | .006* | 15 (83.3) | 6 (35.3) | .006† | 18 (78.3) | 14 (63.7) | NS* | |||
| Treated | 27 (65.9) | 17 (43.6) | .045* | 15 (83.3) | 6 (35.3) | .006† | 12 (52.2) | 11 (50.0) | NS* | |||
| Duration of therapy‡ | 12 (5-45) | 13 (6-28) | NS§ | 13 (6-35) | 15.5 (6-28) | NS§ | 11 (5-45) | 13 (6-20) | NS§ | |||
| No. (%) of relapsing patients: | ||||||||||||
| Detected | 21 of 27 (80.0) | 10 of 17 (58.8) | NS* | 11 of 15 (73.3) | 4 of 6 (66.7) | NS† | 10 of 12 (83.3) | 5 of 11 (45.5) | .089† | |||
| Treated | 16 of 21 (76.2) | 7 of 10 (70.0) | NS† | 10 of 11 (90.9) | 3 of 4 (75.0) | NS† | 6 of 10 (60.0) | 3 of 5 (60.0) | NS† | |||
| Duration of therapy‡ | 16 (6-46) | 16 (7-48) | NS§ | 20.5 (6-46) | 19.5 (13-48) | NS§ | 9.5 (7-17) | 13 (7-16) | NS§ | |||
| Overall duration of therapy‡∥ | 25 (5-68) | 19 (9-76) | NS§ | 30 (7-68) | 25.5 (9-76) | NS§ | 15.5 (5-62) | 17 (9-26) | NS§ | |||
| No. of patients with HCMW disease | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
. | Total patient population in the randomization arm . | . | . | HSCT from UD in the randomization arm . | . | . | HSCT from siblings in the randomization arm . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | IEmRNA . | Antigenemia . | . | IEmRNA . | Antigenemia . | . | IEmRNA . | Antigenemia . | . | |||
| Parameter . | n = 41 . | n = 39 . | P . | n = 18 . | n = 17 . | P . | n = 23 . | n = 22 . | P . | |||
| No. (%) of patients | ||||||||||||
| HCMV-infected (first episode) | 33 (80.5) | 20 (51.3) | .006* | 15 (83.3) | 6 (35.3) | .006† | 18 (78.3) | 14 (63.7) | NS* | |||
| Treated | 27 (65.9) | 17 (43.6) | .045* | 15 (83.3) | 6 (35.3) | .006† | 12 (52.2) | 11 (50.0) | NS* | |||
| Duration of therapy‡ | 12 (5-45) | 13 (6-28) | NS§ | 13 (6-35) | 15.5 (6-28) | NS§ | 11 (5-45) | 13 (6-20) | NS§ | |||
| No. (%) of relapsing patients: | ||||||||||||
| Detected | 21 of 27 (80.0) | 10 of 17 (58.8) | NS* | 11 of 15 (73.3) | 4 of 6 (66.7) | NS† | 10 of 12 (83.3) | 5 of 11 (45.5) | .089† | |||
| Treated | 16 of 21 (76.2) | 7 of 10 (70.0) | NS† | 10 of 11 (90.9) | 3 of 4 (75.0) | NS† | 6 of 10 (60.0) | 3 of 5 (60.0) | NS† | |||
| Duration of therapy‡ | 16 (6-46) | 16 (7-48) | NS§ | 20.5 (6-46) | 19.5 (13-48) | NS§ | 9.5 (7-17) | 13 (7-16) | NS§ | |||
| Overall duration of therapy‡∥ | 25 (5-68) | 19 (9-76) | NS§ | 30 (7-68) | 25.5 (9-76) | NS§ | 15.5 (5-62) | 17 (9-26) | NS§ | |||
| No. of patients with HCMW disease | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
NS indicates not significant.
Chi-square test.
Fisher exact test.
Median number of days (range) of treatment per patient.
Mann Whitney U test.
Primary plus relapse episodes.
The percentage of HCMV relapses, as well as that of treated relapses, was comparable in the 2 arms (Table 5). Similarly, the duration of therapy for the relapse episodes was comparable in the 2 arms. Also, when the total number of days of therapy (first episode of HCMV infection plus relapse episodes) was considered, the difference between the 2 randomization arms was not significant (Table 5).
When stratifying the patient population according to the type of donor used, we found that the significantly greater number of infected and treated patients in the IEmRNA arm with respect to the antigenemia arm consisted of patients who received transplants from a UD, whereas the higher number of relapse episodes was determined mainly by the subgroup of patients who received transplants from an HLA-identical sibling (Table 5). All other therapeutic parameters considered were comparable in the 2 arms of either subgroup (Table 5).
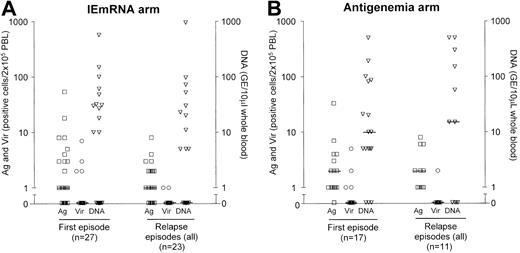
Viral load at treatment onset
To retrospectively verify whether a rationale could be identified for establishing cutoff DNA values for future strategies of pre-emptive therapy, we examined the levels of viral load at treatment inception for both arms. In the IEmRNA arm, upon initiation of therapy for the first HCMV episode in the 27 treated patients, median levels of antigenemia, viremia, and DNAemia were 1 (range, 0-54), 0 (range, 0-7), and 0 (range, 0-571), respectively. The corresponding levels at time of treatment of the 23 episodes of relapse were 1 (range, 0-8), 0 (range, 0-1), and 0 (range, 0-952), respectively, and those of untreated patients (positive once, eg, abortive infection) were negligible (Figure 3A). On the other hand, in the antigenemia arm, upon initiation of treatment of the first HCMV episode in the 17 treated patients, the relevant median levels were 2 (range, 1-33) for antigenemia, 0 (range, 0-5) for viremia, and 10 (range, 0-500) for DNAemia, while at treatment onset of the 11 relapse episodes, median levels were 2 (range, 1-8) for antigenemia, 0 (range, 0-2) for viremia, and 15 (range, 0-600) for DNAemia. Again, for untreated patients, median levels of antigenemia and viremia were negligible, whereas for DNAemia they were 5 (range, 0-208) (Figure 3B).
Individual and median levels of antigenemia, viremia, and DNAemia in treated patients upon onset of first course and subsequent courses of antiviral treatment in the 2 randomization arms. Antigenemia (Ag; left y-axis) is expressed as the number of pp65-positive/2 × 105 PBLs examined on cytospin preparations stained with a pool of 3 pp65-specific monoclonal antibodies. Viremia (Vir; left y-axis) is expressed as the number of fibroblast nuclei positive for the HCMV IE antigen p72 after inoculation of 2 × 105 PBLs onto human embryonic lung fibroblast cell cultures by the shell vial technique. DNAemia (DNA; right y-axis) is expressed as viral genome equivalents (GE)/10 μL whole blood.
Individual and median levels of antigenemia, viremia, and DNAemia in treated patients upon onset of first course and subsequent courses of antiviral treatment in the 2 randomization arms. Antigenemia (Ag; left y-axis) is expressed as the number of pp65-positive/2 × 105 PBLs examined on cytospin preparations stained with a pool of 3 pp65-specific monoclonal antibodies. Viremia (Vir; left y-axis) is expressed as the number of fibroblast nuclei positive for the HCMV IE antigen p72 after inoculation of 2 × 105 PBLs onto human embryonic lung fibroblast cell cultures by the shell vial technique. DNAemia (DNA; right y-axis) is expressed as viral genome equivalents (GE)/10 μL whole blood.
Levels of viral load, as determined by the quantitative assays, were not significantly different between the 2 arms (P = NS). However, a fair number of patients in both arms started treatment concomitantly with levels of DNAemia between 100 and 1000 GE/10 μL blood, thus suggesting that DNA cutoff levels could be considered for initiation of pre-emptive treatment.
Kinetics of HCMV infection and response to treatment
To elucidate the mechanism of response to antiviral treatment in both arms, the kinetics of HCMV infection prior to and during treatment was investigated on a quantitative basis, considering that evaluation of IEmRNA is only a qualitative test.
As shown in Table 3, when considering all blood samples tested, the NASBA assay for detection of IEmRNA gave the highest number of positive samples, followed by antigenemia, which unexpectedly appeared to be more sensitive than DNAemia. However, when blood samples tested were divided into 2 groups (one including pre-therapy samples, the second including blood samples drawn during therapy) in the pre-therapy group, the NASBA assay maintained its higher sensitivity, while there was an overlapping level of sensitivity of antigenemia and DNAemia assays. On the contrary, in the intratherapy group, the antigenemia assay gave by far the highest number of positive samples, followed by NASBA and DNAemia. The apparent increase in sensitivity of the antigenemia assay during therapy was related to the longer persistence of HCMV pp65 in blood (with respect to viral IEmRNA and DNA).
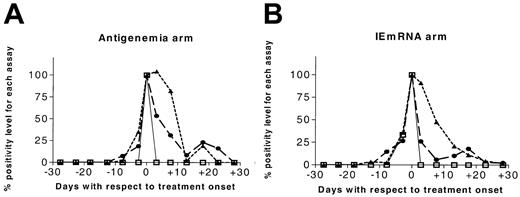
Since determination of IEmRNA was only qualitative, the effect of treatment on viral infection was studied by comparing the kinetics (disappearance) of antigenemia with that of DNAemia. A trend toward a longer persistence of pp65 was shown by (1) the curves illustrating the kinetics of HCMV infection in both arms (Figure 4); and (2) the value of half-time of antigenemia versus DNAemia during treatment, which was significantly longer in both the antigenemia arm (5.0 [range, 1-21] vs 2.3 [range, 1-22] days, P < .01) and the IEmRNA arm (5.3 [range, 1-18] vs 1.5 [range, 1-17] days, P = .04).
Kinetics of HCMV infection before and after treatment onset (set as day 0) in the two randomization arms according to antigenemia, viremia, and DNAemia assays. The value of the parameters at day 0 was arbitrarily set to 100%. ▴ indicates antigenemia; □, viremia; and ▪, DNAemia.
Kinetics of HCMV infection before and after treatment onset (set as day 0) in the two randomization arms according to antigenemia, viremia, and DNAemia assays. The value of the parameters at day 0 was arbitrarily set to 100%. ▴ indicates antigenemia; □, viremia; and ▪, DNAemia.
Discussion
The optimal pre-emptive therapy for HCMV infection in HSCT recipients should allow effective prevention of HCMV disease, short-time administration of antiviral therapy, safe discontinuation of antiviral treatment, and low treatment toxicity. In order to pursue these objectives, HCMV infection must be monitored carefully by a reliable and sensitive assay. Viremia or virus detection in different body sites (eg, broncho-alveolar lavage) have been used in the past, but eventually they have been abandoned due to their low sensitivity, leading to development of HCMV disease in the absence of virus recovery.31,32 The most currently used assays are quantitative or qualitative antigenemia and DNAemia; both have good sensitivity and have provided satisfactory results in terms of prevention of HCMV disease in different transplantation settings.13
The most sensitive assay now available for the detection of a viral product directly related to HCMV replication is NASBA for IEmRNA, which proved to diagnose HCMV infection earlier than antigenemia in a retrospective study on HSCT recipients.18 In addition, antigenemia does not correlate reliably with the actual virus replication, measuring HCMV pp65, which is produced in large excess during virus replication.27,33 After considering that DNAemia also does not necessarily indicate replicating virus, we designed a randomized study to compare antigenemia (a quantitative method in use for more than a decade in our center) with the NASBA assay (a qualitative method) for IEmRNA detection. The study was aimed at testing whether the latter assay, while providing the same efficacy in terms of disease prevention, could also spare some days of antiviral treatment.
Our results confirm the greater sensitivity of the NASBA assay, since both the number of HCMV-infected patients and the number of patients treated for a first episode of HCMV infection was significantly higher in the IEmRNA arm than in the antigenemia arm. The increased sensitivity of the NASBA assay permitted the detection of HCMV infection at an earlier stage, as already reported.18,34 Similarly, seronegative patients receiving the transplant from a seropositive donor had mild HCMV infection, which was detected nearly only by the NASBA assay. In fact, with the limits of the restricted number of cases available, 4 of 4 D+/R– patients were diagnosed in the NASBA arm, whereas only 1 out of 6 cases of infection was detected in the antigenemia arm of this group. This finding also suggests that seropositive donors are capable of transferring specific immunity together with the infection to HSCT recipients in whom the contribution of memory T cells present in the graft is fundamental. In fact, virus reactivation was associated to the highest viral load when deriving from latent infection in seropositive recipients. This hypothesis also is supported by the observation that in the D–/R+ subgroup of patients allocated in the antigenemia arm, the median duration of treatment was longer (although not significantly) than that of the D+/R+ subgroup. This emphasizes the notion that specific immunity plays a pivotal role in the dynamics of HCMV infection in HSCT recipients.
HCMV disease did not develop in any of the patients enrolled in this study, and the cumulative probability of transplantation-related mortality was comparable in the 2 arms. These clinical results compare favorably with those obtained in other previous studies on the use of pre-emptive therapy in HSCT recipients.2,3,9 Thus, our data indicate that with the treatment strategy chosen, IEmRNA detection can safely replace antigenemia in guiding pre-emptive therapy of HCMV infections in HSCT recipients. Moreover, since the NASBA assay can detect HCMV reactivation earlier than antigenemia (30 vs 35 days, respectively), it can anticipate the onset of treatment, thus being potentially useful in avoiding the occurrence of the rare but life-threatening early HCMV disease developing before engraftment and prior to virus detection in blood.12,35
There was no difference in the overall median duration of treatment per patient, the main end point of our study, notwithstanding the greater infection rate detected in the IEmRNA arm in association with a greater treatment rate as compared to antigenemia. This indicates that, despite earlier detection of virus reactivation, patients allocated in the IEmRNA arm required antiviral therapy to clear HCMV infection in duration comparable to those in the antigenemia arm. In fact, even if IEmRNA anticipates antigenemia in detecting HCMV infection, we found no significant difference in viral load at treatment onset between the 2 arms, thus determining similar duration of therapy in the 2 arms. Both the high percentage of relapses in the IEmRNA arm and the comparable duration of the first course of treatment in the 2 arms contributed to the comparable duration of overall antiviral treatment. Thus, from a clinical point of view, the NASBA assay is comparable to but not better than the antigenemia assay. Accordingly, modifications of both the neutrophil and platelet count attributable to the antiviral treatment were not different for patients allocated in the 2 randomization arms (data not shown). Since use of the NASBA assay resulted in first treatment of 50% more patients than with the antigenemia assay, this represents, in terms of patient discomfort and, possibly, of treatment-related costs, a disadvantage. Moreover, the NASBA assay is more expensive than the antigenemia method, as each specific test costs approximately $30 and $10, respectively. However, it should be remarked that determination of antigenemia is more time consuming in terms of working time and requires expertise in reading test results. Advantages of the NASBA assay, with respect to antigenemia, are semiautomation of the test procedure and the need of small blood volumes, sometimes important in small children.
Sensitivity of antigenemia varies according to its determination during treatment or before treatment onset. In fact, antigenemia detected as positive 45% of blood samples among treated patients, compared to 42% of samples positive by NASBA and 29% by DNAemia. On the contrary, in the group of pretherapy blood samples, NASBA for IEmRNA was the most sensitive assay, followed by antigenemia and DNAemia, which showed comparable levels of sensitivity. This finding can be explained by the longer persistence of pp65 in peripheral blood leukocytes compared to infectious virus or viral products such as viral DNA or RNA. This phenomenon, consisting essentially in rising or sustained high levels of pp65-positive leukocytes with concomitantly decreasing levels of viral DNA or RNA or infectious virus, has been observed already during treatment with ganciclovir of primary HCMV infections in solid-organ transplant recipients,36,37 but has also been described for HSCT recipients.2 Its pathophysiologic basis is due to the fact that, during virus replication, pp65 is synthesized largely in excess, accumulating as dense bodies capable of inducing in vivo both humoral and cellular immune response.38 In other words, when viral DNA replication is blocked by a viral DNA inhibitor such as ganciclovir and viral products rapidly decrease in infected cells, pp65 is still detected, partly because of its abundance and partly because it is an early-late viral protein that continues to be synthesized (at a lower rate) when there is a block of viral DNA replication.33,39 It is now well known that detection of virus and virus products in blood of patients with HCMV infection who received transplants is mediated by the presence of PBLs, mostly polymorphonuclear leukocytes and partly monocytes, where the virus is transferred from infected endothelial cells via transient microfusion events.40 It is reasonable to assume that during ganciclovir therapy, in the absence of virus or viral DNA and RNA in infected endothelial cells, only pp65 can be transferred to PBLs. In this case, only antigenemia will be positive concomitantly with negative viremia, DNAemia, and RNAemia. Thus, antigenemia represents only an asynchronous parameter of HCMV infection in blood of patients who received transplants and not a direct marker of virus replication.
Considering that late HCMV disease did not develop in any of our patients, using our strategy of antiviral therapy, the earlier detection and treatment of HCMV infection afforded by the NASBA assay does not seem to alter the long-term recovery of HCMV-specific T cells. Our results cannot answer the crucial question of whether or not it is critical to start therapy upon initial detection of HCMV in blood in the pre-emptive therapy of HSCT recipients.35 If the answer is yes, then the most sensitive assay, the IEmRNA-NASBA, would be advisable as a guiding assay. If not, then a quantitative assay could be used and a cutoff chosen, as is currently done for reactivated HCMV infections of solid-organ transplant recipients in most transplantation centers. This study does not seem to exclude the latter option, considering that some patients have been successfully treated with levels of DNAemia included in the range of 100 to 1000 GE/10 μL blood. A prospective study could answer the question whether, using DNAemia as a guiding assay, a cutoff could be taken as a threshold for starting antiviral treatment. This could avoid treatment of patients with transitory appearance of DNA in blood.
Prepublished online as Blood First Edition Paper, February 13, 2003; DOI 10.1182/blood-2002-12-3636.
Partially supported by Ministero della Salute, Ricerca Corrente Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Policlinico San Matteo, grant 80208; by Istituto Superiore di Sanitá, Progetto Multidisciplinare sulla Terapia delle Malattie da Virus, grant 0AG/F16; and by Ministero dell'Istruzione, Universitá e Ricerca Scientifica, Consiglio Nazionale delle Ricerche, grant 01.00696.PF49.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all technical staff of the Servizio di Virologia for test performance, and Linda D'Arrigo for the revision of the English in this manuscript. We also thank bioMérieux for providing NASBA reagents.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal