Abstract
Metaphase cytogenetic abnormalities (CAs), especially of chromosome 13 (CA 13), confer a grave prognosis in multiple myeloma even with tandem autotransplantations as applied in Total Therapy I, which enrolled 231 patients between 1989 and 1994. With a median follow-up of almost 9 years, the prognostic implications of all individual CAs, detected prior to treatment and at relapse, were investigated. Among all CAs and standard prognostic factors examined prior to therapy, only hypodiploidy and CA 13 (hypo–13 CA), alone or in combination, were associated with shortest event-free survival and overall survival (OS). The shortest postrelapse OS was observed with hypo–13 CA, which was newly detected in 18 of all 28 patients presenting with this abnormality at relapse. Superior prognosis was associated with the absence of any CA at both diagnosis and relapse (10-year OS, 40%). The lack of independent prognostic implications of other CAs points to a uniquely aggressive behavior of hypo–13 CA (present in 16% of patients at diagnosis). With the use of microarray data in 146 patients enrolled in Total Therapy II, overexpression of cell cycle genes distinguished CA from no CA, especially in cases of del(13) detected by interphase fluorescence in situ hybridization (FISH). FISH 13, resulting in a haploinsufficiency of RB1 and other genes mapping to chromosome 13, as well as activation of IGF1R, appears to have an amplifying effect on cell cycle gene expression, thus providing a molecular explanation for the dire outcome of patients with CA 13 compared with those with other CAs.
Introduction
The clinical course of patients with multiple myeloma (MM) is best predicted by the presence of certain cytogenetic abnormalities (CAs), in the context of both standard and high-dose therapies.1-5 A particularly high risk for early treatment failure and short survival has been associated with CA 13, whether detected by traditional metaphase karyotype analysis (present in 15% of patients)1-4,6 or, more recently, by interphase fluorescence in situ hybridization (FISH) (present in 50%).5,7-9 As part of Total Therapy I (TT I),10 a phase 2 pilot program using melphalan-based tandem autotransplantations with curative intent, cytogenetic studies were performed prospectively at baseline and as part of virtually all subsequent bone marrow examinations. In this report, we have investigated, along with standard prognostic factors (SPFs), the individual contributions of every chromosome abnormality present prior to initiation of TT I and at follow-up, especially at relapse. With the use of gene expression profiles (GEPs) and FISH available in 146 patients prospectively enrolled in Total Therapy II (TT II) (successor protocol to TT I),11 the uniquely adverse implications, especially of CA 13, were examined by comparing GEP data in the 4 patient subsets defined by FISH 13 and CA.
Patients and methods
Total Therapy I
As previously published, TT I consisted of induction therapy with mutually non–cross-resistant combinations of 3 cycles of vincristine, doxorubicin, and dexamethasone (VAD); high-dose cyclophosphamide plus granulocyte-macrophage colony-stimulating factor (GM-CSF) with peripheral blood stem cell (PBSC) collection and etoposide, dexamethasone, cytarabine, and cisplatin (EDAP); followed by 2 autotransplantations with melphalan 200 mg/m2 at 3 to 6 months apart; interferon-alpha-2b maintenance was intended until disease recurrence.10 Approval was obtained from the University of Arkansas for Medical Sciences institutional review board (UAMS Human Research Advisory Committee) for these studies. Informed consent was provided according to the Declaration of Helsinki.
Patient demographics.
Between 1989 and 1994, 231 patients with symptomatic or progressive MM were enrolled in TT I; 155 were entirely untreated and the remainder had previously received one cycle of standard chemotherapy or dexamethasone. Patient demographics are summarized in Table 1 along with frequencies of first and second transplantations. Stringently defined complete remission (CR) (intent-to-treat) was achieved by 38% of patients. The median follow-up of living patients approaches 9 years.
Patient characteristics, transplantation completion, and response (n = 231)
| Parameter . | % . |
|---|---|
| Age at least 60 y | 24 |
| CRP level above 4.0 mg/L | 47 |
| LDH level at least 190 U/L | 21 |
| B2M level above 345 nM | 30 |
| Creatinine level above 176.8 μM | 10 |
| Hemoglobin level below 100 g/L | 35 |
| Albumin level at least 35 g/L | 2 |
| Durie-Salmon stage III | 53 |
| IgA isotype | 18 |
| Plasmacytosis BM biopsy at least 50% | 48 |
| Completed HDT-1 | 85 |
| Completed HDT-2 | 72 |
| Cumulative TRM | 9 |
| Post-HDT2 CR (intent to treat) | 38 |
| Parameter . | % . |
|---|---|
| Age at least 60 y | 24 |
| CRP level above 4.0 mg/L | 47 |
| LDH level at least 190 U/L | 21 |
| B2M level above 345 nM | 30 |
| Creatinine level above 176.8 μM | 10 |
| Hemoglobin level below 100 g/L | 35 |
| Albumin level at least 35 g/L | 2 |
| Durie-Salmon stage III | 53 |
| IgA isotype | 18 |
| Plasmacytosis BM biopsy at least 50% | 48 |
| Completed HDT-1 | 85 |
| Completed HDT-2 | 72 |
| Cumulative TRM | 9 |
| Post-HDT2 CR (intent to treat) | 38 |
The median follow-up was 105 months. As part of follow-up evaluations, aliquots of bone marrow aspirates were routinely submitted for cytogenetic analysis, which was performed on unstimulated Giemsa-banded chromosomes.12 CRP indicates C-reactive protein; LDH, lactate dehydrogenase; B2M, β2microglobulin; Ig, immunoglobulin; BM, bone marrow; HDT, high-dosage transplantation; TRM, transplantation-related mortality; CR, complete remission.
Statistical considerations.
The large number of CAs under investigation and the low incidence rate of many of the CAs make it desirable to reduce the list of CAs considered for analysis with outcome variables, so as not to end up with results that may be spurious and irreproducible. To divide the list of specific CAs into subsets, we first calculated the minimum incidence rate necessary for a given CA to achieve adequate statistical power (at least 80%) to distinguish a meaningful effect size in a univariate test for prognostic effect. For this purpose, we chose as an effect size a hazard ratio of at least 3.0 when making comparisons with patients without a given abnormality. Since the hazard ratio for presence of any CA was slightly below 2.0, an increase to 3.0 seemed appropriate, so as to identify specific CAs with comparatively higher prognostic importance. With the use of these assumptions, the minimum incidence rate was calculated to be 5% (approximately 10 or more patients) in the overall population who had baseline cytogenetics (n = 222).
Outcome definitions.
Overall survival (OS) was calculated as the time from initiation of TT I to death from any cause or last contact. Event-free survival (EFS) was calculated as the time from initiation of TT I to either progression of disease, death from any cause, or last contact. Secondary end points of interest were also explored.
Analysis methods.
Survival curves were estimated by the product-limit method13 and compared by means of the log-rank test.14 Cox proportional hazards regression15was used to assess the influence of CAs and SPFs on survival outcomes. Logistic regression was used to relate cytogenetics and SPFs to dichotomous outcomes. Multivariate models were constructed by means of backward stepwise regression methods. All univariately significant covariates were included in the stepwise selection.
Total Therapy II
In an effort to determine the molecular mechanisms underlying the poor prognosis of CA 13 MM, we resorted to data available in 146 patients treated according to TT II. These included conventional cytogenetics, FISH, and GEPs.
Fluorescence in situ hybridization.
Detailed methods for triple-color interphase FISH have been described.11 16 The percentage of clonotypic plasma cells with evidence of monosomy of chromosome 13 was determined for the 146 cases with gene-expression profiling data. For purposes of this study, monosomy of chromosome 13 (referred to as “FISH 13”) was observed in 79 (54%) of 146 cases. Monosomy was defined as at least 50% of cytoplasmic immunoglobulin–positive (cIg+) cells exhibiting only one signal for each of 4 probes, D13S31, D13S221, D13S285, and BRCA2,spanning the long arm of chromosome 13. A total of 67 cases had no evidence of deletion (fewer than 20% cIg+ cells with deletion of any probe). These patients were deemed to lack chromosome 13 deletion (referred to as “no FISH 13”).
Conventional G-band cytogenetics were performed on all 146 samples by means of conventional methodologies.12 CAs were present in 64 cases (44%) and no CAs in 82 cases.
Gene expression profiles.
Detailed protocols for plasma cell isolation and microarray hybridization have been previously described.17 18Briefly, plasma cells were isolated from bone marrow aspirates of 146 newly diagnosed MM patients enrolled into the clinical trial TT II (successor program to TT I) after written informed consent was obtained, in keeping with institutional policies. Total RNA was isolated, converted to biotinylated cRNA, and hybridized to the U95av2 GeneChip microarrays interrogating approximately 12 000 genes (Affymetrix, Santa Clara, CA). Scanned output files were analyzed with GeneChip 5.0.1 software (Affymetrix). Arrays were scaled to an average intensity of 1500 and analyzed independently.
GeneChip data analysis.
All data used in our analysis were derived from Affymetrix Microarray Suite 5.0. GeneChip 5.0 output files are given (1) as a signal that represents the difference between the intensities of the sequence-specific perfect-match probe set and the mismatch probe set, or (2) as a detection of present or absent as determined by the GeneChip 5.0 algorithm. Signals were transformed by the log base 2. Statistical analysis of the data was performed with the SPSS 10.0 (Chicago IL); Significance Analysis of Microarray (SAM) software package,19 and PAM R package.20 To filter genes that tend to be absent in all samples, those with χ2 values greater than 3.84 (P < .05) or with “present” (P) detection in more than half of each sample group were retained. To compare gene expression levels, the nonparametric Wilcoxon rank sum (WRS) test was used to compare 2 groups with the use of Signal. The cutoff was at least a P value less than .001. SAM relies upon the microarray probe set signal values (ie, mean of the signal intensity differences between the 20 matched and mismatched probes arrayed for each gene). Selecting the SAM threshold with the lowest false-discovery rate (FDR) (below 0.01) as a tool for exclusion of any false-positives generated in the χ2 and WRS analysis, we define and report only genes in the intersection of the sequential analysis methods as the differentially expressed genes in these experiments.
Results
Incidence of and associations among CAs
CAs were observed at pretreatment evaluation in 33%. Table2 lists, per individual chromosome, the specific CA encountered along with its frequency. Trisomies present in at least 10% of patients involved chromosomes 3, 5, 7, 9, 11, 15, 19, and 21. Monosomies and deletions, usually of the q arm, affected predominantly chromosomes 6 (6%), 13 (13%), 16 (10%), and 22 (6%). Hypodiploidy, as defined by Smadja et al,5 was present in 11% (Table 3). Abnormalities typical of MDS within the context of a typical MM signature karyotype (MM-MDS) were present in 7%, including −5, 5q−, −7, 7q−, +8, del(20q), and t(1;7)(q10;p10).
CAs considered in analysis (n = 222)
| CA . | Whole chromosome additions, % . | Whole chromosome deletions, % . | Deletions, % . | Translocations, % . | Duplications, % . | Isochromes, % . | Overall, % . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| p-arm . | q-arm . | p-arm . | q-arm . | p-arm . | q-arm . | p-arm . | q-arm . | ||||
| 1 | 0 | 0 | 4 | 0 | 2 | 7 | 0 | 2 | 0 | 0 | 12 |
| 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| 3 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 |
| 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| 5 | 14 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 16 |
| 6 | 0 | 3 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 6 |
| 7 | 10 | 1 | 0 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 14 |
| 8 | 2 | 4 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 8 |
| 9 | 15 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 15 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 13 | 1 | 0 | 1 | 0 | 6 | 0 | 0 | 0 | 0 | 18 |
| 12 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 13 | 0 | 9 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 13 |
| 14 | 0 | 5 | 0 | 1 | 0 | 5 | 0 | 0 | 0 | 0 | 10 |
| 15 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 |
| 16 | 2 | 9 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 10 |
| 17 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| 18 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| 19 | 14 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 15 |
| 20 | 3 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 8 |
| 21 | 11 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 12 |
| 22 | 0 | 5 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 6 |
| CA . | Whole chromosome additions, % . | Whole chromosome deletions, % . | Deletions, % . | Translocations, % . | Duplications, % . | Isochromes, % . | Overall, % . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| p-arm . | q-arm . | p-arm . | q-arm . | p-arm . | q-arm . | p-arm . | q-arm . | ||||
| 1 | 0 | 0 | 4 | 0 | 2 | 7 | 0 | 2 | 0 | 0 | 12 |
| 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| 3 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 |
| 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| 5 | 14 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 16 |
| 6 | 0 | 3 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 6 |
| 7 | 10 | 1 | 0 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 14 |
| 8 | 2 | 4 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 8 |
| 9 | 15 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 15 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 13 | 1 | 0 | 1 | 0 | 6 | 0 | 0 | 0 | 0 | 18 |
| 12 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 13 | 0 | 9 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 13 |
| 14 | 0 | 5 | 0 | 1 | 0 | 5 | 0 | 0 | 0 | 0 | 10 |
| 15 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 |
| 16 | 2 | 9 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 10 |
| 17 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| 18 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| 19 | 14 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 15 |
| 20 | 3 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 8 |
| 21 | 11 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 12 |
| 22 | 0 | 5 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 6 |
Summary of CA analysis (n = 222)
| Chromosomal abnormalities . | % . |
|---|---|
| t(11;14) | 5 |
| Any trisomy | 24 |
| Any deletion | 19 |
| Any translocation | 15 |
| Hyperdiploid3-150 | 20 |
| Hypodiploid3-151 | 11 |
| CA 13 (inclusive of del 13, del 13q, and t13q) | 13 |
| Hypodiploid or CA 13 | 16 |
| MM-MDS3-152 | 7 |
| Any CA | 33 |
| Chromosomal abnormalities . | % . |
|---|---|
| t(11;14) | 5 |
| Any trisomy | 24 |
| Any deletion | 19 |
| Any translocation | 15 |
| Hyperdiploid3-150 | 20 |
| Hypodiploid3-151 | 11 |
| CA 13 (inclusive of del 13, del 13q, and t13q) | 13 |
| Hypodiploid or CA 13 | 16 |
| MM-MDS3-152 | 7 |
| Any CA | 33 |
Abnormalities typical of myelodysplasia (MDS) in the context of myeloma signature karyotype.
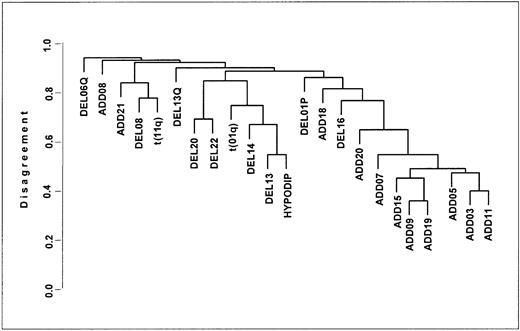
On average, 6 chromosomes were altered in any given CA (median, 6 chromosomes; range, 1-13 chromosomes). We performed clustering analysis of specific CAs present in at least 10 patients prior to the start of protocol therapy (Figure 1).
Cluster tree for pretherapy cytogenetic abnormalities.
Cluster analysis of associations of specific chromosome aberrations present prior to therapy. The ordinate denotes the percentage disagreement in such a way that lower clusters on the tree are more highly correlated with each other: for example, t(11q) and t(14), as well as trisomies (denoted by additions of various chromosome numbers) (add 3, add 5, add 7, add 9, add 11, add 15, add 19).
Cluster tree for pretherapy cytogenetic abnormalities.
Cluster analysis of associations of specific chromosome aberrations present prior to therapy. The ordinate denotes the percentage disagreement in such a way that lower clusters on the tree are more highly correlated with each other: for example, t(11q) and t(14), as well as trisomies (denoted by additions of various chromosome numbers) (add 3, add 5, add 7, add 9, add 11, add 15, add 19).
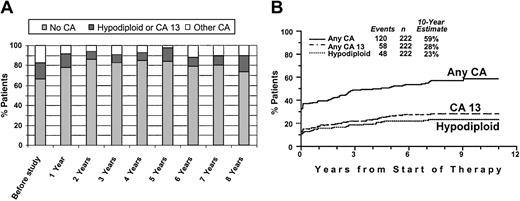
Of patients with hypodiploid CAs at baseline, 58% also had del(13), and 71% had any abnormality of chromosome 13 (including del(13), del(13q), and t(13q)) (CA 13), indicating a high degree of correlation (r = 0.61). Of the total of 222 patients, 16% had either CA 13 or hypodiploidy (“hypo–13 CA”). Translocations of 11q and 14q, representing a t(11;14)(q13;q32) in 10 (59%) patients, clustered closely at diagnosis. The actual frequency of specific CAs (no CA, hypo–13 CA, other CAs [which are prognostically relevant]) during the disease course is depicted in Figure 2A. Presumably as a result of suppression by effective therapy especially of non–CA 13 and nonhypodiploid clones, progressively fewer CAs were noted during the first 2 years after initiation of TT I, and CAs slowly increased thereafter. Eventually, at 8 years, the distribution of CAs was similar to that at baseline. The cumulative CA frequency reached 58% at 10 years, including more than a doubling of hypo–13 CA from 16% to 36% (Figure 2B).
Incidence of cytogenetic abnormalities recorded serially after initiation of treatment.
(A) Incidence of select cytogenetic abnormalities (no cytogenetic abnormalities, no CA; hypodiploid or CA 13; other CAs) before the study and on serial follow-up. Note that, at diagnosis, 33% of patients had CAs, approximately evenly divided between hypodiploid/CA 13 and other CAs. Over time, presumably as a result of effective therapy, the proportion of patients with CAs decreased to a low 15%. Eventually, the incidence of hypodiploid CAs/CA 13 among those with CAs increased. (B) Increase in the cumulative incidence of CAs during the disease course. Cumulative incidence, from start of Total Therapy I, of CAs and adverse prognostic subgroups (CA 13, hypodiploidy).
Incidence of cytogenetic abnormalities recorded serially after initiation of treatment.
(A) Incidence of select cytogenetic abnormalities (no cytogenetic abnormalities, no CA; hypodiploid or CA 13; other CAs) before the study and on serial follow-up. Note that, at diagnosis, 33% of patients had CAs, approximately evenly divided between hypodiploid/CA 13 and other CAs. Over time, presumably as a result of effective therapy, the proportion of patients with CAs decreased to a low 15%. Eventually, the incidence of hypodiploid CAs/CA 13 among those with CAs increased. (B) Increase in the cumulative incidence of CAs during the disease course. Cumulative incidence, from start of Total Therapy I, of CAs and adverse prognostic subgroups (CA 13, hypodiploidy).
CAs and SPFs
Strong associations were noted between some prognostically relevant SPFs and CA 13 as well as hypodiploid CAs, including Durie-Salmon stage III, β-2-microglobulin (B2M) level of at least 345 nM (at least 4 mg/L); lactic dehydrogenase (LDH) level of at least 190 U/L; hypoalbuminemia (albumin level of at least 35 g/L [at least 3.5 g/dL]); hemoglobin level no higher than 100 g/L (no higher than 10 g/dL); and bone marrow plasmacytosis of at least 50%. All of these represent features of high tumor burden or aggressive disease. IgA was more frequent with MM-MDS (odds ratio [OR], 5.4), whereas C-reactive protein (CRP) of at least 4 mg/L was associated with t(1q). Translocations of 11q (t(11q)) predicted for age of at least 65 years and B2M of at least 345 nm (at least 4 mg/L).
CAs and clinical outcome
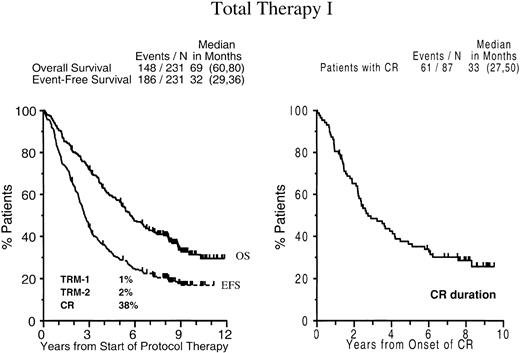
EFSs and OSs of the entire patient population are depicted in Figure 3: 20% of patients remain event-free and 36% alive, with 10-year estimates of EFS and OS of 17% (± 5%) and 31% (± 7%), respectively. The median CR duration is 2.8 years after the onset of CR (median time to CR, 9 months), with 29% (± 10%) of patients in continuous CR at 8 years.
Survival and complete remission duration with total therapy I.
Kaplan-Meier plots of overall survival and event-free survival dated from the initiation of Total Therapy I (left panel) and CR duration dated from the onset of CR (right panel). Numbers in parentheses indicate 95% confidence interval (CI).
Survival and complete remission duration with total therapy I.
Kaplan-Meier plots of overall survival and event-free survival dated from the initiation of Total Therapy I (left panel) and CR duration dated from the onset of CR (right panel). Numbers in parentheses indicate 95% confidence interval (CI).
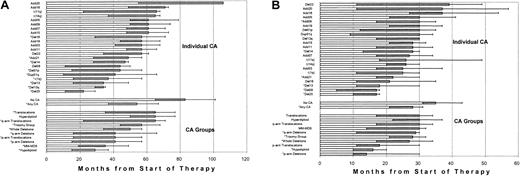
Both EFS and OS differed markedly when examined in the context of individual CAs and of CA groups (Figure4). The poor prognosis of patients presenting with CA 13, and del(20) as well as CA 1 is apparent. When examined in the context of group assignment, hypodiploid CAs had the shortest OS. On multivariate analysis, hypodiploid–13 CA, present in 16% of patients prior to therapy, was the only unfavorable parameter among all CAs that was independently associated with both short EFS and OS (Table4). Both an LDH of at least 190 U/L and renal failure (creatinine level ≥ 2 mg/dL) were associated with inferior OS. Given the potential clinical implications of CR and second autotransplantation, both CAs and SPFs were re-examined in the context of time-dependent parameters. Results indicated superior OS among patients achieving CR promptly and whose relapse occurred late. When considered in the context of SPFs, a CRP level of at least 4 mg/L was the only additional variable that adversely affected both EFS and OS.
Outcomes vary according to specific pretreatment CAs.
Bar chart of median OS (panel A) and EFS (panel B) for specific CAs and derived CAs; 95% CIs are also indicated by the shaded area (lower confidence limit) and tailed lines (upper confidence limit). Asterisks indicate significant differences on univariate analysis (P < .05).
Outcomes vary according to specific pretreatment CAs.
Bar chart of median OS (panel A) and EFS (panel B) for specific CAs and derived CAs; 95% CIs are also indicated by the shaded area (lower confidence limit) and tailed lines (upper confidence limit). Asterisks indicate significant differences on univariate analysis (P < .05).
Multivariate regression results in TTI (n = 222)
| . | Overall survival . | Event-free survival . | ||
|---|---|---|---|---|
| Prognostic variables . | HR (95% CI) . | P4-150 . | HR (95% CI) . | P4-150 . |
| Results for specific pretherapy CAs | ||||
| del(20) | 2.86 (1.3, 6.5) | .012 | — | NS |
| del(8) | — | NS | 3.08 (1.5, 6.3) | .002 |
| Hypodiploid or CA 13 | 2.32 (1.5, 3.5) | < .001 | 1.82 (1.2, 2.7) | .002 |
| Results for specific pretherapy CAs and standard prognostic factors | ||||
| del 8 | — | NS | 3.61 (1.6, 8.4) | .003 |
| Hypodiploid or CA 13 | 2.33 (1.5, 3.5) | < .001 | 1.79 (1.2, 2.7) | .006 |
| CRP level at least 4 mg/L | 1.50 (1.0, 2.2) | .031 | 1.59 (1.1, 2.2) | .006 |
| LDH level at least 190 U/L | 1.70 (1.1, 2.6) | .013 | — | NS |
| Creatinine level above 176.8 μM | 2.52 (1.4, 4.6) | .002 | — | NS |
| B2M level at least 345 nM | — | NS | 1.46 (1.0, 2.1) | .036 |
| Results for specific pretherapy CAs, standard prognostic factors, and time-dependent covariates | ||||
| Time-dependent CR | 0.53 (0.32, 0.86) | .011 | — | NS |
| Time to CR, mo | 1.08 (1.04, 1.01) | < .001 | — | NS |
| Time-dependent relapse | 8.42 (5.26, 3.5) | < .001 | — | NS |
| Time-dependent hypodiploid or CA 13 | 2.75 (1.94, 3.88) | < .001 | 1.39 (1.01, 1.90) | .041 |
| Time-dependent TX2 | — | NS | 0.66 (0.44, 0.99) | .025 |
| CRP level at least 4 mg/L | 1.58 (1.12, 2.23) | .010 | 1.52 (1.12, 2.06) | .007 |
| Creatinine level above 176.8 μM | 1.91 (1.14, 3.22) | .014 | — | NS |
| B2M level at least 345 nM | — | NS | 1.66 (1.20, 2.30) | .002 |
| . | Overall survival . | Event-free survival . | ||
|---|---|---|---|---|
| Prognostic variables . | HR (95% CI) . | P4-150 . | HR (95% CI) . | P4-150 . |
| Results for specific pretherapy CAs | ||||
| del(20) | 2.86 (1.3, 6.5) | .012 | — | NS |
| del(8) | — | NS | 3.08 (1.5, 6.3) | .002 |
| Hypodiploid or CA 13 | 2.32 (1.5, 3.5) | < .001 | 1.82 (1.2, 2.7) | .002 |
| Results for specific pretherapy CAs and standard prognostic factors | ||||
| del 8 | — | NS | 3.61 (1.6, 8.4) | .003 |
| Hypodiploid or CA 13 | 2.33 (1.5, 3.5) | < .001 | 1.79 (1.2, 2.7) | .006 |
| CRP level at least 4 mg/L | 1.50 (1.0, 2.2) | .031 | 1.59 (1.1, 2.2) | .006 |
| LDH level at least 190 U/L | 1.70 (1.1, 2.6) | .013 | — | NS |
| Creatinine level above 176.8 μM | 2.52 (1.4, 4.6) | .002 | — | NS |
| B2M level at least 345 nM | — | NS | 1.46 (1.0, 2.1) | .036 |
| Results for specific pretherapy CAs, standard prognostic factors, and time-dependent covariates | ||||
| Time-dependent CR | 0.53 (0.32, 0.86) | .011 | — | NS |
| Time to CR, mo | 1.08 (1.04, 1.01) | < .001 | — | NS |
| Time-dependent relapse | 8.42 (5.26, 3.5) | < .001 | — | NS |
| Time-dependent hypodiploid or CA 13 | 2.75 (1.94, 3.88) | < .001 | 1.39 (1.01, 1.90) | .041 |
| Time-dependent TX2 | — | NS | 0.66 (0.44, 0.99) | .025 |
| CRP level at least 4 mg/L | 1.58 (1.12, 2.23) | .010 | 1.52 (1.12, 2.06) | .007 |
| Creatinine level above 176.8 μM | 1.91 (1.14, 3.22) | .014 | — | NS |
| B2M level at least 345 nM | — | NS | 1.66 (1.20, 2.30) | .002 |
HR indicates hazard ratio; 95% CI, 95% confidence interval; NS, not significant; TX2, second transplant; and —, not applicable.
Based on regression model.
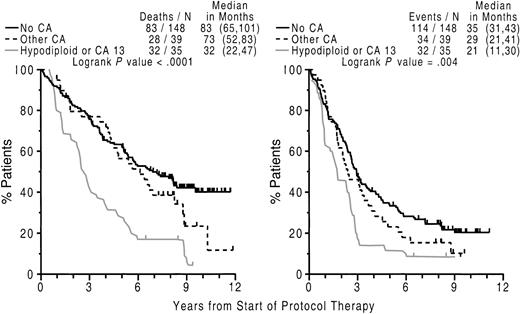
Importantly, hypodiploid–13 CA, also in a time-dependent setting, remained an independent adverse feature for both EFS and OS along with CRP, attesting to its robust clinical implications (Figure5).
Poor outcome with abnormalities of chromosome 13 (CA 13) and hypodiploidy.
Kaplan-Meier plots of OS (left panel) and EFS (right panel) dated from initiation of Total Therapy I, according to the presence or absence of CAs. Superior outcome was noted in the absence of CAs and worst outcome among patients exhibiting hypodiploidy or CA 13; those with other CAs had an intermediate prognosis and their overall survival was not significantly different from that of patients with no CAs. Numbers in parentheses indicate 95% CI.
Poor outcome with abnormalities of chromosome 13 (CA 13) and hypodiploidy.
Kaplan-Meier plots of OS (left panel) and EFS (right panel) dated from initiation of Total Therapy I, according to the presence or absence of CAs. Superior outcome was noted in the absence of CAs and worst outcome among patients exhibiting hypodiploidy or CA 13; those with other CAs had an intermediate prognosis and their overall survival was not significantly different from that of patients with no CAs. Numbers in parentheses indicate 95% CI.
Superior EFS and OS was enjoyed by 67% of patients lacking any CA whereas patients with hypodiploid–13 CA had the worst outcome; those with other CAs had EFS and OS not significantly different from the clinical course of patients without CAs. Thus, median OS exceeded EFS by more than 2-fold, both in the absence of CA and in case of other CAs; in contrast, the hypodiploid–13 CA group showed a median postrelapse OS of less than 1 year. The 7 patients with t(11;14) without concomitant CA 13 or hypodiploid CAs had median EFS and OS of 25 and 54 months, respectively, not different from the entire group of other CAs.
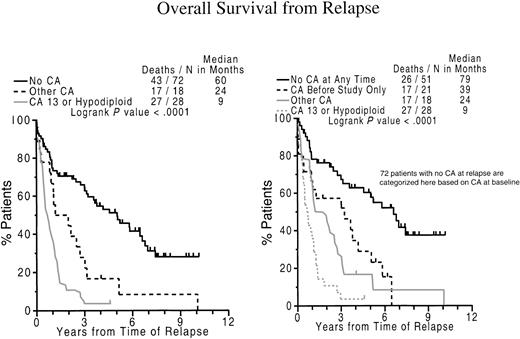
We next investigated the clinical implications of CAs detected on follow-up bone marrow examination. According to univariate time-dependent models to assess the instantaneous hazard of CA detection subsequent to initiation of therapy, both EFS and OS were significantly compromised whenever CA 13 or hypodiploid CAs were detected (data not shown). According to multivariate analysis, the presence of either CA 13 or hypodiploid CAs at relapse was associated with short subsequent OS. Thus, postrelapse OS was longest (5 years) in the absence of CA and progressively shortened to 2 years with other CAs and was only 9 months in case of hypodiploid–13 CA (Figure6).
Relationship of CA 13 or hypodiploidy at relapse and short subsequent survival.
Cytogenetic abnormalities involving chromosome 13 (CA 13) or hypodiploidy at relapse imparts short subsequent survival. Left panel: Kaplan-Meier plots of overall survival dated from the onset of relapse on Total Therapy I. The majority of patients without cytogenetic abnormalities (no CA) had a significantly longer survival than those with other CAs and especially those with hypodiploidy or CA 13. Right panel: additional consideration of pretherapy cytogenetic abnormalities separates, among those without CA at relapse, those with no CAs, also at diagnosis, as having the best survival whereas those without CAs at relapse but CAs at diagnosis had intermediate outcome.
Relationship of CA 13 or hypodiploidy at relapse and short subsequent survival.
Cytogenetic abnormalities involving chromosome 13 (CA 13) or hypodiploidy at relapse imparts short subsequent survival. Left panel: Kaplan-Meier plots of overall survival dated from the onset of relapse on Total Therapy I. The majority of patients without cytogenetic abnormalities (no CA) had a significantly longer survival than those with other CAs and especially those with hypodiploidy or CA 13. Right panel: additional consideration of pretherapy cytogenetic abnormalities separates, among those without CA at relapse, those with no CAs, also at diagnosis, as having the best survival whereas those without CAs at relapse but CAs at diagnosis had intermediate outcome.
Further examination of both baseline and relapse CAs in the 118 patients with available cytogenetic information (Table5) revealed superior postrelapse OS of 79 months among the 51 patients lacking CA at both time points; followed by 39 months in the 21 patients with CAs only at baseline.
Comparison of CAs at relapse and baseline in patients who experienced relapse
| Baseline . | Time of Relapse . | Total no. . | ||
|---|---|---|---|---|
| No CAs . | Hypodilpoid/CA 13 . | Other CAs . | ||
| No CAs | 70% | 16% | 14% | 73 |
| Other CAs | 42% | 25% | 33% | 24 |
| Hypodiploid/CA 13 | 52% | 48% | 0% | 21 |
| Total no. | 72 | 28 | 18 | 118 |
| Baseline . | Time of Relapse . | Total no. . | ||
|---|---|---|---|---|
| No CAs . | Hypodilpoid/CA 13 . | Other CAs . | ||
| No CAs | 70% | 16% | 14% | 73 |
| Other CAs | 42% | 25% | 33% | 24 |
| Hypodiploid/CA 13 | 52% | 48% | 0% | 21 |
| Total no. | 72 | 28 | 18 | 118 |
Of the 28 patients experiencing relapse with hypodiploid–13 CA, 12 had presented with no CA and 6 with other CAs at diagnosis (Table 5). Conversely, of the 21 with baseline hypodiploid–13 CA, 11 had no CA at relapse, none had other CAs, and 10 had experienced a recurrence of hypodiploid–13 CA. The remaining 15 patients of the original 36 who had this abnormality at baseline did not have cytogenetic examinations on follow-up, mostly owing to rapid death upon documentation of relapse.
Gene expression profiles
The data presented so far point to a unique prognostic role of certain baseline CAs (CA 13, hypodiploid CAs) (Figures 4-5). On multivariate analysis of individual CAs and CA groups, CA 13, but not hypodiploidy, was the dominant adverse feature for both OS and EFS (data not shown). Prospective concurrent evaluation of CAs, FISH for detection of del(13) (FISH 13), and GEPs are being conducted as part of TT II. Among approximately 50% of patients with FISH 13, only those also harboring CA 13 had inferior survival whereas those with FISH 13 but without CA 13 fared as well as patients lacking FISH 13; plasma cell–labeling index was not an independent prognosticator.11 We reasoned that in vitro MM cell division, required for metaphase CA to be detected, was a critical biologic feature of stroma cell–independent growth shared by CA 13 and other CAs.11 Availability of GEP data in 146 TT II patients along with CA and FISH 13 status provided the opportunity to determine the molecular basis for the uniquely grave prognosis associated with FISH 13/CA (CA 13) compared with all other groups, especially no FISH 13/CA.
FISH 13 versus no FISH 13.
From a training set of 47 samples with FISH 13 and 51 samples without FISH 13, 36 genes were identified at the intersection of data sets from the χ2, WRS, and SAM statistical analyses (P < .001) (“Patients and methods”). Thirty-five genes (including 32 mapping to chromosome 13) were underexpressed in the FISH 13 group, and only one gene,IGF1R (insulin-like growth factor 1 receptor), was overexpressed in the FISH 13 group. A discriminant analysis was used to find a subset of the 36 differentially expressed genes that could accurately predict del(13) in a set of new patient samples. After the multistepwise discriminant analysis (MSDA) and resulting linear discriminant function created between 2 groups, both forward and backward variable selections were performed. Ten genes, includingRB1, were identified as correctly discriminating 41 (94%) of 48 cases from a held-out validation group in which FISH 13 status was known.
CAs versus no CAs.
The sequential χ2, WRS, and SAM analyses were also performed by comparing 37 MM samples with no FISH 13/no CA and 34 with FISH 13/CA. A total of 157 genes were identified with significantly altered mRNA expression levels (P < .001). Most of the genes (91%) were overexpressed in the FISH 13/CA group, only 14 genes (8 mapping to chromosome 13) were underexpressed in the FISH 13/CA group. The largest class of significant genes coded for proteins involved in different checkpoints of cell cycle progression and DNA replication. Genes included those related to proliferation,PCNA, MKI67, and MCM2; G1/S transition, TK1 and CDI1; G2/M checkpoint, CCNB1, CDC2, UBE2C, and BUB1B; chromosome segregation,CENPA, CENPE, CENPF, KNSL1, MAD2L1, and STK12, STK15,and STK18; and DNA replication, TOP2A,PRKDC, TK1, and TYMS.
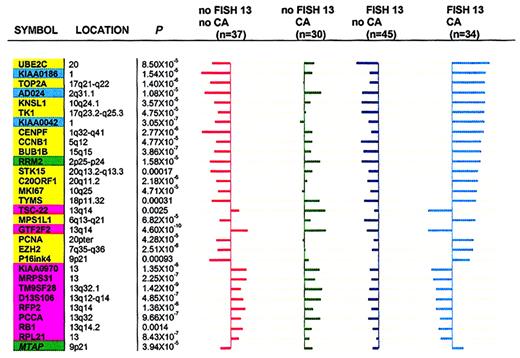
Distinguishing 4 MM subgroups on the basis of FISH 13 and CA.
To demonstrate that expression levels of these genes could discriminate 4 cytogenetic subgroups (no FISH 13/no CA; no FISH 13/CA; FISH 13/no CA; and FISH 13/CA), the so-called “shrunken centroids” method20 was used with 30 genes. These included the top 20 differentially expressed genes in a comparison between FISH 13/CA and no FISH 13/no CA as well as 10 genes identified by MSDA in the comparison of FISH 13 versus no FISH 13. Figure7 shows centroids (average expression of each gene) in each of the 4 patient subgroups.
Cell cycle– and chromosome-related genes in FISH 13/CA subgroups.
Cell cycle– and chromosome-related genes distinguish FISH 13/CA subgroups. The “shrunken centroids” method20 was applied to reveal significant differences across 4 MM subgroups on the basis of 30 genes having at least one nonzero difference (17 related to cell cycle/DNA metabolism, 3 genes of unknown function, and 10 mapping to chromosome 13). The genes with nonzero components in each class are mutually exclusive. The length of horizontal bars for a given gene represents the difference between the overall centroid (vertical axes) and each of the 4 subgroup centroids. Bars to the left indicate lower expression in subgroups relative to the overall centroid; bars to the right indicate higher expression in subgroups relative to the overall centroid. The gene order, from top to bottom, is based on the greatest difference (with the bar longest at top) between the overall centroid and any 1 of the 4 group centroids. Kruskal-Wallis test Pvalues for difference between each group centroid and the overall centroid for all comparisons are indicated. Background colors distinguish different gene groups: pink, chromosome 13 genes; yellow, cell cycle genes; green, DNA metabolism genes; cyan, genes with unknown function. Note: FISH 13/no CA (45 patients) is characterized by uniformly left-sided bars (underexpression of both cell cycle/DNA metabolism– and chromosome 13–related genes). No FISH 13/CA (30 patients) show mainly right-sided bars, indicative of expression of both cell cycle/DNA metabolism– and chromosome 13–related genes. No FISH 13/no CA (37 patients) is characterized by overexpression of chromosome 13 genes (right-sided bars) and underexpression of all 17 cell cycle/DNA metabolism–related and 3 unknown function genes (left-sided bars). FISH/CA (34 patients) shows the mirror image of no FISH/no CA in that all cell cycle/DNA metabolism genes are hyperactivated (more so than in the no FISH 13/CA group) and chromosome 13 genes are inactivated (more so than in the FISH 13/no CA group).
Cell cycle– and chromosome-related genes in FISH 13/CA subgroups.
Cell cycle– and chromosome-related genes distinguish FISH 13/CA subgroups. The “shrunken centroids” method20 was applied to reveal significant differences across 4 MM subgroups on the basis of 30 genes having at least one nonzero difference (17 related to cell cycle/DNA metabolism, 3 genes of unknown function, and 10 mapping to chromosome 13). The genes with nonzero components in each class are mutually exclusive. The length of horizontal bars for a given gene represents the difference between the overall centroid (vertical axes) and each of the 4 subgroup centroids. Bars to the left indicate lower expression in subgroups relative to the overall centroid; bars to the right indicate higher expression in subgroups relative to the overall centroid. The gene order, from top to bottom, is based on the greatest difference (with the bar longest at top) between the overall centroid and any 1 of the 4 group centroids. Kruskal-Wallis test Pvalues for difference between each group centroid and the overall centroid for all comparisons are indicated. Background colors distinguish different gene groups: pink, chromosome 13 genes; yellow, cell cycle genes; green, DNA metabolism genes; cyan, genes with unknown function. Note: FISH 13/no CA (45 patients) is characterized by uniformly left-sided bars (underexpression of both cell cycle/DNA metabolism– and chromosome 13–related genes). No FISH 13/CA (30 patients) show mainly right-sided bars, indicative of expression of both cell cycle/DNA metabolism– and chromosome 13–related genes. No FISH 13/no CA (37 patients) is characterized by overexpression of chromosome 13 genes (right-sided bars) and underexpression of all 17 cell cycle/DNA metabolism–related and 3 unknown function genes (left-sided bars). FISH/CA (34 patients) shows the mirror image of no FISH/no CA in that all cell cycle/DNA metabolism genes are hyperactivated (more so than in the no FISH 13/CA group) and chromosome 13 genes are inactivated (more so than in the FISH 13/no CA group).
The values represent the difference between the individual group centroid and the overall centroid (all 146 cases). The 45 patients with FISH 13/no CA were uniformly characterized by underexpression of both cell cycle– and chromosome 13–related genes. By contrast, the 30 patients in the no FISH 13/CA group showed mainly right-sided bars, indicative of relative overexpression of both cell cycle– and chromosome 13–related genes. The third group of 37 patients with no FISH 13/no CA was characterized by expression of chromosome 13 genes (right-sided bars) and underexpression of all cell cycle genes (left-sided bars). Finally, the 34 patients with FISH 13/CA represented the mirror image of the previous group in that all cell cycle genes were particularly strongly expressed (more so than in the no FISH 13/CA group) and chromosome 13 genes were profoundly underexpressed (more so than in the FISH 13/no CA group). Thus, it appears that, collectively, FISH 13/CA is characterized by a net overexpression of all cell cycle genes and underexpression of chromosome 13 genes. The mirror image is seen in the no FISH 13/no CA group. However, the underexpression of cell cycle–related genes does not match their overexpression in the FISH 13/CA group. In either comparison, FISH 13 appears to be a modulator of cell cycle gene expression.
Discussion
Total Therapy I
The profound CA complexity typical of MM has made it difficult to account for individual prognostic contributions of certain CAs, especially when one considers that 49 specific abnormalities occurred in fewer than 5% of patients and that 93% coexisted. Here, we confirm the adverse implications of CA 13 and hypodiploid CAs, confirming 2 recent other studies.5,21 Thus, among 475 patients enrolled in tandem transplantation trials prior to 2000 (including previously treated and refractory MM), Fassas et al21 noted that CA 13, regardless of ploidy status, and a hypodiploid karyotype without CA 13 both imparted poor outcome after high-dose therapy. The more frequent association of CA 13 with hypodiploidy as compared with other CAs (65% versus 30%;P < .001) is in keeping with results of the cluster analysis of this report (Figure 1). The prognosis of patients with other CAs, including t(11;14)(q13;q32), was superior to that observed with CA 13 or hypodiploidy and, in fact, was not significantly inferior to the outcome of patients with no CAs (Figure 5).
Although perhaps anticipated, this is the first documented observation in MM of an inferior prognosis associated with the detection of CAs, particularly of CA 13 and hypodiploid CAs, at relapse or, according to time-dependent instantaneous hazard models, at any time after initiation of primary therapy. Best outcomes were noted when CAs were absent both before treatment and after transplantation; prognosis was worst when hypodiploidy or CA 13 was present at relapse, regardless of pretreatment findings (Figure 6). Indeed, of the 28 patients with hypodiploid–13 CA at relapse, 18 represented new CAs (12 without CA at baseline, 6 with other CAs) (Table 5). In the first instance (no CA at baseline and hypodiploid–13 CA at relapse), del(13) or hypodiploidy was probably present at diagnosis if interphase FISH had been applied. In the second scenario (hypodiploid–13 CA at relapse with other CAs at diagnosis), CA 13 or hypodiploidy was acquired in addition to other CAs rather than representing a separate clone, consistent with clonal evolution in some patients. Together with nearly doubling by 10 years, in a cumulative account of CA incidence (including hypodiploid–13 CA), these findings are consistent with a progressively more proliferative nature of advancing MM. It is conceivable that eventually all patients with MM develop CA when one considers the universal presence of genetic abnormalities detectable by interphase FISH22 with a 50% incidence of del(13).11 Thus, “no CA” probably reflects normal hematopoiesis, whereas CAs, as a result of successful in vitro MM cell division, distinguish a high-risk subtype that proliferates independently of the bone marrow microenvironment, consistent with an autocrine growth mechanism.11 Associations between the presence of CA and short telomere length/high telomerase activity in highly purified MM cells are consistent with telomere erosion as an important molecular mechanism in the generation of the observed genomic instability.23
Interphase FISH has recently been applied to the setting of high-dose therapy by French MM investigators.24 Applying probes pertinent to the detection of translocations involving the IGHlocus at 14q32 and del(13), the authors reported, among patients with CAs, the worst prognosis in those exhibiting t(4;14)(p16;q32) and superior outcome in cases of t(11;14)(q13;q32). Intermediate prognosis in the remainder was limited to patients without del(13). The median survival of those with FISH del(13), however, almost 4 years after a single transplantation, is significantly longer than the 2.5 years observed after tandem transplantation for patients with CA 13 as reported in this study. A worse prognosis in cases of metaphase-defined CA 13 was noted in a head-to-head comparison of interphase FISH del(13) and standard cytogenetics in patients receiving TT II.11 Indeed, the OS of patients with FISH 13 but without CA 13 was indistinguishable from that of patients without FISH 13 (regardless of the presence or absence of CA).
Total Therapy II
GEP analysis showed significant elevation of a large panel of cell cycle/DNA metabolism genes both in the CA versus no CA and the FISH 13 versus no FISH comparisons (Figure 7). Thus, the expression patterns of these genes could be used to segregate discrete cytogenetic subgroups so that MM with CA and FISH 13 had highly elevated expression levels of cell cycle genes in comparison with CA without FISH 13. The deletion of chromosome 13 not only results in a haploinsufficiency of specific chromosome 13 genes (eg, GTF2F2, TSC-22, and RB1) but also is associated with an amplification of the expression of cell cycle genes. The reason for this effect is not known, but probably reflects a larger proliferative proportion of CD138 cells, thus accounting for the dire outcome of CA 13 (FISH 13/CA) MM in comparison with other CAs (no FISH 13/CA) MM.
On the basis of the integration of molecular and conventional cytogenetics, patient outcome, and global GEPs, we hypothesize that one consequence of del(13) is haploinsufficiency of the tumor suppressor gene RB1. This hypothesis is supported by recent data linking an increased risk of human colon cancer and murine lymphoma to haploinsufficiency of BLM.25,26 Importantly, this model can account for a lack of inactivating mutations in the remaining allele of RB1 and the inability, even after exhaustive searches, to identify a definitive tumor suppressor gene mapping to 13q14.27-29 Given the important influence of DNA repair defects in cancer development, it is noteworthy that 2 excision repair genes, ERCC5 (excision repair cross-complementing rodent repair deficiency, complementation group 5 gene, mapping to chromosome 13) and ERCC1 (mapping to chromosome 19q13.2), were significantly down-regulated in patients with FISH 13 (data not shown), suggesting that loss of these genes could lead to the genomic instability and poor outcome in FISH 13 disease. Interestingly, insulin-like growth factor I receptor(IGF1R), a powerful growth-signaling receptor in MM,30-32 represents the only gene significantly up-regulated (turned on) in FISH 13 MM. We have recently reported that both normal and MM bone marrow plasma cells express IGF1,whereas tonsil-derived plasma cells and tonsillar B cells do not.18 Thus, we speculate that activation ofIGF1R (absent on normal plasma cells) on MM cells could create an autocrine growth–signaling loop in FISH 13 MM.
Elevated expression of cyclin B1 (CCNB1) and the mitotic cyclin–specific ubiquitin-conjugating enzyme E2C (UBE2C) in MM with CAs, and especially in FISH 13/CA, is notable. Since mutantUBE2C results in stabilization of both cyclin A and cyclin B, arrests cells in M phase, and inhibits the onset of anaphase, destruction of mitotic cyclins by ubiquitin-dependent proteolysis seems required for cells to complete mitosis and enter interphase of the next cell cycle.33UBE2C-mediated ubiquitination is also involved in inactivation of CDC2 (hyperactivated in FISH 13/CA) and in sister chromatid separation, 2 processes normally coordinated during exit from mitosis. We speculate that a therapeutic strategy of treating patients with FISH 13/CA might be to inhibitUBE2C function. Overexpression of topoisomerase II(TOP 2A) in the FISH 13/CA MM subgroup suggests that inhibitors of these gene products may be especially effective in this poor prognosis group. In fact, etoposide has been a component of both TT I (EDAP [etoposide, dexamethasone, adriamycin, and cisplatin] regimen) and TT II (DCEP [dexamethasone, cyclophosphamide, etoposide, and cisplatin] for induction and consolidation) (“Patients and methods”). Finally, the recent observation of marked antitumor activity of the proteasome inhibitor PS-34134-36and allogeneic T cells37 justify rapid implementation of clinical trials of these therapeutic approaches in this high-risk MM entity. Thus, molecular profiling may prove useful in moving toward the rational development of new combination therapies.
We conclude that the perceived shortcomings of metaphase karyotyping, that is, its dependence on in vitro mitotic activity in a typically hypoproliferative malignancy such as MM, has helped reveal a possible microenvironment-independent, more aggressive disease subtype with distinct molecular genetic features. Consequently, we strongly recommend that metaphase karyotyping be performed as part of the staging of all patients with MM at both diagnosis and relapse.
The authors gratefully acknowledge the outstanding contributions of Caran Swanson, BS; Bonnie Jenkins, RN, OCN; and the Nursing and Support Staff of the Myeloma Institute for Myeloma and Therapy. This work is dedicated to a lasting memory of Fabio Bertarelli, who died in 1998 at the age of 73 after an 11-year battle with MM, exhibiting high-risk cytogenetics abnormalities. His intellectual, emotional, and fiscal contributions were critical to the establishment of the Arkansas Myeloma Program.
Prepublished online as Blood First Edition Paper, January 16, 2003; DOI 10.1182/blood-2002-09-2873.
Supported in part by CA55819 from the National Cancer Institute, Bethesda, MD.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Bart Barlogie, Director, Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, 4301 West Markham, Slot 816, Little Rock, AR 72205; e-mail:barlogiebart@uams.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal