Abstract
Histone acetylation modulates gene expression, cellular differentiation, and survival and is regulated by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs). HDAC inhibition results in accumulation of acetylated nucleosomal histones and induces differentiation and/or apoptosis in transformed cells. In this study, we characterized the effect of suberoylanilide hydroxamic acid (SAHA), the prototype of a series of hydroxamic acid–based HDAC inhibitors, in cell lines and patient cells from B-cell malignancies, including multiple myeloma (MM) and related disorders. SAHA induced apoptosis in all tumor cells tested, with increased p21 and p53 protein levels and dephosphorylation of Rb. We also detected cleavage of Bid, suggesting a role for Bcl-2 family members in regulation of SAHA-induced cell death. Transfection of Bcl-2 cDNA into MM.1S cells completely abrogated SAHA-induced apoptosis, confirming its protective role. SAHA did not induce cleavage of caspase-8, -9, or -3 in MM.1S cells during the early phase of apoptosis, and the pan-caspase inhibitor ZVAD-FMK did not protect against SAHA. Conversely, poly(ADP)ribose polymerase (PARP) was cleaved in a pattern indicative of calpain activation, and the calpain inhibitor calpeptin abrogated SAHA-induced cell death. Importantly, SAHA sensitized MM.1S cells to death receptor–mediated apoptosis and inhibited the secretion of interleukin 6 (IL-6) induced in bone marrow stromal cells (BMSCs) by binding of MM cells, suggesting that it can overcome cell adhesion–mediated drug resistance. Our studies delineate the mechanisms whereby HDAC inhibitors mediate anti-MM activity and overcome drug resistance in the BM milieu and provide the framework for clinical evaluation of SAHA, which is bioavailable, well tolerated, and bioactive after oral administration, to improve patient outcome.
Introduction
Histone acetylation regulates transcription by altering chromatin structure. Acetylation of core nucleosomal histones neutralizes the positive charge on lysine residues and disrupts nucleosome structure, allowing unfolding of the associated DNA, access by transcription factors, and changes in gene expression.1-5 Chromatin composed of nucleosomes in which the histones have low levels of acetylation on lysine residues of their amino-terminal tails is generally transcriptionally silent. The turnover of histone acetylation is regulated by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs). HDACs catalyze the removal of an acetyl group from the ε-amino group of lysine side chains of the core nucleosomal histones (H2A, H2B, H3, and H4), thereby reconstituting the positive charge on the lysine. HDAC activity is generally associated with transcriptional repression. Importantly, dysregulated HAT or HDAC activity has been associated with the development of certain human cancers. For example, deletions and inactivating mutations have been detected in HAT genes in several types of cancer,6-9 and individuals with the Rubinstein-Taybi syndrome, a developmental disorder caused by germ-line mutations in theCBP gene that inactivate its HAT activity, have an increased risk of cancer.10 In acute promyelocytic leukemia, the oncoprotein produced by the fusion of promyelocytic leukemia protein (PML) and retinoic acid receptor appears to suppress specific gene transcription through aberrant recruitment of HDACs.11 In this manner, the neoplastic cell is unable to undergo differentiation, leading to excess proliferation.
Several compounds have been shown to act as HDAC inhibitors (HDACIs) with low potency, resulting in the accumulation of acetylated histones and an increase in transcriptionally active chromatin.12-15 For example, butyrates induce tumor cell apoptosis and differentiation, albeit at very high (millimolar) concentrations, at which they also may have effects on other systems.12 The anticonvulsant valproic acid also has HDACI activity at high concentrations.16 Trichostatin A (TSA), originally developed as an antifungal agent, has HDACI activity at nanomolar concentrations.12 Depsipeptide (FR901228), a fermentation product isolated from Chromobacterium violaceum, has been used in a phase 2 trial in patients with refractory neoplasms and demonstrated biologic activity.17
Importantly, synthetic hydroxamic acid–based hybrid polar compounds represent novel, highly active HDACIs that cause accumulation of acetylated histones in cultured cells, induce differentiation and/or apoptosis of transformed cells in culture,1-4,9,18,19 and inhibit the growth of tumors in animals.15,19 The prototype of this class of compounds is suberoylanilide hydroxamic acid (SAHA), which binds directly to the catalytic site of HDAC and potently inhibits its enzymatic activity.20 SAHA selectively induces the expression of specific genes such as p21WAF1/CIP1cyclin–dependent kinase inhibitor to effect cell-cycle arrest.21,22 Because the growth-suppressive and apoptotic activity of these agents are restricted to transformed cells,9 HDAC inhibitors represent promising novel anticancer agents.
Ongoing clinical evaluation of SAHA in patients with solid tumors has revealed that orally administered SAHA is bioavailable; biologically active, as evidenced by histone acetylation in vivo; and well tolerated.23-25 In this study we characterized the effect of SAHA on cell lines and freshly isolated tumor cells from patients with B-cell malignancies. SAHA induced apoptosis in all cell lines and primary patient tumor cells tested, associated with accumulation of p21 and p53, and cleavage of Bid. SAHA-induced apoptosis was caspase independent and calpain dependent. Importantly, SAHA also overcame tumor cell adhesion–mediated and cytokine-induced drug resistance in the bone marrow (BM) microenvironment. These studies provide the framework for the clinical evaluation of SAHA to overcome clinical drug resistance and improve patient outcome in multiple myeloma (MM).
Materials and methods
Cell lines and patient cells
Dexamethasone (Dex)–sensitive MM.1S and Dex-resistant MM.1R human MM cell lines were kindly provided by Dr Steven Rosen (Northwestern University, Chicago, IL). RPMI-8226/S cells and its sublines resistant to doxorubicin (Dox40)26 and melphalan (LR5)27 were a kind gift from Dr William Dalton (H. Lee Moffit Cancer Center, Tampa, FL). LP1 cells were a kind gift from Dr P. Leif Bergsagel (Weill Medical College of Cornell University, New York, NY). ARP-1 cells were obtained by Dr O. Bajenova (Boston University, MA). S6B45 cells were kindly provided by Dr T. Kishimoto (Osaka University, Japan). ARH-77 and IM-9 Epstein-Barr virus (EBV)–transformed B-cell lines, MCF7 breast carcinoma cell line, as well as LNCaP and DU145 prostate carcinoma cell lines were obtained from the American Type Culture Collection (Manassas, VA). The WM cell line WSU-WM was obtained from Dr Ayad Al-Katib (Wayne State University, Detroit, MI).
Bone marrow mononuclear cells (BMMCs) of 5 patients with MM and 3 patients with Waldenström macroglobulinemia (WM) were processed by flow cytometric cell sorting in an EPICS cell sorter (Beckman Coulter, Hialeah, FL), and tumor cells (with > 95% purity in CD38+CD138+ for myeloma cells or CD19+CD20+ and light chain–restriction for WM cells, respectively) were obtained.
All cells were cultured in RPMI 1640 medium (GIBCO Laboratories, Grand Island, NY) supplemented with 10% charcoal dextran–treated fetal bovine serum (FBS; Hyclone, Logan, UT) as well asl-glutamine, penicillin, and streptomycin (GIBCO).
Histone acetylation
MM.1S cells were cultured with 5 μM SAHA or vehicle for 4 and 8 hours in medium containing 10% FBS. Histones were then isolated as previously described.19 Briefly, the cells were harvested in ice-cold phosphate-buffered saline (PBS), centrifuged at 1000 rpm for 5 minutes, resuspended in 1 mL histone lysis buffer (8.6% sucrose, 1% Triton X-100, 50 mM sodium bisulfite, 10 mM Tris (tris(hydroxymethyl)aminomethane)–HCl [pH 6.5], and 10 mM MgCl2) and Dounce homogenized. Cell lysates were centrifuged at 700 rpm for 5 minutes. The nuclear pellet then was washed 3 times with the lysis buffer and once with 10 mM Tris-HCl (pH 7.4) and 13 mM EDTA (ethylenediaminetetraacetic acid). Histones were acid extracted from the nuclear pellets as described previously.28 Acetylation of core histones in the cells or tumors was determined by Western blotting using rabbit polyclonal antibodies against acetylated histone H3 and H4 (Upstate Biotechnology, Lake Placid, NY) and visualized using enhanced chemiluminescence (ECL).
Propidium iodide staining.
For cell cycle analysis, 1 × 106 MM cells were incubated with or without 5 μM SAHA in 10% fetal calf serum (FCS) for 24, 48, 72, or 96 hours. The cells were then washed twice with PBS, permeabilized with 70% ethanol in PBS for 30 minutes at 4°C, incubated with 0.5 mL of a 50-μg/mL propidium iodide (PI) solution containing 20 U/mL RNaseA (Boehringer Mannheim, Indianapolis, IN) for 30 minutes, and analyzed by flow cytometry.
BrdU incorporation assay.
Cell proliferation in MM cells treated with SAHA was quantified by measuring the amount of bromodeoxyuridine (BrdU) incorporated into nuclear DNA, using the BrdU Cell Proliferation Assay (QIA58) (Oncogene Research, Cambridge, MA) according to the instructions of the manufacturer. MM.1S cells were plated and treated with SAHA (2.5, 5, and 10 μM for 24 hours). At the end of this incubation, BrdU was added as indicated for 4 additional hours and incorporated into the DNA of dividing cells. Then the cells were fixed and permeabilized using the Fixative/Denaturing Solution (30 minutes at room temperature). The incorporation of BrdU was quantified using an anti-BrdU antibody, followed by a horseradish peroxidase (HRP)–conjugated secondary antibody and exposure to substrate solution in the dark at room temperature for 15 minutes. The absorbance was measured using a plate reader at 450 nm (540-nm reference wavelength). Cell proliferation was expressed as a percentage of the value of control cells (average ± SD).
MTT colorimetric survival assay
Cell survival was examined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma Chemical, St Louis, MO) colorimetric assay, as previously described.29 Cells were plated in 48-well plates at 70% to 80% confluence and then treated as indicated. At the end of each treatment, cells were incubated with 1 mg/mL MTT for 4 hours at 37°C; a mixture of isopropanol and 1 N HCl (23:2, vol/vol) was then added under vigorous pipetting to dissolve the formazan crystals. Dye absorbance (A) in viable cells was measured at 570 nm, with 630 nm as a reference wavelength. Cell viability was estimated as a percentage of the value of untreated controls. All experiments were repeated at least 3 times, and each experimental condition was repeated at least in quadruplicate wells in each experiment. Data reported are average values ± SD of representative experiments.
LDH release assay
Quantification of cell death was also performed by measuring the activity of lactate dehydrogenase (LDH) released from the cytosol of damaged cells into the culture supernatant, using the Cytotoxicity Detection Kit (LDH) (Roche Molecular Biochemicals, Indianapolis, IN), according to the instructions of the manufacturer. MM cells were preincubated with the pan-caspase inhibitor ZVAD-FMK (30 μM; Oncogene Research), or the calpain inhibitor calpeptin (50 μM; Oncogene Research) for 1 hour prior to exposure to SAHA (5 μM for 36 hours).
Immunoblotting analysis
Immunoblotting analysis was performed as previously described.29 The antibodies used were as follows: mouse monoclonal antibodies for Bcl-2 (sc-7382), BclxL (sc-8392), Bax (sc-20067), tubulin (sc-5286), and p53 (sc-98), as well as polyclonal antibodies for p27, A1 (sc-8351), caspase-3 (sc-7148), and caspase-9 (sc-8355) (Santa Cruz Biotechnology, Santa Cruz, CA); monoclonal antibody for caspase-8 (clone 5F7) and polyclonal antibodies for Fas receptor–associated death domain (FADD)–like interleukin 1 (IL-1) β-converting enzyme (FLICE) inhibitory protein (FLIP; no. 06-697) and total retinoblastoma (Rb; no. 05-377) from Upstate Biotechnologies; monoclonal antibody for poly(ADP)ribose polymerase (PARP; no. SA-250) from Biomol (Plymouth Meeting, PA); polyclonal antisera against cellular inhibitor of apoptosis protein 2 (cIAP-2; no. AF817) and X-linked inhibitor of apoptosis protein (XIAP; no. AF822) (R&D Systems, Minneapolis, MN); monoclonal antibody for p21 (no. OP64) and polyclonal antiserum against survivin (no. PC527) (Oncogene Research); polyclonal antiserum against phospho-Rb (no. 9308S) and Bid (no. 2002) (Cell Signaling, Beverly, MA); Complete-TM mixture of proteinase inhibitors and sodium dodecyl sulfate (SDS; Life Technologies, Gaithersburg, MD); and the enhanced chemiluminescence (ECL) kit, which includes the peroxidase-labeled antimouse and antirabbit secondary antibodies (Amersham, Arlington Heights, IL).
Caspase cleavage analysis
The involvement of caspases in SAHA-induced apoptosis was studied by evaluating the levels of procaspases-8, -9, and -3, as well as the emergence of their cleaved active forms, by immunoblotting of lysates of cells treated with SAHA (5 μM) for 0 to 24 hours. Treatment with TRAIL/Apo2L (Immunex, Seattle, WA; 300 ng/mL for 5 hours) served as a positive control.
Effect of SAHA in MM.1S cells overexpressing Bcl-2
To evaluate the role of the antiapoptotic molecule Bcl-2 in SAHA-induced apoptosis, MM.1S cells were stably transfected with vectors carrying the Bcl-2 cDNA (Upstate Biotechnologies), or the empty (neo) vector using Lipofectamine 2000 according to the instructions of the manufacturer. Forty-eight hours later, the cells were incubated in growth medium containing G418 (500 μg/mL; Life Technologies) to select pools of stable clones, that were subsequently treated with SAHA (2.5-10 μM for 36 hours). The overexpression of Bcl-2 in transfected cells has been confirmed by immunoblotting.30
Release of IL-6 from bone marrow mononuclear cells (BMSCs)
MM cell adhesion to BMSCs induces IL-6 secretion from the latter,31 which is an important growth factor for MM cells.32 We therefore investigated the effect of SAHA on this phenomenon. BMSCs were grown on 24-well plates to confluency. Following washings with serum-free medium, MM.1S cells were added to the BMSC-coated or control wells as described previously31and incubated for 24 hours in the presence or absence of SAHA. The supernatants were collected and assayed for IL-6 concentration by enzyme-linked immunosorbent assay (ELISA; R&D Systems).
Statistical analysis
Statistical significance was examined by a 2-way analysis of variance, followed by Duncan post hoc test. In all analyses,P < .05 was considered statistically significant.
Results
SAHA induces accumulation of acetylated histones in MM cells
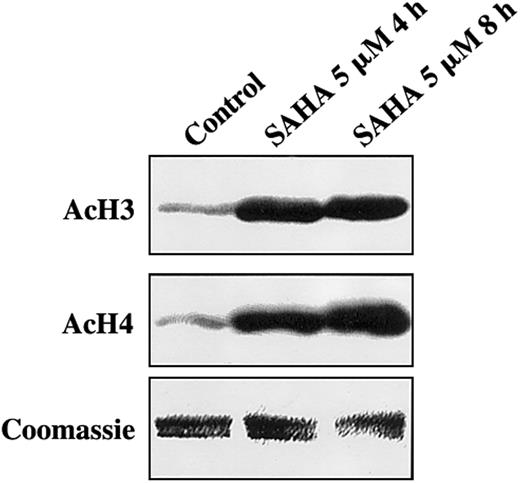
We first investigated the effect of SAHA on histone acetylation status in MM cells. MM.1S cells treated with SAHA for 4 or 8 hours exhibited significantly increased acetylation of histones H3 and H4 than controls (Figure 1). Equal loading of histones was confirmed by Coomassie blue.
SAHA induces accumulation of acetylated histones in MM.1S cells.
MM.1S cells treated with SAHA for 4 or 8 hours exhibited significantly increased acetylation of histones H3 (AcH3) and H4 (AcH4) than untreated controls. Equal loading of histones was confirmed by Coomassie blue.
SAHA induces accumulation of acetylated histones in MM.1S cells.
MM.1S cells treated with SAHA for 4 or 8 hours exhibited significantly increased acetylation of histones H3 (AcH3) and H4 (AcH4) than untreated controls. Equal loading of histones was confirmed by Coomassie blue.
SAHA induces growth arrest and apoptosis in MM and WM cell lines and patient tumor cells, including those resistant to dexamethasone and conventional chemotherapy
We first investigated the effect of SAHA on growth and survival of MM cells. Treatment of MM.1S cells with SAHA (2.5, 5, and 10 μM) for 24 hours potently suppressed cellular proliferation, as quantified by BrdU incorporation (Figure 2A). Cell cycle analysis by PI revealed that SAHA induced early growth arrest and subsequent apoptosis in 8 MM cell lines, 2 EBV-transformed B-cell lines, 1 WM cell line, and tumor cells freshly isolated from 5 patients with MM and 3 patients with WM (Figure 2B-G; Table1). Within this panel, there were lines resistant to dexamethasone (MM.1R), doxorubicin (RPMI-8226-Dox40), melphalan (RPMI-8226-LR5), or thalidomide and its immunomodulatory analogs (RPMI-8226/S) that responded to SAHA.
SAHA induces growth arrest and apoptosis in MM.1S cells.
(A) Quantification of proliferation of MM.1S cells using the BrdU incorporation assay, according to the instructions of the manufacturer. (B-F) PI analysis of MM.1S cells that were treated with SAHA (5 μM) in 10% FCS for 24 (C), 48 (D), 72 (E), or 96 (F) hours or controls untreated (B) for 96 hours. The percentage of cells in the sub-G1 region (horizontal bars) indicates significant SAHA-induced cell death. (G) Survival of MM patient cells (average ± SD), quantified by MTT colorimetric assay, after treatment with (■) or without (▪) SAHA (5 μM) for 48 hours. Error bars represent SD.
SAHA induces growth arrest and apoptosis in MM.1S cells.
(A) Quantification of proliferation of MM.1S cells using the BrdU incorporation assay, according to the instructions of the manufacturer. (B-F) PI analysis of MM.1S cells that were treated with SAHA (5 μM) in 10% FCS for 24 (C), 48 (D), 72 (E), or 96 (F) hours or controls untreated (B) for 96 hours. The percentage of cells in the sub-G1 region (horizontal bars) indicates significant SAHA-induced cell death. (G) Survival of MM patient cells (average ± SD), quantified by MTT colorimetric assay, after treatment with (■) or without (▪) SAHA (5 μM) for 48 hours. Error bars represent SD.
Cell cycle profile and distribution
| . | Sub-G1 . | G0G1 . | S . | G2/M . |
|---|---|---|---|---|
| MM.1S cells (%) | ||||
| Control | 6.0 | 46.5 | 32.1 | 15.4 |
| 24 h | 70.1 | 22.1 | 6.5 | 1.2 |
| 48 h | 91.4 | 5.9 | 2.0 | 0.7 |
| 72 h | 100.0 | 0.0 | 0.0 | 0.0 |
| 96 h | 100.0 | 0.0 | 0.0 | 0.0 |
| MM.1R cells (%) | ||||
| Control | 4.1 | 42.1 | 22.4 | 31.4 |
| 24 h | 50.9 | 26.2 | 11.3 | 11.3 |
| 48 h | 90.4 | 4.6 | 5.0 | 0.0 |
| 72 h | 100.0 | 0.0 | 0.0 | 0.0 |
| 96 h | 100.0 | 0.0 | 0.0 | 0.0 |
| RPMI-8226/S cells (%) | ||||
| Control | 4.8 | 20.9 | 38.8 | 35.5 |
| 24 h | 55.0 | 12.5 | 0.4 | 32.1 |
| 48 h | 98.2 | 0.4 | 0.0 | 1.4 |
| 72 h | 95.9 | 0.9 | 0.3 | 2.8 |
| 96 h | 100.0 | 0.0 | 0.0 | 0.0 |
| RPMI-8226-Dox40 cells (%) | ||||
| Control | 5.3 | 20.8 | 35.6 | 38.3 |
| 24 h | 58.6 | 9.9 | 5.0 | 26.4 |
| 48 h | 86.0 | 1.3 | 12.7 | 0.1 |
| 72 h | 100.0 | 0.0 | 0.0 | 0.0 |
| 96 h | 99.0 | 0.0 | 0.9 | 0.1 |
| RPMI-8226-LR5 cells (%) | ||||
| Control | 5.6 | 27.5 | 66.9 | 0.0 |
| 24 h | 29.0 | 28.5 | 0.0 | 42.5 |
| 48 h | 71.8 | 8.8 | 0.0 | 19.5 |
| 72 h | 99.3 | 0.1 | 0.0 | 0.6 |
| ARP-1 cells (%) | ||||
| Control | 4.5 | 25.0 | 70.4 | 0.1 |
| 24 h | 16.3 | 17.4 | 4.6 | 61.7 |
| 48 h | 95.0 | 2.9 | 0.0 | 2.1 |
| 72 h | 96.0 | 2.4 | 0.0 | 1.6 |
| LP-1 cells (%) | ||||
| Control | 0.0 | 42.6 | 55.0 | 2.4 |
| 24 h | 4.5 | 3.7 | 0.0 | 91.8 |
| 48 h | 61.3 | 1.0 | 2.7 | 35.1 |
| 72 h | 60.3 | 22.7 | 0.0 | 17.1 |
| ARH-77 cells (%) | ||||
| Control | 3.6 | 62.9 | 29.6 | 4.0 |
| 24 h | 12.5 | 40.2 | 25.3 | 22.1 |
| 48 h | 40.5 | 27.3 | 14.4 | 17.8 |
| 72 h | 92.4 | 0.0 | 0.0 | 7.6 |
| 96 h | 95.3 | 0.0 | 0.2 | 4.5 |
| IM-9 cells (%) | ||||
| Control | 0.5 | 44.2 | 40.7 | 14.3 |
| 24 h | 16.3 | 36.6 | 13.2 | 33.8 |
| 48 h | 2.9 | 63.3 | 29.8 | 4.0 |
| 72 h | 99.7 | 0.0 | 0.0 | 0.3 |
| 96 h | 99.5 | 0.0 | 0.0 | 0.5 |
| . | Sub-G1 . | G0G1 . | S . | G2/M . |
|---|---|---|---|---|
| MM.1S cells (%) | ||||
| Control | 6.0 | 46.5 | 32.1 | 15.4 |
| 24 h | 70.1 | 22.1 | 6.5 | 1.2 |
| 48 h | 91.4 | 5.9 | 2.0 | 0.7 |
| 72 h | 100.0 | 0.0 | 0.0 | 0.0 |
| 96 h | 100.0 | 0.0 | 0.0 | 0.0 |
| MM.1R cells (%) | ||||
| Control | 4.1 | 42.1 | 22.4 | 31.4 |
| 24 h | 50.9 | 26.2 | 11.3 | 11.3 |
| 48 h | 90.4 | 4.6 | 5.0 | 0.0 |
| 72 h | 100.0 | 0.0 | 0.0 | 0.0 |
| 96 h | 100.0 | 0.0 | 0.0 | 0.0 |
| RPMI-8226/S cells (%) | ||||
| Control | 4.8 | 20.9 | 38.8 | 35.5 |
| 24 h | 55.0 | 12.5 | 0.4 | 32.1 |
| 48 h | 98.2 | 0.4 | 0.0 | 1.4 |
| 72 h | 95.9 | 0.9 | 0.3 | 2.8 |
| 96 h | 100.0 | 0.0 | 0.0 | 0.0 |
| RPMI-8226-Dox40 cells (%) | ||||
| Control | 5.3 | 20.8 | 35.6 | 38.3 |
| 24 h | 58.6 | 9.9 | 5.0 | 26.4 |
| 48 h | 86.0 | 1.3 | 12.7 | 0.1 |
| 72 h | 100.0 | 0.0 | 0.0 | 0.0 |
| 96 h | 99.0 | 0.0 | 0.9 | 0.1 |
| RPMI-8226-LR5 cells (%) | ||||
| Control | 5.6 | 27.5 | 66.9 | 0.0 |
| 24 h | 29.0 | 28.5 | 0.0 | 42.5 |
| 48 h | 71.8 | 8.8 | 0.0 | 19.5 |
| 72 h | 99.3 | 0.1 | 0.0 | 0.6 |
| ARP-1 cells (%) | ||||
| Control | 4.5 | 25.0 | 70.4 | 0.1 |
| 24 h | 16.3 | 17.4 | 4.6 | 61.7 |
| 48 h | 95.0 | 2.9 | 0.0 | 2.1 |
| 72 h | 96.0 | 2.4 | 0.0 | 1.6 |
| LP-1 cells (%) | ||||
| Control | 0.0 | 42.6 | 55.0 | 2.4 |
| 24 h | 4.5 | 3.7 | 0.0 | 91.8 |
| 48 h | 61.3 | 1.0 | 2.7 | 35.1 |
| 72 h | 60.3 | 22.7 | 0.0 | 17.1 |
| ARH-77 cells (%) | ||||
| Control | 3.6 | 62.9 | 29.6 | 4.0 |
| 24 h | 12.5 | 40.2 | 25.3 | 22.1 |
| 48 h | 40.5 | 27.3 | 14.4 | 17.8 |
| 72 h | 92.4 | 0.0 | 0.0 | 7.6 |
| 96 h | 95.3 | 0.0 | 0.2 | 4.5 |
| IM-9 cells (%) | ||||
| Control | 0.5 | 44.2 | 40.7 | 14.3 |
| 24 h | 16.3 | 36.6 | 13.2 | 33.8 |
| 48 h | 2.9 | 63.3 | 29.8 | 4.0 |
| 72 h | 99.7 | 0.0 | 0.0 | 0.3 |
| 96 h | 99.5 | 0.0 | 0.0 | 0.5 |
Cell cycle profile and distribution were defined by PI analysis of cells that were either treated with SAHA (5 μM) in 10% FCS for 24, 48, 72, or 96 hours or untreated controls. The percentage of cells in the sub-Gt region indicates significant SAHA-induced cell death.
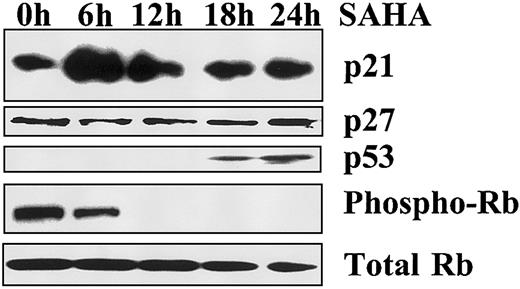
SAHA increases p21 and p53 protein levels and decreases phosphorylation of Rb
To detect the mediators of SAHA-induced growth arrest, we evaluated the levels of p21 and p53 by immunoblotting analysis. SAHA rapidly increased p21 protein levels in MM.1S cells (peak at 6 hours) but not p27 levels. We also detected an increase in p53 levels, which occurred later than p21 (Figure 3). These events suggest that p21 is the most likely mediator of SAHA-induced growth arrest in our model and that its up-regulation is p53 independent. Because p21 induces growth arrest by inhibiting the ability of the cyclin-cdk complex to phosphorylate the cell cycle regulator Rb, we investigated the effect of SAHA on the phosphorylation status of Rb. We found that SAHA profoundly and rapidly decreased the phosphorylation of Rb (Figure 3). These data suggest that SAHA-induced growth arrest occurs via p21-mediated dephosphorylation of Rb.
Mechanism of SAHA-induced growth arrest.
Growth arrest induced by SAHA (5 μM) in MM.1S cells is associated with early up-regulation of p21, but not p27, and dephosphorylation of Rb, as evidenced by immunoblotting. p53 is also up-regulated, but at a later time point.
Mechanism of SAHA-induced growth arrest.
Growth arrest induced by SAHA (5 μM) in MM.1S cells is associated with early up-regulation of p21, but not p27, and dephosphorylation of Rb, as evidenced by immunoblotting. p53 is also up-regulated, but at a later time point.
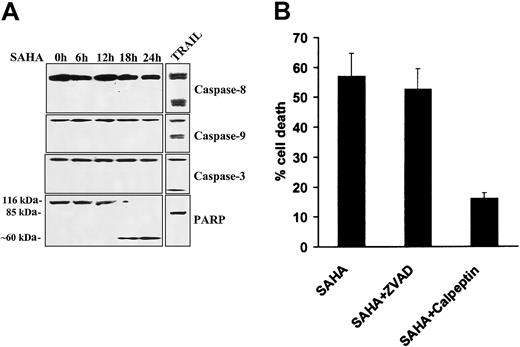
Lack of involvement of caspases in SAHA-induced apoptosis
We subsequently assessed the functional role of caspases in SAHA-induced apoptosis in MM cells. We found by immunoblotting that SAHA did not induce cleavage of caspases-8, -9, or -3 in our model, whereas treatment with TRAIL/Apo2L readily induced caspase cleavage and served as a positive control (Figure 4A). We then investigated for possible cleavage of PARP, a protein well known to be enzymatically cleaved during apoptosis. PARP cleavage was detected after 18 hours of SAHA treatment in MM.1S cells. However, in contrast to TRAIL, which induced the classical 85-kDa PARP fragment, SAHA triggered an atypical pattern of cleavage (∼ 60 kDa) (Figure4A), similar to that induced by the proteolytic enzyme calpain when activated by dysfunctioning mitochondria.33
Lack of involvement of caspases in SAHA-induced apoptosis.
(A) Immunoblotting for caspases-8, -9, and -3 in MM.1S cells treated with SAHA (5 μM) failed to detect caspase cleavage within the first 24 hours of treatment, arguing against the role of caspases in SAHA-induced apoptosis. Treatment with TRAIL/Apo2L (300 ng/mL for 5 hours) served as a positive control, as previously described.29 SAHA induced cleavage of PARP into an atypical approximate 60-kDa fragment, reminiscent of apoptosis mediated by the protease calpain,33 whereas TRAIL/Apo2L resulted in “classic” cleavage of PARP to an approximate 85-kDa fragment. (B) Quantification of SAHA-induced cell death (average ± SD as indicated by error bars), in the absence or presence of caspase and calpain inhibitors, using the LDH release assay. The pan-caspase inhibitor ZVAD-FMK had no protective effect on SAHA-induced cell death, confirming the lack of involvement of caspases in this model. In contrast, the calpain inhibitor calpeptin attenuated SAHA-induced cell death, further supporting the role of the protease calpain in SAHA-induced apoptotic signaling.
Lack of involvement of caspases in SAHA-induced apoptosis.
(A) Immunoblotting for caspases-8, -9, and -3 in MM.1S cells treated with SAHA (5 μM) failed to detect caspase cleavage within the first 24 hours of treatment, arguing against the role of caspases in SAHA-induced apoptosis. Treatment with TRAIL/Apo2L (300 ng/mL for 5 hours) served as a positive control, as previously described.29 SAHA induced cleavage of PARP into an atypical approximate 60-kDa fragment, reminiscent of apoptosis mediated by the protease calpain,33 whereas TRAIL/Apo2L resulted in “classic” cleavage of PARP to an approximate 85-kDa fragment. (B) Quantification of SAHA-induced cell death (average ± SD as indicated by error bars), in the absence or presence of caspase and calpain inhibitors, using the LDH release assay. The pan-caspase inhibitor ZVAD-FMK had no protective effect on SAHA-induced cell death, confirming the lack of involvement of caspases in this model. In contrast, the calpain inhibitor calpeptin attenuated SAHA-induced cell death, further supporting the role of the protease calpain in SAHA-induced apoptotic signaling.
We next further evaluated the functional role of caspases and calpain in our model. We found that the pan-caspase inhibitor ZVAD-FMK had no inhibitory effect on SAHA-induced cell death (Figure 4B), confirming that caspases do not play a major role in mediating SAHA-induced apoptosis in MM cells. In contrast, the calpain inhibitor calpeptin protected against SAHA-induced apoptosis (Figure 4B), confirming that SAHA-induced apoptosis involves activation of the protease calpain.
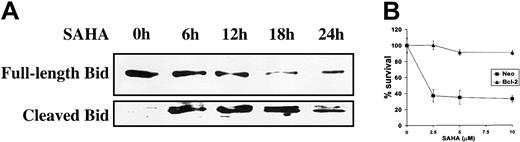
SAHA-induced apoptosis is regulated by members of the Bcl-2 family
Subsequently, we investigated the involvement of members of the Bcl-2 family in SAHA-induced apoptosis. SAHA treatment promotes cleavage of the Bcl-2 family member BH3-interacting domain death agonist (Bid) (Figure 5A). Cleavage of Bid results in a truncated form (tBid), which translocates to the mitochondria and results in an allosteric activation of Bak and Bax, inducing their intramembranous oligomerization that leads to mitochondrial dysfunction.34 These events are counteracted by the antiapoptotic members of the Bcl-2 family, such as Bcl-2 and Bcl-xL, and we, therefore, hypothesized that inhibition of Bid-induced mitochondrial events would protect from SAHA-induced apoptosis. Indeed, overexpression of Bcl-2 in MM.1S cells protected against SAHA-induced apoptosis (Figure 5B). These data support the pivotal role of mitochondria in SAHA-induced apoptotic signaling.
Involvement of mitochondrial events in SAHA-induced apoptosis.
(A) SAHA (5 μM) induced cleavage of Bid, a member of the Bcl-2 family. Cleaved Bid translocates to the mitochondria to promote apoptosis. (B) Overexpression of Bcl-2, which stabilizes the mitochondria and antagonizes the effects of Bid, protected MM.1S cells against SAHA-induced cell death. Cell survival (average ± SD) was quantified by MTT. Neo indicates neomycin, and refers to the empty vector that carries only the neomycin resistance gene but no Bcl-2 cDNA.
Involvement of mitochondrial events in SAHA-induced apoptosis.
(A) SAHA (5 μM) induced cleavage of Bid, a member of the Bcl-2 family. Cleaved Bid translocates to the mitochondria to promote apoptosis. (B) Overexpression of Bcl-2, which stabilizes the mitochondria and antagonizes the effects of Bid, protected MM.1S cells against SAHA-induced cell death. Cell survival (average ± SD) was quantified by MTT. Neo indicates neomycin, and refers to the empty vector that carries only the neomycin resistance gene but no Bcl-2 cDNA.
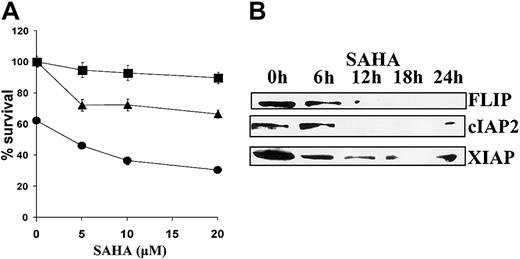
SAHA sensitizes MM cells to death receptor–induced apoptosis
Finally, we investigated the effect of SAHA on apoptosis induced by cell surface death receptors. SAHA sensitized MM.1S cells to apoptosis mediated by cross-linking Fas with the CH11 antibody and to apoptosis induced by TRAIL/Apo2L (Figure6A). This sensitizing effect was associated with decreased expression of the antiapoptotic proteins FLIP, cIAP2, and XIAP (Figure 6B).
Sensitizing effect of SAHA to death receptor–induced apoptosis.
(A) MM.1S cells were pretreated with the indicated concentrations of SAHA for 6 hours and then treated with low concentrations of TRAIL/Apo2L (50 ng/mL, ●), the Fas-activating antibody CH-11 (25 ng/mL, ▴), or vehicle (▪) for additional 18 hours. Error bars represent SD. (B) SAHA down-regulated the antiapoptotic proteins FLIP, cIAP2, and XIAP, which regulate death receptor–mediated apoptotic signaling.
Sensitizing effect of SAHA to death receptor–induced apoptosis.
(A) MM.1S cells were pretreated with the indicated concentrations of SAHA for 6 hours and then treated with low concentrations of TRAIL/Apo2L (50 ng/mL, ●), the Fas-activating antibody CH-11 (25 ng/mL, ▴), or vehicle (▪) for additional 18 hours. Error bars represent SD. (B) SAHA down-regulated the antiapoptotic proteins FLIP, cIAP2, and XIAP, which regulate death receptor–mediated apoptotic signaling.
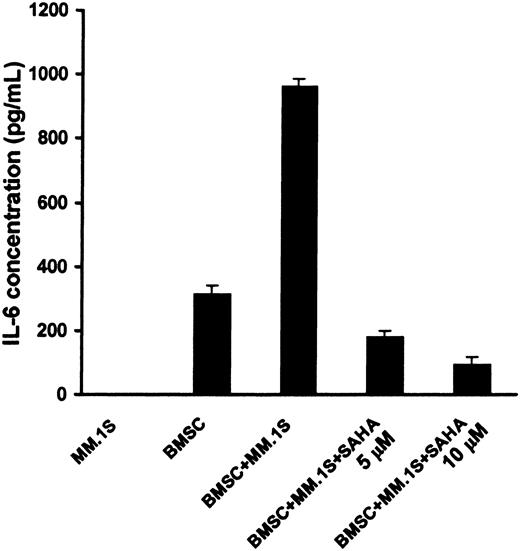
SAHA blocks IL-6 secretion from BMSCs
MM cell adhesion to BMSCs induces IL-6 synthesis and secretion in BMSCs,35 which mediates paracrine growth, survival, and drug resistance of MM cells in the BM milieu.32 We found that SAHA suppressed the secretion of IL-6 by BMSCs triggered by binding of MM cells (Figure 7). The viability of BMSCs during this treatment was found to be more than 96%, as quantified by MTT, confirming that the effect of SAHA on IL-6 secretion is not due to BMSC death. These findings suggest that SAHA acts both directly on MM cells and indirectly against tumor cells via its inhibitory effect on the production of IL-6 in the BM microenvironment.
SAHA suppresses the stimulation of production of IL-6 by BMSCs triggered by binding of MM cells.
MM.1S cells were cocultured with or without BMSCs for 24 hours, in the presence or absence of SAHA (5-10 μM). During this incubation period, the viability of BMSCs was maintained. The presence of IL-6 in the supernatants was quantified by ELISA. Error bars represent SD.
SAHA suppresses the stimulation of production of IL-6 by BMSCs triggered by binding of MM cells.
MM.1S cells were cocultured with or without BMSCs for 24 hours, in the presence or absence of SAHA (5-10 μM). During this incubation period, the viability of BMSCs was maintained. The presence of IL-6 in the supernatants was quantified by ELISA. Error bars represent SD.
Comparison of sensitivity of malignant B cells versus solid tumor cells to SAHA
SAHA has demonstrated in vitro activity against a broad variety of transformed cells, including prostate, breast, colon, and bladder carcinoma and neuroblastoma cells.9,19 36 In this study, we investigated the relative sensitivity to treatment with SAHA (36 hours) in prostate and breast carcinoma cells compared with B-cell malignancies. We found that the MM cell line MM.1S and the WM cell line WM-WSU were very sensitive to SAHA (IC50 [concentration that inhibits 50%] was 0.16 and 0.14 μM, respectively). For comparison, the LNCaP, DU145, and MCF7 lines had IC50s of 12, 2.32, and 4.2 μM, respectively (Figure8).
Comparison of sensitivity of malignant B cells versus solid tumor cells to SAHA.
MM.1S (MM, ♦), WM-WSU (WM, ■), LNCaP (prostate carcinoma, ▵), DU145 (prostate carcinoma, ⋄), and MCF7 (breast carcinoma, ▪) cells were treated with SAHA for 36 hours. Cell survival was quantified with the MTT assay. We found that the MM cell line MM.1S and the WM cell line WM-WSU were very sensitive (IC50 of 0.16 and 0.14 μM, respectively) to SAHA. For comparison, the IC50s of LNCaP, DU145, and MCF7 lines were 12, 2.32, and 4.2 μM, respectively. A570 indicates absorbance at 570 nm.
Comparison of sensitivity of malignant B cells versus solid tumor cells to SAHA.
MM.1S (MM, ♦), WM-WSU (WM, ■), LNCaP (prostate carcinoma, ▵), DU145 (prostate carcinoma, ⋄), and MCF7 (breast carcinoma, ▪) cells were treated with SAHA for 36 hours. Cell survival was quantified with the MTT assay. We found that the MM cell line MM.1S and the WM cell line WM-WSU were very sensitive (IC50 of 0.16 and 0.14 μM, respectively) to SAHA. For comparison, the IC50s of LNCaP, DU145, and MCF7 lines were 12, 2.32, and 4.2 μM, respectively. A570 indicates absorbance at 570 nm.
Discussion
We have evaluated the effects of HDAC inhibition in cell lines and primary tumor cells from patients with B-cell malignancies. The HDACI SAHA potently induced growth arrest and subsequent apoptosis in all cell lines and patient cells tested. It induced cleavage of Bid in our model and apoptosis that was caspase independent, calpain dependent, and was completely abrogated by overexpression of Bcl-2. Within the BM milieu, SAHA inhibited the production of IL-6 triggered by binding of MM cells to BMSCs. Finally, SAHA increased the sensitivity of MM.1S cells to death receptor–induced apoptosis. These findings provide the preclinical rationale for clinical studies of SAHA, alone and in combination with other therapies, to improve patient outcome.
Hydroxamic acid–based hybrid polar compounds that inhibit HDACs result in accumulation of acetylated core nucleosomal histones in cultured cells and induction of differentiation and/or apoptosis of transformed cells. These agents, therefore, represent promising novel anticancer agents. SAHA, the prototype member of this family of compounds, suppresses the growth of LNCaP, PC-3, and TSU-Pr1 cell lines at micromolar concentrations (2.5-7.5 μM) in vitro19 and induces accumulation of acetylated histones in tumor tissue from xenografts of human prostate carcinomas and neuroblastomas37 in nude mice. In this study of B-cell malignancies, SAHA induced accumulation of acetylated histones, growth arrest, and apoptosis. Early growth arrest was associated with potent and early up-regulation of the cdk inhibitor p21 and associated decrease in phosphorylation of the cdk substrate Rb. P27 expression was not altered, in agreement with a previous report in bladder carcinoma cells.21 The peak in p21 expression preceded the increase in p53 expression and is probably p53 independent, consistent with studies by Richon et al,21 Huang et al,22 and Vrana et al,38 showing that SAHA-induced up-regulation of p21 is mediated by Sp1 sites in the p21 promoter and is p53 independent. This observation has clinical application, because HDACIs like SAHA are expected to be active even against malignant cells with defects in the p53 pathway.
We further characterized the mechanism of SAHA-induced apoptosis. Treatment with SAHA for 48 or more hours potently induced apoptosis in our model, as evidenced by sub-G1 peak in cell cycle analysis. The role of caspases in HDACI-induced apoptosis appears to be model dependent. Amin et al39 have reported caspase-dependent apoptosis in acute promyelocytic leukemia (APL) cells, whereas Ruefli et al40 have demonstrated that SAHA induces caspase-independent apoptosis in acute T-cell leukemia cells. Because in our model the pan-caspase inhibitor ZVAD-FMK exerted no protective effect, SAHA-induced cell death was caspase independent. In agreement, caspase cleavage was not detected during the early (0-24 hour) time points of SAHA treatment, when commitment to cell death occurs. Furthermore, in contrast to the death ligand TRAIL/Apo2L that activates caspases-8, -9, and -329 and results in classic cleavage of PARP to an approximate 85-kDa fragment, SAHA resulted in the appearance of an atypical, approximately 60-kDa fragment. This SAHA-induced pattern of PARP cleavage is reminiscent of that observed in human breast carcinoma cells treated with the cytotoxic agent β-lapachone, which is mediated by the protease calpain.33 β-Lapachone is reduced by the mitochondrial enzyme NADPH:quinone oxidoreductase (NQO1) into a hydroquinone form, which is unstable and spontaneously reverts to the oxidized form, thereby depleting mitochondrial reduced nicotinamide adenine dinucleotide phosphate (NADPH) levels and resulting in mitochondrial dysfunction that ultimately activates calpain-mediated cell death.33 In our study, the calpain inhibitor calpeptin blocked SAHA-induced cell death. Calpain exists in the cytosol of cells as an inactive proenzyme, procalpain,41that translocates to the cell membrane where it is activated autocatalytically in the presence of micromolar concentrations of Ca++. Mitochondria play a major role in Ca++homeostasis, and their dysfunction results in increased cytosolic Ca++ and activation of calpain, which, in turn, proteolytically cleaves substrate proteins and functions as an executional death protease. Activation of calpain induces morphologic changes in thrombocytes that resemble caspase-mediated apoptotic death, including exposure of phosphatidylserine on the outer plasma membrane, microvesiculation, and cell shrinkage.42 Our studies support a role for calpain mediating caspase-independent tumor cell death induced by SAHA treatment. Furthermore, we show that the trigger for SAHA-induced calpain activation appears to be mitochondrial in origin, because we detected cleavage of Bid induced by treatment with SAHA and hypothesized that this leads to mitochondrial dysfunction and downstream activation of the described calpain pathway. Our findings agree with Ruefli et al,40 who reported cleavage of Bid and caspase-independent cell death in SAHA-treated leukemic T cells. In support of these findings, overexpression of Bcl-2 in MM.1S cells, which restores the balance between proapoptotic and antiapoptotic members of the Bcl-2 family toward the antiapoptotic side, also blocks SAHA-induced cell death.
We next investigated the effect of SAHA on apoptosis induced by cell surface death receptors, because we have previously reported that MM cells are sensitive to apoptosis induced by death receptors, such as Fas and the TRAIL receptors.43 Because both FasL and TRAIL are expressed by activated immune effector cells and participate in cell-mediated cytotoxicity and antitumor surveillance, modulation of cancer cell sensitivity to the Fas and TRAIL apoptotic pathways could affect their response to immune-based therapeutic approaches. SAHA sensitized MM.1S cells to a subtoxic concentration of a Fas-activating monoclonal antibody CH-11 and to a low concentration of recombinant TRAIL. This sensitizing effect was associated with decreased expression of the antiapoptotic proteins FLIP, cIAP2, and XIAP. FLIP and cIAP-2 inhibit death receptor–induced caspase-8 activation in MM cells,29 whereas XIAP inhibits caspases-3, -7, and -9.44 45 These data, therefore, indicate that HDACIs modulate immune responsiveness of tumor cells and could be useful to overcome refractoriness to immune-based therapies. Furthermore, they indicate that HDACIs can influence the interaction of the tumor cells with their local microenvironment.
We also demonstrated that SAHA suppressed the stimulation of IL-6 secretion in BMSCs triggered by MM cell adhesion, without significantly affecting BMSC viability. This finding suggests that SAHA acts both directly on MM cells and via its inhibitory effect on IL-6 secretion in the BM milieu. SAHA possesses anti-inflammatory activity, as it suppresses tumor necrosis factor α (TNF-α), IL-1-β, IL-12, and interferon γ (IFN-γ) release in human peripheral blood mononuclear cells stimulated with lipopolysaccharide (LPS), as well as circulating TNF-α, IL-1-β, IL-6, and IFN-γ levels in mice induced by LPS.46 Also, SAHA may inhibit nuclear factor κB (NF-κB) activity (C.S.M., manuscript in preparation), an effect that would also suppress IL-6 production. These data suggest that SAHA can overcome cell adhesion–mediated and cytokine-induced drug resistance in the BM microenvironment.
Although we cannot exclude the possibility that some of the effects of SAHA in our model are not mediated by its HDACI activity, other HDACIs have also been reported to have anti-MM activity, thus confirming the importance of this pathway. For example, the HDACIs sodium butyrate, trichostatin A, and LAQ824 have been reported to induce growth arrest and apoptosis in human MM cell lines.47 48
SAHA is currently being evaluated clinically as an antitumor agent in a variety of cancer types. Oral administration of SAHA resulted in good bioavailability; biologic activity (accumulation of acetylated histone); an acceptable toxicity profile; and evidence of anticancer activity in several types of cancer, including B-cell malignancies.23,25 This result provides strong evidence for the existence of specific anticancer activity of HDACIs. Breast cancer cells are also more sensitive to SAHA than normal breast epithelial cells and fibroblasts, although the underlying mechanism is not known.49
Our studies further demonstrate that B-cell malignancies exhibit high sensitivity to SAHA-induced cell death, because malignant B cells had IC50s less than 0.2 μM. For comparison, the prostate (LNCaP, DU145) and breast (MCF7) carcinoma cell lines had IC50s more than 2 μM. Although other solid tumor cell lines may also have very high sensitivity to SAHA, such as the melanoma cell line MALME-3M (IC50 = 106 nM; V.M.R. et al, unpublished data), it is clear that B-cell malignancies lie on the high end of the spectrum of sensitivity among various malignancies. The explanation for this phenomenon may be related to the ability of SAHA to readily induce apoptosis in hematopoietic cell lines, such as those used in our experiments, whereas growth arrest is the most prominent effect in some solid tumor cell lines. This observation suggests that SAHA is expected to be a particularly effective agent for the treatment of plasma cell dyscrasias, including MM.
In conclusion, we have evaluated the effects of histone deacetylase inhibition in a panel of B-cell–derived cell lines and primary patient tumor cells, using the HDACI SAHA. We found that SAHA up-regulated p21 and induced growth arrest, as well as promoted cleavage of Bid and caspase-independent, calpain-dependent apoptosis. SAHA sensitized MM cells to apoptosis mediated by death receptors and suppressed IL-6 production by BMSCs. Our data, therefore, provide the framework for clinical evaluation of HDACIs to overcome clinical drug resistance and improve patient outcome.
Prepublished online as Blood First Edition Paper, January 16, 2003; DOI 10.1182/ blood-2002-11-3514.
Supported by the Multiple Myeloma Research Foundation (N.M., C.S.M.), the Laurie Strauss Leukemia Foundation (N.M., C.S.M.), and the Doris Duke Distinguished Clinical Research Scientist Award (K.C.A.).
Memorial Sloan-Kettering Cancer Center and Columbia University jointly hold patents on the hydroxamic acid–based hybrid polar compounds, including SAHA, which are exclusively licensed to Aton Pharma, Inc (Tarrytown, NY), of which P.A.M. is a founder and member of the Board of Directors. Both institutions and founder have an equity position in Aton Pharma.
N.M. and C.S.M. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kenneth C. Anderson, Jerome Lipper Multiple Myeloma Center, Dana-Farber Cancer Institute, Mayer Bldg, Rm M557, 44 Binney St, Boston, MA 02115; e-mail:kenneth_anderson@dfci.harvard.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal