Abstract
In B cells, somatic hypermutation (SHM) and class switch recombination (CSR) depend on the activation-induced cytidine deaminase (AID) gene product, although the precise mode of action of AID remains unknown. Because some chronic lymphocytic leukemia (CLL) B cells can undergo CSR without SHM, it constitutes a useful model to dissect AID function. In this work, we have studied AID expression, the presence of mutations in the preswitch μ DNA region, CSR, and the SHM in 65 CLL patients. Our results demonstrate that unmutated CLL B cells can constitutively express AID and that AID expression is associated with the presence of mutations in the preswitch region and in clonally related isotype-switched transcripts. They also demonstrate that in CLL without constitutive AID expression, AID induction on stimulation results in preswitch mutations and the CSR process. Our results show a dissociation between SHM and CSR in CLL and suggest that, in this disease, AID would require additional help for carrying out the SHM process.
Introduction
After gene rearrangement, immunoglobulin (Ig) variable genes are diversified by somatic hypermutation (SHM), whereas the effector function of the constant domain is modified by class switch recombination (CSR). These processes depend on activation-induced cytidine deaminase (AID), a putative RNA-editing enzyme expressed in B cells from secondary lymphoid organs on CD40 ligand (CD40L) stimulation.1 Given that the absence of AID expression in one form of the hyper-IgM syndrome in humans2 and in AID-targeted mice abolishes CSR and SHM, this protein is thought to play a major role in both processes.3
Fifty percent of patients with chronic lymphocytic leukemia (CLL) display mutated VH genes.4The mutational profile of immunoglobulin genes represents an important prognostic factor5,6 because patients expressing unmutatedVH genes exhibit poor prognoses. In addition, previous reports7-9 have demonstrated that CSR frequently occurs in CLL and predominates in unmutated B-cell CLLs (B-CLLs).
In this work, we have examined the expression of AID transcripts, SHM, and CSR in 65 patients with CLL expressing either unmutated (33 of 65) or mutated (32 of 65) VH genes. Our results show that patients with unmutated B-CLL can constitutively express AID transcripts. This fact is associated with the presence of mutations in the preswitch μ DNA region and with an active CSR, but it is not related to the SHM process. Additionally, in patients with mutated CLL without constitutive AID expression, AID induction on stimulation results in preswitch mutations and the CSR process.
Study design
Healthy and CLL samples
Blood was collected from 4 healthy controls and 65 typical CLL patients from Hôtel-Dieu, Pitié-Salpêtrière, and Pasteur hospitals (Paris, France), Sao Paulo and Servidor Publico Estadual hospitals (Sao Paulo, Brazil), and Hospital Maciel (Montevideo, Uruguay). B cells were purified through negative depletion by using RosetteSep antibody cocktail (StemCell Technologies, Vancouver, BC, Canada) and were stimulated in vitro for 5 days with 1 μg/mL recombinant soluble CD40L (Immunex, Seattle, WA) and 1000 U/mL interleukin (IL)–4 (PharMingen, San Diego, CA).
Analysis of AID transcripts
Polymerase chain reaction (PCR) amplification of CLL cDNA was carried out as described previously,2 and a semiquantitative protocol for AID expression was performed. Briefly, cDNA obtained from 5 × 106 B cells was amplified by 20 cycles of PCR using AID2 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase)10 primers. The fragments obtained were transferred and hybridized with specific AID and GAPDH α-[32P] dCTP (deoxycytidine-5′-triphosphate)–labeled probes.
Analysis of CSR process
Mutation analysis of VHand preswitch regions
Sequences of VH genes were determined as previously described.12 Mutations in the Iμ/Sμ region were studied by PCR using Taq High Fidelity polymerase (Roche Diagnostics, Mannheim, Germany) with the following primers: (A) 5′-GGC TGA CCG AAA CTG AAA AGG C-3′; and (D) 5′-GAA AGC TGG ATG AGT GTC ATG GCC-3′.
Results and discussion
CLL B cells can constitutively express AID transcripts, which predominate among unmutated cases and are associated with the expression of additional clonally related transcripts
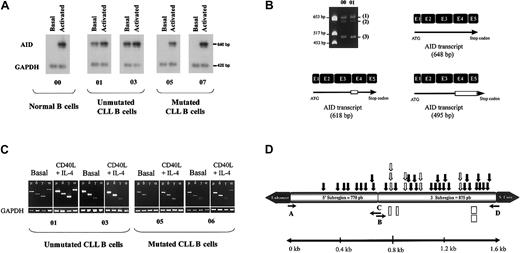
Unmutated cases predominated among aggressive forms of CLL (25 of 34, stages B and C) whereas mutated cases predominated in stage A (23 of 31). By using a stringent semiquantitative reverse transcription (RT)–PCR, we could substantiate constitutive AID expression in 10 of 65 patients, which might have accounted for a lower incidence when compared with another series13 (Figure1A). Interestingly, all these patients expressed unmutated rearranged VHgenes and displayed either advanced disease (stage B in 4) or progressive disease (stage A in 6). As shown in Figure 1B, different AID transcripts were amplified by RT-PCR, corresponding, respectively, to the complete sequence of AID transcript previously reported by Revy et al,2 a fragment displaying a 10–amino acid deletion in the initial portion of exon 4, and a fragment with a 51–amino acid deletion including exons 4 and 5. Different AID-splicing variants have also been reported by Noguchi et al.14
Expression of AID RNA transcripts in CLL and their relation in CSR and SHM process.
(A) Semiquantitative AID expression. The expression of AID transcripts was monitored by semiquantitative RT-PCR using AID and GAPDH-specific primers in the same RT-PCR tube reaction. Representative amplification for healthy B cells (00) and either unmutated (01 and 03) or mutated (05 and 07) B-CLLs are shown. Amounts of AID transcripts were determined by normalization with internal GAPDH expression. Relative units corresponding to AID and GAPDH transcript amplification levels were quantified by Quant software (Molecular Dynamics). (B) Presence of different AID RNA transcripts. Normal cDNA (00) and CLL cDNA (01) were amplified and migrated. Three different RNA forms of the AID gene were found (1, 2, 3). The figure depicts a schematic sequence of AID mRNA previously reported by Schroeder et al,4 corresponding to 198 amino acids. The other 2 variants are spliced forms, one consisting of 618 base pair (bp) with an open-reading frame containing a deletion of 10 amino acids and the other consisting of 495 bp containing a complete deletion of exons 4 and 5 (51 amino acids). Deletions are depicted as unfilled rectangles. (C) Clonal isotype switch transcripts. mRNA transcript amplifications with tumor-relatedVH primers in 5′ and Cμ, Cδ, Cγ, and Cα in 3′ from patients 1 and 3 with unmutated and patients 5 and 7 with mutated disease. Patient 1 expresses μ, δ, γ, and α transcripts, and patient 3 expresses μ, δ, and γ tumorally related transcripts, whereas patients 5 and 7 only express μ and δ transcripts related to the tumoral clone. The smearlike amplification observed for patient 7 corresponds to a polyclonal amplification of different γ transcripts as confirmed by sequence. After stimulation patient 5 expressed a tumorally related γ transcript, and patient 7 acquired tumorally related γ and α transcripts. (D) Distributions of mutations in the Iμ/Sμ region. A 1625-bp genomic fragment between the enhancer and the Sμ switch core was amplified with primers A and D for 4 healthy controls and 7 patients with CLL (Table1). Closed and open arrows indicate point mutations (30 in the 3′ subregion, 9 in the 5′ subregion). In addition, 4 deletions in the 3′ subregion, depicted as rectangles, were observed. Open arrows illustrate where repeated mutations took place. Primers B (5′-TGC CTG TCT CTT ACC ATG TCG GG-3′) and C (5′-GAC ATG GTA AGA GAC AGG CAG CCG-3′) were used as internal primers for sequencing reaction. Given that in all cases 3 independent sequences obtained from different PCRs with a high-fidelity Taq DNA polymerase (10−6 expected rate mutation) were carried out, Taq infidelity should play a minor, if any, role in the appearance of mutations.
Expression of AID RNA transcripts in CLL and their relation in CSR and SHM process.
(A) Semiquantitative AID expression. The expression of AID transcripts was monitored by semiquantitative RT-PCR using AID and GAPDH-specific primers in the same RT-PCR tube reaction. Representative amplification for healthy B cells (00) and either unmutated (01 and 03) or mutated (05 and 07) B-CLLs are shown. Amounts of AID transcripts were determined by normalization with internal GAPDH expression. Relative units corresponding to AID and GAPDH transcript amplification levels were quantified by Quant software (Molecular Dynamics). (B) Presence of different AID RNA transcripts. Normal cDNA (00) and CLL cDNA (01) were amplified and migrated. Three different RNA forms of the AID gene were found (1, 2, 3). The figure depicts a schematic sequence of AID mRNA previously reported by Schroeder et al,4 corresponding to 198 amino acids. The other 2 variants are spliced forms, one consisting of 618 base pair (bp) with an open-reading frame containing a deletion of 10 amino acids and the other consisting of 495 bp containing a complete deletion of exons 4 and 5 (51 amino acids). Deletions are depicted as unfilled rectangles. (C) Clonal isotype switch transcripts. mRNA transcript amplifications with tumor-relatedVH primers in 5′ and Cμ, Cδ, Cγ, and Cα in 3′ from patients 1 and 3 with unmutated and patients 5 and 7 with mutated disease. Patient 1 expresses μ, δ, γ, and α transcripts, and patient 3 expresses μ, δ, and γ tumorally related transcripts, whereas patients 5 and 7 only express μ and δ transcripts related to the tumoral clone. The smearlike amplification observed for patient 7 corresponds to a polyclonal amplification of different γ transcripts as confirmed by sequence. After stimulation patient 5 expressed a tumorally related γ transcript, and patient 7 acquired tumorally related γ and α transcripts. (D) Distributions of mutations in the Iμ/Sμ region. A 1625-bp genomic fragment between the enhancer and the Sμ switch core was amplified with primers A and D for 4 healthy controls and 7 patients with CLL (Table1). Closed and open arrows indicate point mutations (30 in the 3′ subregion, 9 in the 5′ subregion). In addition, 4 deletions in the 3′ subregion, depicted as rectangles, were observed. Open arrows illustrate where repeated mutations took place. Primers B (5′-TGC CTG TCT CTT ACC ATG TCG GG-3′) and C (5′-GAC ATG GTA AGA GAC AGG CAG CCG-3′) were used as internal primers for sequencing reaction. Given that in all cases 3 independent sequences obtained from different PCRs with a high-fidelity Taq DNA polymerase (10−6 expected rate mutation) were carried out, Taq infidelity should play a minor, if any, role in the appearance of mutations.
The expression of constitutive AID transcripts in 10 unmutated patients led us to examine the CSR process by analysis of clonal isotype switch transcripts and the presence of γ-CTs as recently reported in CLL by Cerutti et al.15 Seven of these 10 CLLs expressed μ, δ, γ, and α transcripts (Figure 1C), which were substantiated to display VH sequences identical to those expressed by the tumoral clone (data not shown). In addition, γ-CTs were observed in all these unmutated patients, indicating that the initiation of a CSR process11 is accounted for in these B-CLLs (data not shown).
AID expression enables CLL-B cells to carry out CSR somatic-like mutations but not SHM
To further investigate the dissociation between SHM and CSR in B-CLLs cells, taking into account that upon stimulation in the conditions that induce CSR, AID induction is associated with hypermutation in the Sμ region under the conditions that induce CSR.16 We have studied B cells from 4 healthy donors and 7 patients with CLL (4 unmutated and 3 mutated) before and after stimulation with CD40L + IL-4 and have considered AID expression by semiquantitative RT-PCR and CSR by searching of clonally related isotype variants, CTs, and somatic-like mutations in the preswitch μ DNA region. Figure 1D depicts the schematic amplified sequence of the preswitch region. Consistent with previous findings,2CD40L + IL-4 stimulation induced the expression of AID transcripts (Figure 1A). After the exclusion of well-known polymorphisms, comparison of the sequences before and after stimulation demonstrated that AID induction was associated with a significant mutation rate (3 × 10−3) in the preswitch region. In agreement with previous reports,16 a high number of these mutations were accumulated mainly in the 3′ region of the amplified fragment (Figure1D), indicating that a hypermutation process took place in the human Ig preswitch region and that this process was AID dependent. CLL patients 1, 2, 3, and 4 without SHM in VH genes were found to constitutively express AID, clonally related isotype variants, and a high rate of mutations in the preswitch region (Table1). In addition, neither AID transcripts nor additional tumorally related transcripts were observed in CLL patients 5, 6, and 7 with VH mutations (Table 1) before CD40L + IL-4 stimulation. However, AID transcripts and clonally related isotype variants were found for patients 5 and 7 on stimulation (Figure 1C).
Implication of AID expression in mutational profile of Iμ/Sμ region, CSR, and SHM
| Sample DNA and RNA stimulation, CD40L + IL4 for 5 days . | AID transcripts, RT-PCR . | SHM homology to germline, % . | Iμ/Sμ region . | Clonal isotype switched transcripts, RT-PCR . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ region . | 3′ region . | Complete region . | |||||||||
| Mutations . | Deletions . | Mutations . | Deletions . | Mutations/bp sequences . | VH/μ . | VH/δ . | VH/γ . | VH/α . | |||
| Healthy B cells | |||||||||||
| No | − | N/C | 0 of 769 | 0 of 769 | 0 of 875 | 0 of 875 | 0 of 1625 | − | − | − | − |
| Yes | + | N/C | 0 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 1.2 × 10−3 | − | − | − | − |
| B-CLLs no. 1 | |||||||||||
| No | + | 100 | 0 of 769 | 0 of 769 | 4 of 875 | 0 of 875 | 2.4 × 10−3 | + | + | + | + |
| Yes | + | 100 | 1 of 769 | 0 of 769 | 4 of 875 | 1 of 875 | 3.0 × 10−3 | + | + | + | + |
| B-CLLs no. 2 | |||||||||||
| No | + | 100 | 2 of 769 | 0 of 769 | 1 of 875 | 0 of 875 | 1.8 × 10−3 | + | + | + | − |
| Yes | + | 100 | 3 of 769 | 0 of 769 | 2 of 875 | 1 of 875 | 3.0 × 10−3 | + | − | + | − |
| B-CLLs no. 3 | |||||||||||
| No | + | 99 | 0 of 769 | 0 of 769 | 3 of 875 | 1 of 875 | 1.8 × 10−3 | + | + | + | − |
| Yes | + | 99 | 0 of 769 | 0 of 769 | 5 of 875 | 1 of 875 | 3.1 × 10−3 | + | + | + | − |
| B-CLLs no. 4 | |||||||||||
| No | + | 99 | 2 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 2.4 × 10−3 | + | + | + | + |
| B-CLLs no. 5 | |||||||||||
| No | − | 95 | 0 of 769 | 0 of 769 | 0 of 875 | 0 of 875 | 0 of 1625 | + | + | − | − |
| Yes | + | 95 | 0 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 1.2 × 10−3 | + | + | − | + |
| B-CLLs no. 6 | |||||||||||
| No | − | 92 | 0 of 769 | 0 of 769 | 1 of 875 | 0 of 875 | 6.1 × 10−4 | − | − | + | − |
| Yes | + | 92 | 1 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 1.8 × 10−3 | − | − | + | − |
| B-CLLs no. 7 | |||||||||||
| No | − | 93 | 0 of 769 | 0 of 769 | 0 of 875 | 0 of 875 | 0 of 1625 | + | + | − | − |
| Yes | + | 93 | 0 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 1.2 × 10−3 | + | + | + | + |
| Sample DNA and RNA stimulation, CD40L + IL4 for 5 days . | AID transcripts, RT-PCR . | SHM homology to germline, % . | Iμ/Sμ region . | Clonal isotype switched transcripts, RT-PCR . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ region . | 3′ region . | Complete region . | |||||||||
| Mutations . | Deletions . | Mutations . | Deletions . | Mutations/bp sequences . | VH/μ . | VH/δ . | VH/γ . | VH/α . | |||
| Healthy B cells | |||||||||||
| No | − | N/C | 0 of 769 | 0 of 769 | 0 of 875 | 0 of 875 | 0 of 1625 | − | − | − | − |
| Yes | + | N/C | 0 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 1.2 × 10−3 | − | − | − | − |
| B-CLLs no. 1 | |||||||||||
| No | + | 100 | 0 of 769 | 0 of 769 | 4 of 875 | 0 of 875 | 2.4 × 10−3 | + | + | + | + |
| Yes | + | 100 | 1 of 769 | 0 of 769 | 4 of 875 | 1 of 875 | 3.0 × 10−3 | + | + | + | + |
| B-CLLs no. 2 | |||||||||||
| No | + | 100 | 2 of 769 | 0 of 769 | 1 of 875 | 0 of 875 | 1.8 × 10−3 | + | + | + | − |
| Yes | + | 100 | 3 of 769 | 0 of 769 | 2 of 875 | 1 of 875 | 3.0 × 10−3 | + | − | + | − |
| B-CLLs no. 3 | |||||||||||
| No | + | 99 | 0 of 769 | 0 of 769 | 3 of 875 | 1 of 875 | 1.8 × 10−3 | + | + | + | − |
| Yes | + | 99 | 0 of 769 | 0 of 769 | 5 of 875 | 1 of 875 | 3.1 × 10−3 | + | + | + | − |
| B-CLLs no. 4 | |||||||||||
| No | + | 99 | 2 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 2.4 × 10−3 | + | + | + | + |
| B-CLLs no. 5 | |||||||||||
| No | − | 95 | 0 of 769 | 0 of 769 | 0 of 875 | 0 of 875 | 0 of 1625 | + | + | − | − |
| Yes | + | 95 | 0 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 1.2 × 10−3 | + | + | − | + |
| B-CLLs no. 6 | |||||||||||
| No | − | 92 | 0 of 769 | 0 of 769 | 1 of 875 | 0 of 875 | 6.1 × 10−4 | − | − | + | − |
| Yes | + | 92 | 1 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 1.8 × 10−3 | − | − | + | − |
| B-CLLs no. 7 | |||||||||||
| No | − | 93 | 0 of 769 | 0 of 769 | 0 of 875 | 0 of 875 | 0 of 1625 | + | + | − | − |
| Yes | + | 93 | 0 of 769 | 0 of 769 | 2 of 875 | 0 of 875 | 1.2 × 10−3 | + | + | + | + |
Results of 3 independent colony experiments are shown together.
N/C indicates no clonal VDJ gene
AID may act as an RNA-editing enzyme involved in the edition of endonucleases responsible for CSR and SHM,3,17 either by editing a single substrate or separate pre-mRNAs for CSR and SHM.18 On the other hand, recent evidence suggests that AID triggers antibody diversification by the deamination of nucleotides within the immunoglobulin locus itself.19,20 Different results suggest that AID is sufficient to activate the SHM and CSR processes and that its activity does not depend on other centroblast-specific factors.19,21,22 Our results show, however, that AID expression is associated with CSR but not with SHM. This dissociation observed in CLL, combined with previous work in a mouse model16 and in the BL2 Burkitt cell line,23 favors the view that AID may act differentially on CSR and SHM. In agreement with recent work24 demonstrating that with strong stimulation through the B-cell receptor (BCR), patients with CLL are able to acquire somatic mutations, our results suggest that AID is necessary but not sufficient for the SHM process, and that additional levels of regulation could be required for the efficient function of this protein.25
We thank Drs Mirta Giordano, Michelle Goodhardt, and Frédéric Davi for review and discussion of this manuscript.
Prepublished online as Blood First Edition Paper, January 9, 2003; DOI 10.1182/blood-2002-10-3175.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
G. Dighiero, Unitéd'Immuno-Hématologie et d'Immunopathologie, Institut Pasteur, 28 Rue Docteur Roux, 75015 Paris, France; e-mail:dighiero@pasteur.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal