Abstract
Although multiple myeloma (MM) is a unique entity, a marked heterogeneity is actually observed among the patients, which has been first related to immunoglobulin (Ig) types and light chain subtypes and more recently to chromosomal abnormalities. To further investigate this genetic heterogeneity, we analyzed gene expression profiles of 92 primary tumors according to their Ig types and light chain subtypes with DNA microarrays. Several clusters of genes involved in various biologic functions such as immune response, cell cycle control, signaling, apoptosis, cell adhesion, and structure significantly discriminated IgA- from IgG-MM. Genes associated with inhibition of differentiation and apoptosis induction were up-regulated while genes associated with immune response, cell cycle control, and apoptosis were down-regulated in IgA-MM. According to the expression of the 61 most discriminating genes, BJ-MM represented a separate subgroup that did not express either the genes characteristic of IgG-MM or those of IgA-MM at a high level. This suggests that transcriptional programs associated to the switch could be maintained up to plasma cell differentiation. Several genes whose products are known to stimulate bone remodeling discriminate between κ- and λ-MM. One of these genes, Mip-1α, was overexpressed in the κ subgroup. In addition, we established a strong association (P = .0001) between κ subgroup expressing high levels of Mip-1α and active myeloma bone disease. This study shows that DNA microarrays enable us to perform a molecular dissection of the bioclinical diversity of MM and provide new molecular tools to investigate the pathogenesis of malignant plasma cells.
Introduction
Multiple myeloma (MM) is characterized by the accumulation of malignant plasma cells (PCs), usually within the bone marrow. Besides the demonstration of this excess of PCs, the diagnosis of MM is usually supported by the finding of lytic bone lesions on x-rays and the presence of a monoclonal immunoglobulin (Ig) in the serum and/or urine. The monoclonal Ig allows us to define several types of MM, depending on the Ig heavy chain (IgH) isotype and light chain subtype (IgL). Most MMs are characterized by the excretion of a complete monoclonal Ig, easily detectable on the serum electrophoresis. The most frequent Ig is IgG (about 60% of the patients), followed by IgA (about 25%). In a few cases, other Ig classes are observed—that is, IgD, IgM, or IgE (less than 2% of the patients). In other instances, malignant PCs do not excrete any Ig chain, representing about 1% of the patients. Finally, approximately 15% of the patients excrete only light chains: the so-called Bence Jones MM (BJ-MM). Apart from these IgH characteristics, the M component may be further classified upon the light chain subtype—that is, either the κ chain or the λ chain. Roughly two thirds of the patients present a κ-type MM and one third a λ-type MM, a proportion similar to that observed in normal PCs. These light chains are often produced in excess, and even in common IgG- or IgA-MM, free light chains are detected in the serum and/or urine.
Of note, these different Ig types and light chain subtypes have been associated with specific clinical or biologic presenting features, such as bone involvement and renal impairment, and with different clinical outcome.1 However, so far, no published study has addressed the question of possible different biologic behaviors in these different types of MMs—that is, IgG-versus IgA-MM, IgG- and IgA-MM versus BJ-MM, or κ-type versus λ-type MM. The recent development of the microarray technology has opened new windows on the way to approach specific intracellular biologic pathways2,3 and to improve tumor classifications.4-9 To address this issue, we have analyzed a large series of patients with MM using gene expression profiling, focusing our analysis on the differences in gene activation (or repression) associated with the different Ig types—that is, IgG versus IgA, and κ light chain versus λ light chain subtypes.
Patients, materials, and methods
Patients
The diagnosis of MM was done according to the criteria of the Southwest Oncology Group.10 From a total of 105 patients analyzed in the present study, high-quality gene expression data were obtained on 88 MMs and 4 primary cell leukemia (PCLs) defined by more than 20% of malignant PCs in the peripheral blood. Newly diagnosed untreated patients were referred to one of the clinical centers of the Intergroupe Francophone du Myélome (IFM). Written informed consent was obtained from all the patients according to the Declaration of Helsinki. The median age of the patients was 60 years (range, 31-78 years). The clinical staging was established according to standard criteria of Durie and Salmon11 : 14 stage I, 7 stage II, 67 stage III, and 4 PCL. The monoclonal Ig was IgGκ in 28 patients, IgGλ in 22 patients, IgAκ in 16 patients, IgAλ in 6 patients, BJPκ in 12 patients, and BJPλ in 8 patients. Correlations of the 2 strongest prognostic factors, β2-microglobulin (β2M) and chromosome 13 abnormalities, across the isotypes and subtypes subgroups were analyzed. Serum β2M levels and chromosome 13 deletions (fluorescent in situ hybridization analysis) were not significantly different among these MM subgroups.
PC purification and total RNA isolation
Mononuclear bone marrow cells were separated using gradient density centrifugation (lymphocyte separation medium, Eurobio, Les Ulis, France), and plasmacytosis was evaluated by morphology in these mononuclear cell suspensions. PCs were then positively selected using anti-CD138–coated microbeads (Miltenyi, Paris, France), because CD138 is expressed only on PCs (normal and malignant) within the bone marrow. Purity and viability of the positively selected cell suspension was assessed by morphology and was above 96% in all the cases. One million PCs were used to prepare total RNA using the guanidinium thiocyanate–phenol method.12 The RNA integrity was randomly verified by using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA). Half the preparation was used to generate a complex probe.
Gene expression profiling procedures
Vector oligomer labeling and hybridization conditions. One microgram of vector oligonucleotide was labeled at the 5′ end with 30 μCi (1.11 MBq) [γ-33P]adenosine triphosphate ([γ-33P]ATP) and 10 units of T4 polynucleotide kinase (Invitrogen, Cergy Pontoise, France) for 45 minutes at 37°C; unincorporated nucleotides were removed by purification on a Sephadex G-25 column (Roche, Meylan, France). The vector oligonucleotide sequence used was 5′-ACTGGCCGTCGTTTTACA. Nylon microarrays were prehybridized in hybridization mix (5 × SSC, 5 × Denhardt solution, 0.5% sodium dodecyl sulfate [SDS]) for 4 hours at 42°C and then hybridized with the vector probe for 2 hours at 42°C. After hybridization, filters were washed in 2 × SSC, 0.1% SDS at room temperature for 10 minutes and at 42°C for 5 minutes. After phosphor screen acquisition (Fuji BAS 5000; Fuji, Tokyo, Japan), filters were stripped in 0.1 × SSC, 0.1% SDS at 68°C for 3 hours before hybridization with a complex probe. Quantification of the vector probe hybridization signal provided a value corresponding to the amount of DNA fixed in each spot of the microarray.
Preparation and labeling of complex probes from total RNA and hybridization conditions. Aliquots of 2.5 μg total RNA, 8 μg oligo(dT)25 to saturate long polyA tails, and 0.3 ng in vitro–synthesized Arabidopsis thaliana cytochrome c554 allowing interfilter normalization were mixed, heated to 70°C for 8 minutes, and cooled to 42°C before reverse transcription in a reaction mixture containing 50 mM Tris (tris(hydroxymethyl)aminomethane)–HCl (pH 8.3), 75 mM KCl, 10 mM dithiothreitol (DTT), 3 mM MgCl2, 5 units RNase inhibitor (GIBCO BRL, Cergy Pontoise, France), 0.4 mM each of deoxyuridine triphosphate (dATP), deoxyguanosine triphosphate (dGTP), and deoxythymidine triphosphate (dTTP), 240 nM deoxycytidine triphosphate (dCTP), 30 μCi (1.11 MBq) [α-33P]dCTP, and 400 units Superscript RNAse H reverse transcriptase (GIBCO BRL) for 2 hours at 42°C. After alkali treatment and neutralization, unincorporated nucleotides were removed by purification on a Sephadex G-50 column (Roche). The complex probe was then incubated for 2 hours at 65°C with 2 μg poly(dA)80 to eliminate spurious hybridization via the polyA tail present in some clones before hybridization. Nylon microarrays were prehybridized in 1 mL hybridization mix for 6 hours at 68°C and then hybridized with the complex probe in 0.4 mL hybridization mix for 48 hours at 68°C. After hybridization, filters were washed twice in 0.1 × SSC, 0.1% SDS at 68°C for 90 minutes.
Nylon microarray technology. The feasibility, reproducibility, and sensitivity of spotting procedures onto nylon membrane currently used in our laboratory to produce cDNA arrays have been previously described.13-16
All cDNA clones were chosen using the expressed sequence tag (EST) database from the NCBI: http://www.ncbi.nlm.nih.gov. The clones were selected from libraries constructed with cloning vectors pT7T3D or Lafmid BA, same host bacteria, and an insert size of approximately 1 kb. Clones were provided by the Human Genome Mapping Project Resource Centre (Hinxton, United Kingdom). From our 7200 cDNA library, preferentially composed of genes expressed in carcinogenesis and immune response, we used 5376 cDNA clones to design our nylon microarrays. The selection of these clones was based on preliminary DNA microarray expression data obtained with a cDNA probe prepared from a pool of total RNA isolated from 4 different MM cell lines and 2 MM patients hybridized to a microarray containing 7200 cDNAs.
The cDNA clones were amplified in 96-well microtiter plates with 5′-GTGGAATTGTGAGCGGATAAC and 5′-GCAAGGCGATTAAGTTGGG. Polymerase chain reaction (PCR) products with more than one band or with an unexpected size were rejected. PCR product concentration was adjusted to 0.3 μg/μL. PCR products were spotted onto Hybond N+ filter (Amersham, Orsay, France) using GMS 417 arrayer. DNA spotted was then denatured and UV–cross-linked onto nylon filter. All membranes contained a set of control spots. The pT7T3D vector, poly(dA)80, vector oligonucleotide, and 50 PCR reactions without template were used as negative controls. Many genes were spotted in duplicate spots to assess the reproducibility. Arabidopsis thaliana cytochrome c554 clone that is devoid of similarities to human DNA sequences is used to normalize the differences in labeling of each complex probe.
The reproducibility of complex probe hybridization was verified first by analyzing a variation in average intensity difference. Up to 3% of the clones showed a 2-fold difference in signal intensity.15 Secondly, we compared 2 microarrays hybridized with complex probes prepared with 2 RNA samples extracted separately from the same patient. The samples were always found clustered in directly adjacent columns. We have demonstrated previously that the hybridization signal is proportional to the abundance of individual species in the complex probe and to the amount of PCR products spotted onto the microarray. In our hybridization conditions the minimum sample for detection is 0.2 × 106 molecules. In addition, the amount of PCR products per spot being 1500 times more than the detection limit, the signal measured for highly expressed genes is not saturated.
Sample quality standard. In this study we obtained high-quality gene expression data on 92 of 105 samples (88%). The quality is estimated by measuring the signal intensity of all clones in a microarray and the number of spots detected; if these 2 parameters are too low the microarray is not considered for the study.
Data acquisition and normalization. DNA microarrays were scanned at 25-μm resolution in an image plate system (Fuji BAS 5000; Fuji). The hybridization signals were quantified using ArrayGauge software v.1.3 (Fuji). A background value for each membrane hybridization was calculated from negative controls and subtracted to each expression value. The data were corrected for the amount of PCR product detected by the vector probe and normalized using Arabidopsis thaliana cytochrome c554 control clone. Sets of genes that were not measured on at least 60 of the 92 samples were removed. Statistical analyses were performed using the remaining set of 2600 genes.
Statistical analysis
Samples and genes were median centered and log transformed before data analysis. Gene and sample classifications were obtained by unsupervised hierarchic clustering using uncentered correlation distance and average linkage aggregative method with the Cluster and Tree View softwares (M. Eisen, http://www.microarrays.org/software).17
Gene-discriminating particular subgroups of MM were searched using a signal-to-noise calculation: DS = (μ1 – μ2)/(σ1 + σ2); where μ1 and σ1, respectively, represent mean and standard deviation of the expression levels of the gene in subgroup 1, and μ2 and σ2 represent mean and standard deviation of the same gene in subgroup 2.4 A total of 200 random permutations of the samples were used to calculate significance level at 1:10 000 risk, giving less than 1 gene found by chance (false-positive gene). The data sets used for the identification of genes that distinguish Ig subtypes or isotypes are available at our website: http://tagc.univ-mrs.fr/pub/.
RT-PCR procedures
The first cDNA strand was synthesized using total RNA (2.5 μg) at 37°C for 1 hour in a 50-μL reaction mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 10 mM DTT, 3 mM MgCl2, 5 units RNase inhibitor (GIBCO BRL), 0.5 mM of each deoxynucleotide triphosphate (dNTP), 400 units Moloney murine leukemia virus (M-MLV) reverse transcriptase (Gibco), and 0.5 μg oligo(dT) 15 mer. Five microliters of the reaction mixture was made up to 50 μL using Taq polymerase buffer (10 mM Tris-HCl [pH 9], 50 mM KCl, 1.5 mM MgCl2) containing 25 pmol of each primer, 1 mM of each deoxynucleotide triphosphate (dNTP), and 1 unit of AmpliTaq DNA polymerase (Amersham). Amplifications were performed using a thermal cycler for 20 cycles under the following conditions: denaturation for 1 minute at 94°C, annealing for 1 minute at 55°C, and elongation for 1 minute at 72°C. Mip-1α was amplified using the following primers: sense primer: 5′-CGAGCCCACATTCCGTCACC-3′ and antisense primer: 5′-CCATGACTGCCTACACAGGC-3′. PCR products were separated in a 1% agarose gel and directly visualized after ethidium-bromide staining. Expression of β-actin using primers 5′-ATCTGGACCACACCTTCTACAATGAGCTGCG-3′ and antisense 5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′ was assessed to ensure uniformity of amplification.18
Results
In this study, we determined the gene expression profiles of 88 newly diagnosed MMs and 4 primary PCLs according to their Ig types and light chain subtypes because M component is the major source of biologic heterogeneity in MM patients.
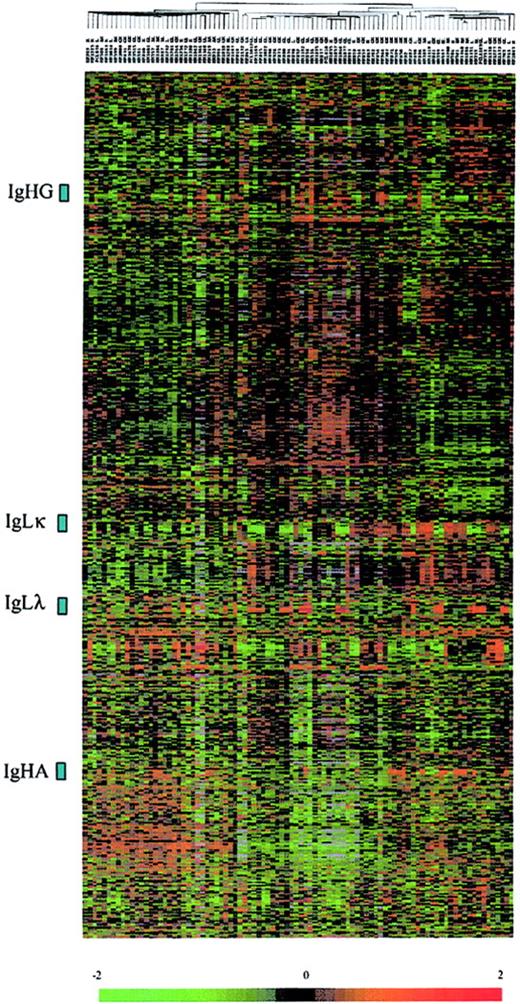
From highly enriched CD138+ cells of each tumor sample, 2.5 μg total RNA was extracted and used to prepare radioactively labeled cDNA complex targets. Hybridizations were carried out on DNA microarrays containing 5376 genes. Radioactive dot intensities of scanned images were measured and normalized to yield a ratio of background-corrected single-dot intensity to background-corrected median-dot intensity (see “Patients, materials, and methods”). The study was performed using a set of 2600 genes that were significantly expressed across the MM patients. In the initial analysis of the gene expression data, we applied an unsupervised hierarchic clustering algorithm to group the myeloma samples on the basis of similarities in their expression of these genes. The same clustering method was used to group genes on the basis of similarity in their pattern of expression over all the samples. This analysis revealed that the patients were not grouped according their Ig light chain subtypes or Ig heavy chain isotypes, and genes encoding Ig were found in 4 highly contrasted clusters (Figure 1). We next analyzed gene expression profiles of the patients by using a discriminating score (DS) based on signal-to-ratio calculation4 to identify and rank the differentially expressed genes among biologic subtypes of MM. The higher score denotes the greater ability to differentiate the 2 MM groups. We used a random permutation test, patients were randomly permutated (200 times) into 2 groups, and for each gene a DS was calculated. A gene significantly distinguishes the 2 groups of patients if it passed a 99.99% significance threshold (α less than 0.0001).
Hierarchic clustering of 92 diagnostic MM and PCL samples (columns) versus 2600 genes (rows). The normalized expression value for each gene is depicted according to the scale at the bottom; red indicates expression levels greater than the median, and green indicates levels less than the median (–2 to 2 in log base 2 units). Gray indicates excluded values.
Hierarchic clustering of 92 diagnostic MM and PCL samples (columns) versus 2600 genes (rows). The normalized expression value for each gene is depicted according to the scale at the bottom; red indicates expression levels greater than the median, and green indicates levels less than the median (–2 to 2 in log base 2 units). Gray indicates excluded values.
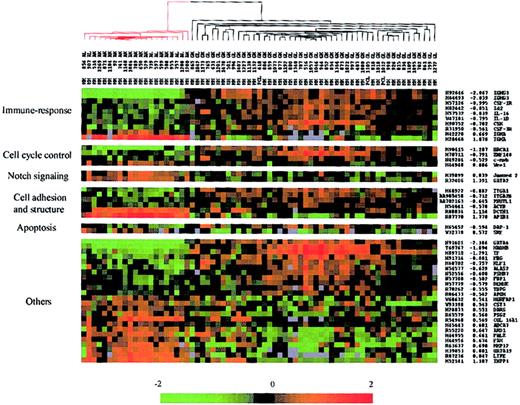
First, we compared expression data of one subgroup including 45 IgG-MMs and 3 IgG-PCLs with a second subgroup including 21 IgA-MMs. The DS test yielded 61 unique cDNA sequences from 58 different genes whose change in expression among all the patients best distinguished IgG-MM from IgA-MM (Table 1). Hierarchic cluster analysis of the 69 MM samples was performed using expression data of the 61 selected genes; the algorithm perfectly segregated IgG-MM from IgA-MM (data not shown). Figure 2 shows the matrix depicting gene expression values of the MM samples with rows representing characterized genes (49 elements) grouped according to putative biologic functions including immune response, cell cycle control, Notch signaling, cell adhesion and structure, and apoptosis. Most of these genes were up-regulated in IgG-MM versus IgA-MM. The highest differentiating score, except for IgH genes, was found for GATA6, a member of the GATA transcription factor family, which has recently been shown to regulate a WNT family member, WNT7b.19 Immune response–associated genes were up-regulated in IgG-MM (eg, the cytokines IL-1β and IL-16, the cytokine receptors CSF-1R and CSF-3R, and the B-cell development regulators Id2 and CSK). Three transcription factors (BRCA1, ZNF148, c-myb) involved in cell cycle control were also up-regulated in IgG-MM, whereas 2 genes involved in Notch signaling in hematopoietic cells (Jagged2, GATA2) were down-regulated in this subgroup of MM. Among the 6 genes potentially associated with cell adhesion and structure, 2 integrins (ITGA1 and ITGA2) as well as ACTB were up-regulated in IgG-MM, whereas AP1B1 and DCTN1 were significantly up-regulated in IgA-MM. Only 2 members of the apoptosis class were significantly differentially expressed in these subgroups of MM. The first gene, DAP-1, a proapoptotic factor, was overexpressed in IgG-MM, while the second, SRF, a regulator of the antiapoptotic molecule Mcl-1, was down-regulated in the same subgroup of MM.
Statistically different genes between IgG-MM and IgA-MM
Gene symbol . | Gene description . | Accession no. . | Discriminating score . |
|---|---|---|---|
| IGHG3 | IgG | H68233 | -2.75 |
| GATA6 | GATA-binding protein 6 | N91601 | -2.39 |
| IGHG3 | IgG | N92646 | -2.07 |
| IGHG3 | IgG | H64493 | -2.04 |
| HADHB | Hydroxyacyl-CoA dehydrogenase, beta subunit | T69767 | -1.89 |
| IF | I factor (complement) | H89710 | -1.29 |
| BRCA1 | Breast cancer 1, early onset | H90415 | -1.29 |
| ESTs | Unknown | W46567 | -1.10 |
| CSF-1R | Colony-stimulating factor 1 receptor, v-fms oncogene homolog | H57126 | -1.00 |
| ESTs | Unknown | H93533 | -0.98 |
| ITGA1 | Integrin, alpha 1 | H68922 | -0.88 |
| FBG | Fibrinogen, B beta polypeptide | H91714 | -0.88 |
| Id2 | Inhibitor of DNA binding 2, dominant-negative helix-loop-helix | H82442 | -0.85 |
| IL-16 | Interleukin-16 (lymphocyte chemoattractant factor) | H57532 | -0.84 |
| IL-1B | Interleukin-1, beta | W47101 | -0.80 |
| ZNF148 | Zinc finger protein 148 (pHZ-52) | H70711 | -0.79 |
| KLF1 | Kruppel-like factor 1 (erythroid) | H60702 | -0.76 |
| ITGA2B | Integrin, alpha 2b (antigen CD41B) | AA905458 | -0.71 |
| ESTs | Unknown | AA279804 | -0.71 |
| CSK | c-src tyrosine kinase | H90752 | -0.70 |
| ESTs | Unknown | H22578 | -0.70 |
| PNUTL1 | Peanut (Drosophila)-like 1 | AA702163 | -0.65 |
| ALAS2 | Aminolevulinate, delta-, synthase 2 | N54577 | -0.63 |
| PSMB7 | Proteasome subunit, beta type, 7 (large multifunctional protease 7) | N52556 | -0.61 |
| ESTs | Unknown | N52646 | -0.61 |
| DAP-I | Death-associated protein 1 | H65452 | -0.59 |
| FBP1 | Fructose-1,6-bisphosphatase 1 | N57708 | -0.58 |
| DEHUE | Glutamate dehydrogenase | N57779 | -0.58 |
| ACTB | Actin beta | H54441 | -0.58 |
| CSF-3R | CSF3R: colony-stimulating factor 3 receptor (granulocyte) | R31950 | -0.56 |
| TBPG | Trophoblaste glycoprotein | R70262 | -0.56 |
| APOH | Apolipoprotein H | R06433 | -0.54 |
| MYB | v-myb avian myeloblastosis viral oncogene homolog | N49284 | -0.53 |
| NGRFAP1 | Nerve growth factor receptor (TNFRSF16)-associated protein 1 | W68632 | 0.54 |
| CST3 | Cystatin C | W93398 | 0.54 |
| DARS | Aspartyl t-RNA synthetase | H28873 | 0.55 |
| ESTs | Unknown | AA626888 | 0.56 |
| PSG2 | Pregnancy-specific beta-1—glycoprotein 2 | R65579 | 0.57 |
| COL 16A1 | Collagen type XVI alpha 1 | R54968 | 0.57 |
| SRF | Serum response factor (c-fos SRE-binding transcription factor) | W32378 | 0.57 |
| ABCA7 | ATP-binding cassette, subfamily A, member 7 | H45443 | 0.60 |
| ESTs | Unknown | H51280 | 0.61 |
| ARD1 | N acetyltransferase | R55220 | 0.65 |
| FALZ | Fetal Alzheimer antigen | H44955 | 0.66 |
| IGHA | IgA | H42228 | 0.67 |
| FAH | Fumarylacetoacetate hydrolase (fumarylacetoacetase) | H44956 | 0.67 |
| MMP12 | Matrix metalloproteinase 12 (macrophage elastase) | R63637 | 0.70 |
| OR2A19 | Olfactory receptor, family 2, subfamily A, member 19 | H39853 | 0.80 |
| JAG2 | Jagged2 | H39899 | 0.84 |
| ESTs | Unknown | H43142 | 0.84 |
| LIPE | Lipase, hormone sensitive | R87236 | 0.85 |
| WEE1 | Wee 1 like | H44948 | 0.89 |
| ESTs | Unknown | H29761 | 0.97 |
| INPP1 | Inositol polyphosphate-1-phosphatase | H52141 | 1.11 |
| DCTN1 | Dynactin 1 | R88834 | 1.11 |
| ESTs | Unknown | Ai201426 | 1.13 |
| GATA2 | GATA-binding protein 2 | R32406 | 1.39 |
| ESTs | Unknown | H28469 | 1.65 |
| ESTs | Unknown | H43035 | 1.72 |
| AP1B1 | Adaptor-related protein complex 1, beta 1 subunit | R87770 | 1.78 |
Gene symbol . | Gene description . | Accession no. . | Discriminating score . |
|---|---|---|---|
| IGHG3 | IgG | H68233 | -2.75 |
| GATA6 | GATA-binding protein 6 | N91601 | -2.39 |
| IGHG3 | IgG | N92646 | -2.07 |
| IGHG3 | IgG | H64493 | -2.04 |
| HADHB | Hydroxyacyl-CoA dehydrogenase, beta subunit | T69767 | -1.89 |
| IF | I factor (complement) | H89710 | -1.29 |
| BRCA1 | Breast cancer 1, early onset | H90415 | -1.29 |
| ESTs | Unknown | W46567 | -1.10 |
| CSF-1R | Colony-stimulating factor 1 receptor, v-fms oncogene homolog | H57126 | -1.00 |
| ESTs | Unknown | H93533 | -0.98 |
| ITGA1 | Integrin, alpha 1 | H68922 | -0.88 |
| FBG | Fibrinogen, B beta polypeptide | H91714 | -0.88 |
| Id2 | Inhibitor of DNA binding 2, dominant-negative helix-loop-helix | H82442 | -0.85 |
| IL-16 | Interleukin-16 (lymphocyte chemoattractant factor) | H57532 | -0.84 |
| IL-1B | Interleukin-1, beta | W47101 | -0.80 |
| ZNF148 | Zinc finger protein 148 (pHZ-52) | H70711 | -0.79 |
| KLF1 | Kruppel-like factor 1 (erythroid) | H60702 | -0.76 |
| ITGA2B | Integrin, alpha 2b (antigen CD41B) | AA905458 | -0.71 |
| ESTs | Unknown | AA279804 | -0.71 |
| CSK | c-src tyrosine kinase | H90752 | -0.70 |
| ESTs | Unknown | H22578 | -0.70 |
| PNUTL1 | Peanut (Drosophila)-like 1 | AA702163 | -0.65 |
| ALAS2 | Aminolevulinate, delta-, synthase 2 | N54577 | -0.63 |
| PSMB7 | Proteasome subunit, beta type, 7 (large multifunctional protease 7) | N52556 | -0.61 |
| ESTs | Unknown | N52646 | -0.61 |
| DAP-I | Death-associated protein 1 | H65452 | -0.59 |
| FBP1 | Fructose-1,6-bisphosphatase 1 | N57708 | -0.58 |
| DEHUE | Glutamate dehydrogenase | N57779 | -0.58 |
| ACTB | Actin beta | H54441 | -0.58 |
| CSF-3R | CSF3R: colony-stimulating factor 3 receptor (granulocyte) | R31950 | -0.56 |
| TBPG | Trophoblaste glycoprotein | R70262 | -0.56 |
| APOH | Apolipoprotein H | R06433 | -0.54 |
| MYB | v-myb avian myeloblastosis viral oncogene homolog | N49284 | -0.53 |
| NGRFAP1 | Nerve growth factor receptor (TNFRSF16)-associated protein 1 | W68632 | 0.54 |
| CST3 | Cystatin C | W93398 | 0.54 |
| DARS | Aspartyl t-RNA synthetase | H28873 | 0.55 |
| ESTs | Unknown | AA626888 | 0.56 |
| PSG2 | Pregnancy-specific beta-1—glycoprotein 2 | R65579 | 0.57 |
| COL 16A1 | Collagen type XVI alpha 1 | R54968 | 0.57 |
| SRF | Serum response factor (c-fos SRE-binding transcription factor) | W32378 | 0.57 |
| ABCA7 | ATP-binding cassette, subfamily A, member 7 | H45443 | 0.60 |
| ESTs | Unknown | H51280 | 0.61 |
| ARD1 | N acetyltransferase | R55220 | 0.65 |
| FALZ | Fetal Alzheimer antigen | H44955 | 0.66 |
| IGHA | IgA | H42228 | 0.67 |
| FAH | Fumarylacetoacetate hydrolase (fumarylacetoacetase) | H44956 | 0.67 |
| MMP12 | Matrix metalloproteinase 12 (macrophage elastase) | R63637 | 0.70 |
| OR2A19 | Olfactory receptor, family 2, subfamily A, member 19 | H39853 | 0.80 |
| JAG2 | Jagged2 | H39899 | 0.84 |
| ESTs | Unknown | H43142 | 0.84 |
| LIPE | Lipase, hormone sensitive | R87236 | 0.85 |
| WEE1 | Wee 1 like | H44948 | 0.89 |
| ESTs | Unknown | H29761 | 0.97 |
| INPP1 | Inositol polyphosphate-1-phosphatase | H52141 | 1.11 |
| DCTN1 | Dynactin 1 | R88834 | 1.11 |
| ESTs | Unknown | Ai201426 | 1.13 |
| GATA2 | GATA-binding protein 2 | R32406 | 1.39 |
| ESTs | Unknown | H28469 | 1.65 |
| ESTs | Unknown | H43035 | 1.72 |
| AP1B1 | Adaptor-related protein complex 1, beta 1 subunit | R87770 | 1.78 |
Hierarchic clustering of IgH-secreting MM based on the identified genes (49 members) that best distinguished IgG-MM from IgA-MM. Red-colored branches represent IgA-MM, and black branches represent IgG-MM. Genes were grouped in potential functional categories. Each column represents an MM sample, and each row represents an individual gene. A pseudocolor representation of gene expression is shown according to the scale at the bottom; red indicates expression levels greater than the median, and green indicates levels less than the median. Gene accession numbers, discriminating scores, and gene symbols are labeled on the right. Gray indicates excluded values.
Hierarchic clustering of IgH-secreting MM based on the identified genes (49 members) that best distinguished IgG-MM from IgA-MM. Red-colored branches represent IgA-MM, and black branches represent IgG-MM. Genes were grouped in potential functional categories. Each column represents an MM sample, and each row represents an individual gene. A pseudocolor representation of gene expression is shown according to the scale at the bottom; red indicates expression levels greater than the median, and green indicates levels less than the median. Gene accession numbers, discriminating scores, and gene symbols are labeled on the right. Gray indicates excluded values.
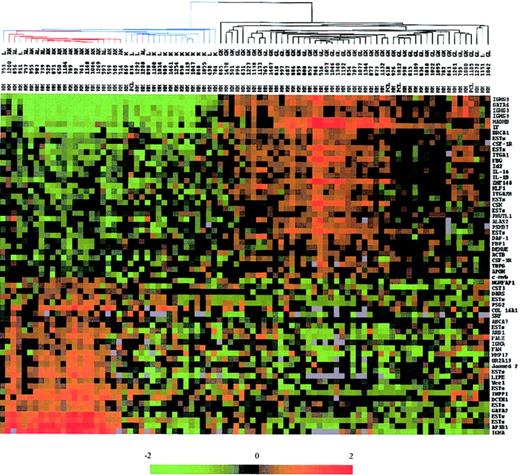
Second, we used the 61 genes that best discriminated IgG- and IgA-MM to investigate whether BJ-MM could represent a distinct group or not. Using hierarchic clustering algorithm, MM samples were organized on the basis of overall resemblance in their gene expression patterns restricted to these genes (Figure 3). The measure of these similarities was provided by a dendrogram that clearly separated the 3 subgroups of MM. The colored gene expression matrix showed that IgA and IgG transcripts were absent in most of the BJ-MMs, indicating that the lack of IgH protein synthesis observed in these patients was due to either abnormalities at the DNA level or RNA level. A recent study combining Southern blot and PCR analysis demonstrated that defects at the DNA level are responsible for the lack of IgH protein production in at least 75% of the BJ-MMs studied.20 Given that 17 of 19 BJ-MM patients we examined have illegitimate rearrangements of the IgH gene (data not shown), our results are also in favor of DNA defects as a major cause of failure to synthesize IgH proteins. Expression level of the other discriminating genes appeared to be globally diminished in almost all BJ-MMs (see colored matrix in Figure 2).
Dendrogram and color matrix representing the hierarchic clustering of the 88 myeloma samples versus 61 genes. The genes used in this analysis were chosen by a discriminating score statistic that is most highly correlated with the 2 IgH isotypes. Blue-colored branches represent BJ-MM, red branches represent IgA-MM, and black branches represent IgG-MM. Columns represent individual MM samples, and rows represent individual genes on the microarray. A pseudocolor representation of gene expression is shown according to the scale at the bottom; red indicates expression levels greater than the median, and green indicates levels less than the median. Gray indicates excluded values.
Dendrogram and color matrix representing the hierarchic clustering of the 88 myeloma samples versus 61 genes. The genes used in this analysis were chosen by a discriminating score statistic that is most highly correlated with the 2 IgH isotypes. Blue-colored branches represent BJ-MM, red branches represent IgA-MM, and black branches represent IgG-MM. Columns represent individual MM samples, and rows represent individual genes on the microarray. A pseudocolor representation of gene expression is shown according to the scale at the bottom; red indicates expression levels greater than the median, and green indicates levels less than the median. Gray indicates excluded values.
Third, to determine whether IgL subtypes could be phenotypically different, we used discriminating score methods to compare gene expression profiles of 73 patients (13 stage I, 7 stage II, 49 stage III, and 4 PCL). Statistical analysis identified 80 different genes (P < .0001) between Igλ- and Igκ-MM (Table 2). As expected, the top-ranked discriminatory genes were Igλ and Igκ. Detailed analysis revealed the presence of several genes essential for bone remodeling, mainly osteoclastogenesis, that were up-regulated in Igκ-MM: Mip-1α, TGFβ3, and BMP2. Conversely, one gene corresponding to a protein that negatively regulates TGFβ activity (LTBP4) was overexpressed in Igλ-MM cases.
Statistically different genes between Igλ-MM and Igκ-MM
Gene symbol . | Gene description . | Accession no. . | Discriminating score . |
|---|---|---|---|
| IGL@ | Ig lambda | H14524 | -2.74 |
| ESTs | Unknown | H26676 | -2.56 |
| ESTs | Unknown | H26661 | -1.94 |
| IGL@ | Ig lambda | R83196 | -1.81 |
| ESTs | Unknown | H15899 | -1.80 |
| CDSN | Corneodesmosin | W95594 | -1.77 |
| MCP | Membrane cofactor protein (CD46) | H26673 | -1.66 |
| ESTs | Unknown | R83001 | -1.63 |
| IGL@ | Ig lambda | H15030 | -1.62 |
| ESTs | Unknown | R74030 | -1.33 |
| ABCD4 | ATP-binding cassette, subfamily D, member 4 | H51632 | -1.22 |
| SAMHD1 | SAM domain and HD domain, 1 | H47862 | -1.07 |
| LTBP4* | Latent transforming growth factor-beta-binding protein 4 | R73631 | -1.02 |
| ESTs | Unknown | R91051 | -1.02 |
| ESTs | Unknown | R89772 | -0.99 |
| LMNA | LaminA/C | H26659 | -0.98 |
| IGL@ | Ig lambda | H41911 | -0.95 |
| ESTs | Unknown | R16095 | -0.91 |
| ZNF361 | Zinc finger protein 361 | R73795 | -0.90 |
| LTA | Lymphotoxin-alpha | AA910185 | -0.81 |
| RPL4 | Ribosomal protein L4 | N35801 | -0.77 |
| ESTs | Unknown | N35710 | -0.66 |
| FMO5 | Flavin-containing monooxygenase 5 | H51750 | -0.65 |
| ESTs | Unknown | R72642 | -0.64 |
| MXI1 | Max-interacting protein 1 | AA115514 | -0.63 |
| SCN1B | Sodium channel, voltage-gated, type 1, beta subunit | R74526 | -0.61 |
| ARHG | Ras homolog gene family, member G | H45512 | -0.58 |
| NTE | Neuropathy target esterase | H09751 | -0.57 |
| HVEB | Herpesvirus entry mediator B | R73625 | -0.56 |
| AXL | AXL receptor tyrosine kinase | H09737 | -0.52 |
| GAR22 | GAS2-related on chromosome 22 | R72509 | -0.51 |
| PBX1 | Pre-B-cell leukemia transcription factor 1 | AA403031 | -0.51 |
| HYAL1 | Hyaluronoglucosaminidase 1 | W84634 | -0.50 |
| PRKCI | Protein kinase C, iota form | AA062633 | 0.50 |
| PLXNB3 | Plexin B 3 | R41456 | 0.50 |
| CTSB | Cathepsin B | H98635 | 0.51 |
| CSNK2A2 | Casein kinase II, alpha-2 | AA099405 | 0.53 |
| MMP12 | Matrix metalloproteinase 12 | R63637 | 0.54 |
| ACK1 | Human activated p21 cdc42Hs kinase | R44803 | 0.54 |
| ESTs | Unknown | AA883518 | 0.55 |
| EDN3 | Endothelin 3 | AA553611 | 0.58 |
| ESTs | Unknown | W74500 | 0.59 |
| CUL4A | Cullin 4A | AA100650 | 0.59 |
| DDR1 | Discoidin domain receptor family, member 1 | AA574033 | 0.60 |
| Mip-1α* | Macrophage inflammatory protein 1-alpha (SCYA3) | W74288 | 0.64 |
| ESTs | Unknown | T86845 | 0.64 |
| NRID1 | Nuclear receptor subfamily 1, group D | R85515 | 0.66 |
| UBE4A | Ubiquitination factor E4A | R45238 | 0.66 |
| FGR | Gardner-Rasheed feline sarcoma viral oncogene homolog | W81586 | 0.66 |
| SIAH2 | Seven in absentia, drosophila, homolog of 2 | R61685 | 0.68 |
| ESTs | Unknown | AA548364 | 0.71 |
| TGFβ3* | Transforming growth factor, beta-3 | W80655 | 0.73 |
| ZNF198 | Zinc finger protein 198 | N71855 | 0.81 |
| TJP2 | Tight junction protein 2 (zona occludens 2) | H05082 | 0.82 |
| CEP3 | cdc42 effector protein 3 | R43949 | 0.86 |
| ESTs | Unknown | H89735 | 0.87 |
| GSTM5 | Glutathione S-transferase, mu-5 | R40442 | 0.89 |
| ESTs | Unknown | N58796 | 0.91 |
| IGKC | Ig kappa | AA284839 | 0.92 |
| UBE3A | Ubiquitin-protein ligase E3A | AA126792 | 0.97 |
| DSG1 | Desmoglein 1 | W72927 | 0.97 |
| RECQL5 | RECQ protein-like 5 | R32075 | 1.04 |
| BPHL | Biphenyl hydrolase-like | AA167197 | 1.04 |
| ELAVL4 | Embryonic lethal, abnormal vision, drosophila, homolog-like 4 | R55730 | 1.06 |
| TNFRSF6 | Tumor necrosis factor receptor superfamily, member 6 (CD95) | AA031300 | 1.06 |
| NR2C1 | Nuclear receptor subfamily 2, group C, member 1 | H68838 | 1.10 |
| SMARCA1 | Actin-dependent regulator of chromatin, subfamily A, member 1 | AA057875 | 1.12 |
| NFE2L3 | Nuclear factor erythroid 2-like 3 | R43198 | 1.17 |
| CYB5 | Methemoglobinemia due to deficiency of cytochrome b5 | N23249 | 1.19 |
| LAMB3 | Laminin, beta-3 | AA622206 | 1.19 |
| SH3GL2 | SH3 domain, GRB2-like 2 (endophilin 1) | R20729 | 1.20 |
| HSD17B2 | 17-@beta-hydroxysteroid dehydrogenase II | N23665 | 1.30 |
| BMP2* | Bone morphogenetic protein 2 | AA114112 | 1.34 |
| PRKCABP | PRKCA-binding protein | AA641722 | 1.54 |
| HOX11 | Homeobox 11 | AA007444 | 1.61 |
| ESTs | Unknown | N30860 | 1.66 |
| ESTs | Unknown | AA826328 | 1.69 |
| EXT1 | Exostoses, multiple, type 1 | R13402 | 1.70 |
| ESTs | Unknown | W92915 | 1.79 |
| IGKC | Ig kappa | R71916 | 1.82 |
Gene symbol . | Gene description . | Accession no. . | Discriminating score . |
|---|---|---|---|
| IGL@ | Ig lambda | H14524 | -2.74 |
| ESTs | Unknown | H26676 | -2.56 |
| ESTs | Unknown | H26661 | -1.94 |
| IGL@ | Ig lambda | R83196 | -1.81 |
| ESTs | Unknown | H15899 | -1.80 |
| CDSN | Corneodesmosin | W95594 | -1.77 |
| MCP | Membrane cofactor protein (CD46) | H26673 | -1.66 |
| ESTs | Unknown | R83001 | -1.63 |
| IGL@ | Ig lambda | H15030 | -1.62 |
| ESTs | Unknown | R74030 | -1.33 |
| ABCD4 | ATP-binding cassette, subfamily D, member 4 | H51632 | -1.22 |
| SAMHD1 | SAM domain and HD domain, 1 | H47862 | -1.07 |
| LTBP4* | Latent transforming growth factor-beta-binding protein 4 | R73631 | -1.02 |
| ESTs | Unknown | R91051 | -1.02 |
| ESTs | Unknown | R89772 | -0.99 |
| LMNA | LaminA/C | H26659 | -0.98 |
| IGL@ | Ig lambda | H41911 | -0.95 |
| ESTs | Unknown | R16095 | -0.91 |
| ZNF361 | Zinc finger protein 361 | R73795 | -0.90 |
| LTA | Lymphotoxin-alpha | AA910185 | -0.81 |
| RPL4 | Ribosomal protein L4 | N35801 | -0.77 |
| ESTs | Unknown | N35710 | -0.66 |
| FMO5 | Flavin-containing monooxygenase 5 | H51750 | -0.65 |
| ESTs | Unknown | R72642 | -0.64 |
| MXI1 | Max-interacting protein 1 | AA115514 | -0.63 |
| SCN1B | Sodium channel, voltage-gated, type 1, beta subunit | R74526 | -0.61 |
| ARHG | Ras homolog gene family, member G | H45512 | -0.58 |
| NTE | Neuropathy target esterase | H09751 | -0.57 |
| HVEB | Herpesvirus entry mediator B | R73625 | -0.56 |
| AXL | AXL receptor tyrosine kinase | H09737 | -0.52 |
| GAR22 | GAS2-related on chromosome 22 | R72509 | -0.51 |
| PBX1 | Pre-B-cell leukemia transcription factor 1 | AA403031 | -0.51 |
| HYAL1 | Hyaluronoglucosaminidase 1 | W84634 | -0.50 |
| PRKCI | Protein kinase C, iota form | AA062633 | 0.50 |
| PLXNB3 | Plexin B 3 | R41456 | 0.50 |
| CTSB | Cathepsin B | H98635 | 0.51 |
| CSNK2A2 | Casein kinase II, alpha-2 | AA099405 | 0.53 |
| MMP12 | Matrix metalloproteinase 12 | R63637 | 0.54 |
| ACK1 | Human activated p21 cdc42Hs kinase | R44803 | 0.54 |
| ESTs | Unknown | AA883518 | 0.55 |
| EDN3 | Endothelin 3 | AA553611 | 0.58 |
| ESTs | Unknown | W74500 | 0.59 |
| CUL4A | Cullin 4A | AA100650 | 0.59 |
| DDR1 | Discoidin domain receptor family, member 1 | AA574033 | 0.60 |
| Mip-1α* | Macrophage inflammatory protein 1-alpha (SCYA3) | W74288 | 0.64 |
| ESTs | Unknown | T86845 | 0.64 |
| NRID1 | Nuclear receptor subfamily 1, group D | R85515 | 0.66 |
| UBE4A | Ubiquitination factor E4A | R45238 | 0.66 |
| FGR | Gardner-Rasheed feline sarcoma viral oncogene homolog | W81586 | 0.66 |
| SIAH2 | Seven in absentia, drosophila, homolog of 2 | R61685 | 0.68 |
| ESTs | Unknown | AA548364 | 0.71 |
| TGFβ3* | Transforming growth factor, beta-3 | W80655 | 0.73 |
| ZNF198 | Zinc finger protein 198 | N71855 | 0.81 |
| TJP2 | Tight junction protein 2 (zona occludens 2) | H05082 | 0.82 |
| CEP3 | cdc42 effector protein 3 | R43949 | 0.86 |
| ESTs | Unknown | H89735 | 0.87 |
| GSTM5 | Glutathione S-transferase, mu-5 | R40442 | 0.89 |
| ESTs | Unknown | N58796 | 0.91 |
| IGKC | Ig kappa | AA284839 | 0.92 |
| UBE3A | Ubiquitin-protein ligase E3A | AA126792 | 0.97 |
| DSG1 | Desmoglein 1 | W72927 | 0.97 |
| RECQL5 | RECQ protein-like 5 | R32075 | 1.04 |
| BPHL | Biphenyl hydrolase-like | AA167197 | 1.04 |
| ELAVL4 | Embryonic lethal, abnormal vision, drosophila, homolog-like 4 | R55730 | 1.06 |
| TNFRSF6 | Tumor necrosis factor receptor superfamily, member 6 (CD95) | AA031300 | 1.06 |
| NR2C1 | Nuclear receptor subfamily 2, group C, member 1 | H68838 | 1.10 |
| SMARCA1 | Actin-dependent regulator of chromatin, subfamily A, member 1 | AA057875 | 1.12 |
| NFE2L3 | Nuclear factor erythroid 2-like 3 | R43198 | 1.17 |
| CYB5 | Methemoglobinemia due to deficiency of cytochrome b5 | N23249 | 1.19 |
| LAMB3 | Laminin, beta-3 | AA622206 | 1.19 |
| SH3GL2 | SH3 domain, GRB2-like 2 (endophilin 1) | R20729 | 1.20 |
| HSD17B2 | 17-@beta-hydroxysteroid dehydrogenase II | N23665 | 1.30 |
| BMP2* | Bone morphogenetic protein 2 | AA114112 | 1.34 |
| PRKCABP | PRKCA-binding protein | AA641722 | 1.54 |
| HOX11 | Homeobox 11 | AA007444 | 1.61 |
| ESTs | Unknown | N30860 | 1.66 |
| ESTs | Unknown | AA826328 | 1.69 |
| EXT1 | Exostoses, multiple, type 1 | R13402 | 1.70 |
| ESTs | Unknown | W92915 | 1.79 |
| IGKC | Ig kappa | R71916 | 1.82 |
Genes known to be involved in bone remodeling.
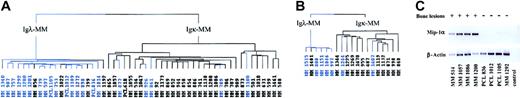
Because we found several factors known to stimulate osteoclast formation, we wanted to determine whether we could distinguish MM with an increased osteoclast activity from the others. Among the 73 patients previously analyzed, we selected patients with high myeloma cell mass and known score of bone lesions (49 stage III and 4 PCL) and applied hierarchic clustering algorithm according to their expression of the 80 genes that significantly discriminated between Igκ-MM and Igλ-MM. The sample dendrogram distinguished 2 branches (Figure 4A): the left branch grouped 16 of the 21 MMs showing absence or limited lesions (ie, score 0, 1, or 2 according to Durie and Salmon11 staging system), and the right branch captured 28 of the 32 MMs having multiple bone lesions (ie, score 3 according to Durie and Salmon staging system). Thus, the IgL signature clearly separated MM presenting aggressive bone destruction from the others (χ2 test, P < .001). This gene signature was then tested in a new group of 19 patients (validation group) with stage III MM (6 Igλ-MM and 13 Igκ-MM). The dendrogram (Figure 4B) assigned 5 of 7 MMs without bone disease on the left branch and 11 of 12 MMs with bone disease on the right branch (P < .001). This result validates the relationship between bone disease and light chain cluster.
Dendrogam representing the hierarchic clustering of MM patients in stage III and PCL with known bone lesions; score based on the genes of Igλ expression signature. (A) Hierarchic tree of 53 MMs (preliminary group). (B) Hierarchic tree of 19 MMs (validation group). Blue-colored branches represent MMs showing absence or limited bone lesions; black branches represent MMs displaying multiple bone lesions. (C) RT-PCR experiments showing Mip-1α mRNA expression in 8 patients, labeled (+) for multiple bone lesions or (–) for absence of or limited bone lesions. Control indicates no template in the PCR reaction.
Dendrogam representing the hierarchic clustering of MM patients in stage III and PCL with known bone lesions; score based on the genes of Igλ expression signature. (A) Hierarchic tree of 53 MMs (preliminary group). (B) Hierarchic tree of 19 MMs (validation group). Blue-colored branches represent MMs showing absence or limited bone lesions; black branches represent MMs displaying multiple bone lesions. (C) RT-PCR experiments showing Mip-1α mRNA expression in 8 patients, labeled (+) for multiple bone lesions or (–) for absence of or limited bone lesions. Control indicates no template in the PCR reaction.
In addition, Mip-1α gene expression data obtained with DNA microarray experiments were confirmed by reverse transcriptase–polymerase chain reaction (RT-PCR) analysis (Figure 4C). Interestingly, IL-1β, another factor involved in the increase of osteoclast formation and bone destruction in MM,21 is differentially expressed according to Ig isotype but not according to Ig light chain subtype, supporting the view that IL-1β protein could not be produced by malignant PCs as previously emphasized.22-24
Discussion
Gene expression profiling represents a novel molecular approach to examine MM pathogenesis. Using this technology, we addressed questions regarding the marked heterogeneity of MM: do different biologic phenotypes explain divergent clinical courses? Expression profiles of highly purified malignant PCs from 88 MM and 4 PCL patients revealed that specific transcriptional programs are associated with the Ig types and light chain subtypes. Careful analysis of the differentially expressed genes revealed molecular portraits related to disease presentation.
The most discriminating gene between IgG- and IgA-MM, apart from IgH genes, is the transcription factor GATA6, which belongs to the GATA zinc finger family and plays an important role in lung epithelium development.25 Recently it has been demonstrated that GATA6 regulates WNT7B,19 a member of the WNT family that is up-regulated in malignant breast tissue26 and in bladder tumors.27 Several members of the WNT signaling pathway (WNT5A, WNT10B, FRZB) have been shown to be deregulated in MM.28,29 Given the key role played by the WNT signaling pathway in carcinogenesis and embryology,30,31 we can hypothesize that over-expression of these growth factors in MM may play a role in the pathogenesis of MM.
Analysis of genes included in the immune response cluster is of particular interest. One of the most discriminating genes is Id2, a member of the Id class of the helix-loop-helix proteins. Id proteins can act as negative regulators of the class I helix-loop-helix proteins (E proteins), which are known to play a crucial role in lymphocyte development and activity.32,33 Furthermore, ectopic expression of Id3 in mature activated B cells interferes with IgA expression.34 These data are in agreement with our results that showed a relative underexpression of Id2 in IgA-MM. Because the ratio of E proteins to Id proteins appears to be important for later stages of B-cell development, it is important to determine their respective expression in MM. Another gene of interest in this cluster is IL-16. Our DNA microarray data showed that IL-16 was up-regulated in IgG-MM patients. Its expression in purified myeloma cells has been recently reported.35 IL-16 has been previously described in a variety of cells including CD4+, CD8+ T cells, dendritic cells, and CD19+ B cells.36,37 IL-16 is a ligand of CD4 and induces in vitro chemotaxis of CD4+ T cells,38 and CD4 dendritic cells,36 and CD8+ T cells.39 Furthermore, IL-16 markedly inhibits CD3-induced T-cell activation, but this effect is limited to CD4+ T cells.40 Recently, Koike et al41 have shown that IL-16 serum level of stage III MM was significantly higher than that of normal controls. In addition, the authors found a correlation between IL-16 levels and CD4/CD8 ratio. Taken together, these observations suggest that malignant PCs could be a source of IL-16 and that deregulated expression of IL-16 could contribute to change of the T-cell phenotype in this disease.42,43
Among the 3 cell cycle regulators (c-myb, BRCA1, and ZNF148/ZBP-89) overexpressed in IgG-MM, 2 of them, BRCA1 and ZNF148, have been shown to bind to the C terminus of p53 and stabilized specifically the wild-type form.44-46 Furthermore, overex-pression of ZNF148 induced growth arrest and apoptosis.44 It may be highly interesting to investigate the functional role of BRCA1 and ZNF148 in the pathogenesis of MM. We can speculate that accumulation of wild-type p53 in malignant PCs could increase efficacy to radiotherapy and chemotherapy, explaining in part the better prognosis of IgG-MM compared with IgA-MM.47,48
Two genes involved in Notch signaling are overexpressed in IgA-MM samples compared with IgG-MM samples, GATA2 and Jagged2. Maintenance of GATA2 expression is necessary for Notch signaling in hematopoietic cells.49 Jagged2, the ligand for Notch1,50 was previously found to be up-regulated in MM cell lines as compared with EBV (Epstein-Barr virus)–immortalized polyclonal B cells derived from the same patient.29 Because Notch signaling is known to inhibit differentiation of hematopoietic cells,51 overexpression of GATA2 and Jagged2 could favor the maintenance of malignant immature cells in IgA-MM. Another gene overexpressed in IgA-MM, SRF, is of interest because it regulates an antiapoptotic member of the Bcl-2 family, Mcl-1.52 Very recent results from our laboratory and others demonstrated that Mcl-1 is required for the survival of MM cells.53,54 Conversely, an inducer of apoptosis, DAP-1, is down-regulated in the same group of patients. DAP-1 is a member of the ubiquitin-homology proteins that specifically interact with the death domain of TNF-R1 and induce apoptosis in a variety of cell lines.55 This deregulated balance between proapoptotic and antiapoptotic molecules (in favor of antiapoptotic signals) may explain, at least in part, the lesser chemosensitivity of IgA-MM.
Our analysis of gene expression data of IgH-MM subtypes revealed that genes associated with inhibition of differentiation and apoptosis were up-regulated in IgA-MM, while genes associated with immune response, cell cycle control, and apoptosis induction were down-regulated in this subgroup of MM. These findings were consistent with previous studies demonstrating that the IgA isotype is significantly associated with a shorter survival.47,48,56 In addition, the fact that BJ-MM patients have strongly diminished IgA and IgG gene signatures suggests that transcriptional programs related to IgH class switch recombination are maintained up to differentiation of activated B cells into PCs in the bone marrow.
The presence of several genes known to be involved in bone remodeling, mainly Mip-1α 24,57-59 but also TGFβ3,60-62 BMP2,63 and LTBP4,64 in the cluster of genes differentiating Igκ-MM and Igλ-MM (indicated by asterisks in Table 2) led us to focus our analysis on clinical presentation and IgL subtypes. Using the IgL discriminating cluster of genes to classify the MM patients, we found a strong association between patients lacking bone lesions and Igλ subtype. Our results confirmed previous studies showing that (1) almost all osteosclerotic MM, without or with the paraneoplastic syndrome (POEMS [polyneuropathy, organomegaly, endocrinopathy, monoclonal gammopathy, and skin lesions]), are Igλ-MM65 ; (2) patients unable to destroy bones even at the end stage of the disease are osteoblastic MM at the histologic level and are Igλ-MM66 ; and (3) two thirds of Igλ-MMs reach high cell mass at diagnosis without lytic bone involvement in contrast to Igκ-MM.66 Of major interest, our DNA microarray and RT-PCR data revealed a correlation between increase of Mip-1α mRNA and severity of bone destruction associated with Igκ subtype. Similarly, very recent results reported by Abe et al59 showed an increase of Mip-1α secretion in highly purified MM plasma cells compared with normal PCs. Given that Mip-1α induced in vitro osteoclast formation in human bone marrow cultures58 and that highly purified PCs of MM patients with active bone disease induced bone resorption in vitro,59 the overall data support the view that Mip-1α plays a major role in vivo in MM-induced bone resorption and that osteolytic activity found in Igκ-MM is more likely related to an excessive bone resorption than to a decrease of bone formation. The next step will be the identification of specific transcription factors able to induce Mip-1α expression in Igκ-MM rather than in Igλ-MM. Because Mip-1α promoter contains GATA2 plus AML-1 plus C/EBPα regulatory regions,67 these transcription factors appear as good candidates to regulate Mip-1α expression in MM.
In conclusion, our attempt to examine heterogeneity of MM by using the DNA microarray approach led us to identify genes whose deregulated expression was associated with the pathogenesis of MM. In addition, our results provide an explanation for the association between Igκ-MM and bone destruction. The major goal of this global approach is to establish a gene expression–based survival predictor for the newly diagnosed patients included within clinical trials of the Intergroupe Francophone du Myélome.
Prepublished online as Blood First Edition Paper, March 6, 2003; DOI 10.1182/blood-2002-11-3385.
S.M. and R.H. equally supervised this work.
Supported by grants from the International Myeloma Foundation and from the Ligue Nationale contre le Cancer (Equipe labellisée 2001 et Progamme Carte d'Identité des Tumeurs).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal