Abstract
The transcription factor PU.1 is required for normal blood cell development. PU.1 regulates the expression of a number of crucial myeloid genes, such as the macrophage colony-stimulating factor (M-CSF) receptor, the granulocyte colony-stimulating factor (G-CSF) receptor, and the granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor. Myeloid cells derived from PU.1−/− mice are blocked at the earliest stage of myeloid differentiation, similar to the blast cells that are the hallmark of human acute myeloid leukemia (AML). These facts led us to hypothesize that molecular abnormalities involving the PU.1 gene could contribute to the development of AML. We identified 10 mutant alleles of the PU.1 gene in 9 of 126 AML patients. The PU.1 mutations comprised 5 deletions affecting the DNA-binding domain, and 5 point mutations in 1) the DNA-binding domain (2 patients), 2) the PEST domain (2 patients), and 3) the transactivation domain (one patient). DNA binding to and transactivation of the M-CSF receptor promoter, a direct PU.1 target gene, were deficient in the 7 PU.1 mutants that affected the DNA-binding domain. In addition, these mutations decreased the ability of PU.1 to synergize with PU.1-interacting proteins such as AML1 or c-Jun in the activation of PU.1 target genes. This is the first report of mutations in the PU.1 gene in human neoplasia and suggests that disruption of PU.1 function contributes to the block in differentiation found in AML patients.
Introduction
Although a number of oncogenes that affect proliferation and cell death have been identified in leukemias, only a few differentiation genes, such as AML1 or C/EBPα, have been implicated in the malignant phenotype.1-9 As transcription factors play a major role in cell differentiation, including the development of specific hematopoietic lineages from stem cells,2-4 they represent targets for disruption in acute myeloid leukemia (AML), a disease characterized by a block in differentiation of white blood cells. Heterozygous germ-line mutations of the AML1 gene cause a congenital platelet defect and a propensity to develop AML.6 Sporadic heterozygous and biallelic point mutations in the runt domain of the AML1 gene were recently reported in 6 of 123 AML patients.5,9 In addition, biallelic mutations in AML1 were found at an increased frequency in AML-M0 and in myeloid malignancies with acquired trisomy 21.7 Similarly, our group and others have identified heterozygous mutations in the transcription factor C/EBPα, which is crucial for the granulocytic lineage, in patients with AML.8 10 However, in many other cases of AML, the genetic basis for this differentiation block remains poorly understood.
The transcription factor PU.1 represents a unique transcriptional regulator within the hematopoietic system.3,4 PU.1 is a member of the Ets transcription family and is predominantly expressed in hematopoietic cells.11-14 Ets factors contain a characteristic DNA-binding domain of approximately 80 amino acids.15 The PU.1 protein consists of 264 amino acids, with the DNA-binding domain located in the carboxyl terminal part of the protein, whereas the amino terminus contains the activation domain.16 PU.1 is required for the proper generation of both myeloid (macrophages and neutrophils) and lymphoid lineages (B- and T- lymphocytes).14,17,18 PU.1 regulates the expression of a number of myeloid genes, such as CD11b, the macrophage colony-stimulating factor (M-CSF) receptor, the granulocyte colony-stimulating factor (G-CSF) receptor, and the granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor.19-24 PU.1−/− mice completely lack macrophages, as well as B cells, and show impaired granulopoiesis and T-cell development.17,18,24,25 Myeloid cells derived from PU.1−/− mice are blocked at the earliest stage of myeloid differentiation, similar to the blast cells in human AML.18,23,24,26,27 We therefore wondered whether molecular abnormalities involving the PU.1 gene could contribute to the development of AML. Here, we demonstrate that PU.1 is mutated in 7% of all AML patients, predominantly in undifferentiated AML (M0) or in AML of the monocytic lineage (M4/M5). We show that mutations in the DNA-binding domain result in a loss of the ability to activate important target genes, such as the M-CSF receptor. As cancer in general represents a block in differentiation and PU.1 is crucial for proper blood development, our findings support a model in which a mutated PU.1 protein disrupts the normal differentiation process and leads to a block in differentiation. The finding in AML patients of mutations in other genes that are important for myeloid development (such as AML1 and C/EBPα) further supports such a model, as these transcription factors interact at various stages of normal myeloid development.21,22,28 29 This is the first report of mutations of the Ets transcription factor PU.1 in the context of a malignant human disease.
Patients, materials, and methods
Patients
Patient samples, most of which were previously screened for AML1 mutations,5 were diagnosed according to the French-American-British (FAB) criteria. The patient samples were collected at the time of diagnosis with informed consent before the initiation of treatment. Mononuclear cells were isolated from bone marrow or peripheral blood samples by Ficoll density gradient centrifugation and cryopreserved in liquid nitrogen until molecular analysis.
Mutational analysis
Total RNA was isolated from mononuclear cells and reverse transcribed using oligo(dT) primers. For analyzing cDNA, primers were designed from the PU.1 sequence in GenBank accession number X52056; primer sequences are provided in Table 1. To analyze DNA, exon-specific primer pairs were designed (GenBank accession numbers AC019059 and AC018410) (Table 1). Polymerase chain reaction (PCR) products were electrophoresed on 1% agarose gels, gel purified (Qiagen, Santa Clarita, CA), and sequenced using BigDye Terminators and AmpliTaq FS (Applied Biosystems, Foster City, CA). Sequencing results containing mutations were repeated 3 times, including repetitions of the PCR and sequencing with an alternative primer. Mutated PCR products were subcloned into the pGEM-T vector (Promega, Madison, WI) and subsequently sequenced.
Sequences of the primers used
| Primers . | Direction . | Exon . |
|---|---|---|
| cDNA | ||
| 5′-TCACCCAGGGCTCCTGTAGCTCA-3′ | Sense | |
| 5′-CCGGGAGCGTCCTCCCTGTGTCCG-3′ | Antisense | |
| 5′-AGCAGATGCACGTCCTCGATACC-3′ | Sense | |
| 5′-TCGCCCTCCTCCTCATCTGAGCT-3 | Antisense | |
| Genomic DNA | ||
| 5′-TCACCCAGGGCTCCTGTAGCTCA-3′ | Sense | Exon 1 |
| 5′-TCGTGGGCAGGCAGGCAGGCGTCC-3′ | Antisense | Exon 1 |
| 5′-TGATGGGGACCAGCGTGCGGGGT-3′ | Sense | Exon 2 |
| 5′-TCTCTCCAGACCCCAGGACCAGGC-3′ | Antisense | Exon 2 |
| 5′-ACTATAACCTTTTCCTGCCCTGCC-3′ | Sense | Exon 3 |
| 5′-AGCCTGTGTCAGCTTCCTGTGAAG-3′ | Antisense | Exon 3 |
| 5′-TGCACTCCTTCTCTCCCCAGCTGACC-3′ | Sense | Exon 4 |
| 5′-ACACACACGCGACTCGGTGGCGTG-3′ | Antisense | Exon 4 |
| 5′-CCGGGCCCCTGTGCGTACGCAAGG-3′ | Sense | Exon 5 |
| 5′-CCGGGAGCGTCCTCCCTGTGTCCG-3′ | Antisense | Exon 5 |
| Primers . | Direction . | Exon . |
|---|---|---|
| cDNA | ||
| 5′-TCACCCAGGGCTCCTGTAGCTCA-3′ | Sense | |
| 5′-CCGGGAGCGTCCTCCCTGTGTCCG-3′ | Antisense | |
| 5′-AGCAGATGCACGTCCTCGATACC-3′ | Sense | |
| 5′-TCGCCCTCCTCCTCATCTGAGCT-3 | Antisense | |
| Genomic DNA | ||
| 5′-TCACCCAGGGCTCCTGTAGCTCA-3′ | Sense | Exon 1 |
| 5′-TCGTGGGCAGGCAGGCAGGCGTCC-3′ | Antisense | Exon 1 |
| 5′-TGATGGGGACCAGCGTGCGGGGT-3′ | Sense | Exon 2 |
| 5′-TCTCTCCAGACCCCAGGACCAGGC-3′ | Antisense | Exon 2 |
| 5′-ACTATAACCTTTTCCTGCCCTGCC-3′ | Sense | Exon 3 |
| 5′-AGCCTGTGTCAGCTTCCTGTGAAG-3′ | Antisense | Exon 3 |
| 5′-TGCACTCCTTCTCTCCCCAGCTGACC-3′ | Sense | Exon 4 |
| 5′-ACACACACGCGACTCGGTGGCGTG-3′ | Antisense | Exon 4 |
| 5′-CCGGGCCCCTGTGCGTACGCAAGG-3′ | Sense | Exon 5 |
| 5′-CCGGGAGCGTCCTCCCTGTGTCCG-3′ | Antisense | Exon 5 |
In order to analyze DNA, exon-specific genomic primer pairs were designed (GenBank accession numbers AC019059 and AC018410). One cDNA primer pair (the first pair) was designed to amplify the entire coding region of the human PU.1 cDNA, including 143 bp of the 5′UTR and 66 bp of the 3′UTR (GenBank accession number X52056). The second two cDNA primers correspond to sequences in the middle of the coding region and were used to sequence these amplified PCR products.
Plasmids
PU.1 wild-type and mutants were subcloned between theBamHI and EcoRI sites of the pcDNA3 expression vector, and a FLAG sequence (ATG GAC TAC AAA GAC GAT GAC GAC AAG) was added in frame at the 5′ end. PU.1 wild-type and the mutant G208fsX were fused in frame at the carboxyl end to the ligand-binding domain of the estrogen receptor alpha (ERα) in the retroviral pBabePuro vector.30 As a control, the ERα sequence alone was also subcloned into the pBabePuro vector.
Immunoblotting
Cells were lysed in RIPA buffer, and protein extracts were fractionated on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to nitrocellulose membranes by electroblotting. PU.1 was detected with rabbit anti-rat PU.1 polyclonal serum (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, catalog #sc-352) followed by an anti–rabbit IgG-horseradish peroxidase (HRP)–conjugated secondary antibody (Santa Cruz, catalog #sc-2004). A monoclonal FLAG-M2 antibody (Sigma, St Louis, MO, catalog #F-3165) was used at a concentration of 10 μg/mL and detected with an anti–mouse IgG-HRP–conjugated secondary antibody (Santa Cruz, catalog #sc-2055). A monoclonal anti–mouse β-tubulin antibody served as a loading control (Boehringer Mannheim, Indianapolis, IN, catalog #1111876) and was detected with an anti–mouse IgG-HRP–conjugated secondary antibody (Santa Cruz, catalog #sc-2005).
Electrophoretic mobility shift assay
Nuclear extracts were prepared after lysing cells with a small-gauge syringe as previously described.31,32 The M-CSF receptor promoter oligonucleotide (base pair [bp] −53 to −36 containing the PU.1 binding site) had the sequence 5′-TAAAAGGGGAAGAAGAGG-3′.20 For supershift experiments, 2 μl of PU.1 polyclonal rabbit serum were added using either a commercially available PU.1 antibody (Santa Cruz, catalog #sc-352X) directed against amino acids 251 to 271 of the murine PU.1 protein or an antiserum directed against the amino terminus of the PU.1 protein.19
Flow cytometry
1 × 105 cells were incubated with 2 μL of phycoerythrin (PE)-conjugated mouse anti-human monoclonal CD11b antibody (PharMingen, San Diego, CA, catalog #30455X) or isotype control and analyzed on a FACScan flow cytometer (Becton Dickinson, Mountain View, CA) using Cellquest software. Human recombinant G-CSF (Pharmacia) was biotinylated using N-hydrosuccinimide ester (NHS-LC)-biotin (Pierce, Rockford, IL) following the manufacturer's procedure and utilized to measure G-CSF receptor levels as previously described.33
Transient transfections
NIH-3T3 cells at 70% confluency were transfected using Superfect Transfection Reagent (Qiagen) with 1 μg of PU.1 reporter plasmid with either wild-type or mutant PU.1 sites inserted into the promoterless luciferase vector pXP2,34,35 500 ng of expression vector, and 100 ng of cytomegalovirus (CMV)-LacZ construct. For experiments including PU.1 expression vectors, either 500 ng of a single PU.1 allele or 250 ng each of 2 PU.1 alleles were transfected. Luciferase activities were normalized for transfection efficiency with the cotransfected CMV-LacZ construct, using a chemiluminescent reporter gene assay for β-galactosidase (Tropix, Foster City, CA). F9 cells were transfected as described previously.35 All transfection experiments were repeated 3 times with different preparations of each plasmid. Equal expression levels of PU.1 derivatives in transfected cells were confirmed by Western blotting.
In vitro protein-protein binding assays
Glutathione-S-transferase (GST) pull-down assays were performed as previously described.36 All GST proteins were quantitated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining.[35]S-methionine–labeled proteins were prepared using 1 μg of plasmid DNA as template for coupled in vitro transcription-translation (TNT; Promega, Madison, WI). For the in vitro binding assays, equal amounts of all GST proteins were incubated with 2 μL of [35]S-methionine–labeled proteins. The bead volume of all samples was adjusted to 50 μL with GST beads alone. Bound proteins were resolved on 10% SDS-PAGE gels and autoradiography, and the percentage of in vitro translated protein complexed with GST fusion proteins on beads was calculated with a phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Cell lines with conditional PU.1 expression
Phoenix cells, a human packaging cell line,37 were transiently transfected with either the pBabePuro-estrogen receptor (ER) vector alone, the pBabePuro-PU.1 wild-type-ER, or the pBabePuro-PU.1 G208fsX mutant-ER plasmid using lipofectamine (Gibco BRL, Grand Island, NY). Supernatant containing viral particles was harvested after 4 days. PU.1−/− cells (line 50324) were incubated in 4 mL of viral supernatant and 5 μg/mL of polybrene for 4 hours. A second infection cycle was performed after 24 hours. PU.1−/− cells were then grown in 96-well plates, and selection was started 48 hours after the first infection cycle in 0.5 μg/mL of puromycin. Clones were screened for the presence of the PU.1 fusion gene by Western blot analysis using PU.1 and/or FLAG antibody.
Results
Detection of heterozygous mutations of the transcription factor PU.1 in AML
The entire coding region of the PU.1 gene was amplified by PCR using cDNA (99 patients) or genomic DNA (27 patients). PCR products were directly sequenced to screen for mutations. FAB subtypes of the patients are shown in Table 2, and the karyotypes are described in Table 3. Of the 126 AML patients, 9 demonstrated at least one mutation of the PU.1 gene (7%). Subcloning of PCR products revealed that the wild-type sequence was present in all samples with PU.1 mutations, with the exception of patients #54 and #70. Since the percentage of wild-type clones was approximately 50% (ranging from 33% to 71%; Table 3), we therefore conclude that PU.1 mutations in AML patients generally are heterozygous.
Summary of the AML patients screened for PU.1 mutations
| AML phenotype . | FAB . | n2-155 . | Mutations . |
|---|---|---|---|
| Undifferentiated | M0 | 13 | 3 |
| Myeloblastic | M1 | 8 | 12-153 |
| M2 | 23* | 0 | |
| Promyelocytic | M3 | 11‡ | 0 |
| Myelomonocytic | M4 | 49† | 3 |
| Monoblastic | M5 | 14 | 1 |
| Erythroleukemia | M6 | 6 | 1 |
| AUL | 2 | 0 | |
| Total | 126 |
| AML phenotype . | FAB . | n2-155 . | Mutations . |
|---|---|---|---|
| Undifferentiated | M0 | 13 | 3 |
| Myeloblastic | M1 | 8 | 12-153 |
| M2 | 23* | 0 | |
| Promyelocytic | M3 | 11‡ | 0 |
| Myelomonocytic | M4 | 49† | 3 |
| Monoblastic | M5 | 14 | 1 |
| Erythroleukemia | M6 | 6 | 1 |
| AUL | 2 | 0 | |
| Total | 126 |
Subtypes according to the French-American-British (FAB) classification.
Of the 23 M2 patients, 3 were positive for the AML1-ETO translocation.
Of the 49 M4 patients, 10 carried the inv(16).
All 11 M3 patients had the PML-RARA translocation.
This patient was originally classified as M4.
n indicates the number of patients with this subtype.
Summary of the patients with PU.1 mutations
| Patient . | Subtypes . | Karyotypes . | bp . | aa . | Mut alleles (%) . |
|---|---|---|---|---|---|
| #57 | M0 | 46,XX,20q− | 618delC | P136fsX179 | 11/27 (41) |
| #104 | M0 | 46,XY | G524-1003del | V105-H264del | 8/12 (67) |
| #54 | M0 | NA | G335-1003del | D42-H264del | 3-151 |
| #683-150,3-152 | M1 | 46,XY | 833-834delGG | G208fsX | 8/22 (36) |
| #383-150 | M4 | 46,XY (11/20) | 659G>A | G150R | 7/14 (50) |
| 47,XY,− 7,+ 8,+ 8 (5/20) | |||||
| 47,XY,+ 5,− 7,+ 8 (4/20) | |||||
| #703-150 | M4 | 46,XY | 968delC | 253fsX | 4/7 (57) |
| 971G>A | G254R | 3/7 (43) | |||
| #109 | M4 | 46,XX | 841G>C | Q210H | 3/9 (33) |
| #63 | M5b | NA | 659G>A | G150R | 5/7 (71) |
| #115 | M6 | 46,XX | 222T>C | F4S | 3/8 (38) |
| Patient . | Subtypes . | Karyotypes . | bp . | aa . | Mut alleles (%) . |
|---|---|---|---|---|---|
| #57 | M0 | 46,XX,20q− | 618delC | P136fsX179 | 11/27 (41) |
| #104 | M0 | 46,XY | G524-1003del | V105-H264del | 8/12 (67) |
| #54 | M0 | NA | G335-1003del | D42-H264del | 3-151 |
| #683-150,3-152 | M1 | 46,XY | 833-834delGG | G208fsX | 8/22 (36) |
| #383-150 | M4 | 46,XY (11/20) | 659G>A | G150R | 7/14 (50) |
| 47,XY,− 7,+ 8,+ 8 (5/20) | |||||
| 47,XY,+ 5,− 7,+ 8 (4/20) | |||||
| #703-150 | M4 | 46,XY | 968delC | 253fsX | 4/7 (57) |
| 971G>A | G254R | 3/7 (43) | |||
| #109 | M4 | 46,XX | 841G>C | Q210H | 3/9 (33) |
| #63 | M5b | NA | 659G>A | G150R | 5/7 (71) |
| #115 | M6 | 46,XX | 222T>C | F4S | 3/8 (38) |
Subtypes are according to the French-American-British (FAB) classification. The location of the mutations is described for the position of the bp or the amino acids (aa) that are mutated. The last column represents the ratio of mutant (mut) sequences among all subcloned PCR products (mut/mut + wt).
NA indicates not available.
AML in these patients evolved from myelodysplastic syndromes (MDS).
In patient #54, only the mutant allele was detected.
Patient #68 was originally classified as M4.
We detected PU.1 mutations in the myelomonocytic or monocytic subtypes (M4, M5), in undifferentiated (M0) AML, and in one patient with erythroleukemia. One patient (#68) was originally diagnosed as M4, and subsequently reclassified as M1. However, no mutation was observed in 34 AML patients of the granulocytic lineage with the phenotypes M2 (23 patients) or M3 (11 patients).
Karyotype analysis revealed that PU.1 mutations were not observed in the 10 M4 patients with inv(16), the 3 M2 patients with the t(8;21) AML1/ETO translocation, or in 11 M3 patients with the PML-RARα translocation. Although the number of patients analyzed so far is limited, our data suggest that PU.1 mutations are not associated with one of the common translocations cited above. Furthermore, PU.1 mutations represented the only genomic abnormalities identified so far in 5 AML patients with normal cytogenetics (Table 3).
To assess the possibility that the abnormal PU.1 sequences detected in some AML patients represented polymorphisms, we sequenced DNA from peripheral blood leukocytes of 43 healthy volunteers, and we did not detect any abnormalities in the coding region of PU.1. In addition, where possible we analyzed cells from patients with PU.1 mutations at remission to distinguish between germ-line or sporadic mutations. We obtained paraffin-embedded material at remission from one patient (#109); in this remission sample, we did not detect the Q210H mutation observed in the blasts of this AML patient at diagnosis. In a second patient (#104), we established Epstein-Barr virus (EBV)–immortalized B-cell lines and found only PU.1 wild-type sequences could be detected. We therefore conclude that the sequence variations observed in AML patients likely represent mutations rather than polymorphisms.
Molecular anatomy of the PU.1 mutations
The 10 mutations in the coding region of PU.1 comprised 5 deletions and 5 point mutations. Further details and the precise location of the mutations are presented in Table 3. There is no defined region with a strikingly increased frequency for mutational events. In the M0/M4/M5 patients, 8 of the 9 mutations occurred either in the PEST domain (between amino acids 105 and 150) or in the DNA-binding domain (between amino acids 208 and 254).
Frame shift mutations in the PEST domain
We identified 2 AML-M0 patients with deletions in the PEST domain that caused a frame shift (#57: P136fsX179 and #104: V105-H264del) with consequent loss of parts of the PEST domain and of the entire DNA-binding domain (Figure 1). Since these deletions involved the entire DNA-binding domain, we predicted that binding of these peptides to PU.1 target gene promoters would be abolished. Thus, the effect of these 2 mutations likely results in a significant decrease in the amount of functional PU.1 protein in a particular leukemic cell. To support this hypothesis, we were fortunate to obtain cells at diagnosis from one of these 2 patients (#104: V105-H264del) whose mutation was confirmed in both a cDNA sample and in genomic DNA. We determined that the amount of wild-type PU.1 protein in leukemic cells from this AML-M0 patient (#104) is reduced by at least 50% as compared to other AML-M0 patients without PU.1 mutations (#97 or #103 in Figure 2). We therefore confirmed that this mutation led to a significant decrease in the amount of functional PU.1 protein in leukemic cells of patient #104. Such a decrease in functional PU.1 protein may contribute to the early block in differentiation observed in malignant cells of this particular AML-M0 patient.
Schematic representation of the PU.1 mutations found in AML patients.
PU.1 wild-type consists of 2 transactivation domains (TAD), a PEST domain, and a DNA-binding domain (DBD); the numbers refer to the location of the amino acids of the human PU.1 protein. Mutated sequences or frame shift sequences downstream of the mutation are depicted with hatched bars. The FAB subtypes are shown in the second column. 9P-10PinsQ represents a 3 base pair insertion splice variant found equally in healthy volunteers as well as in AML patients, which demonstrated no difference in DNA binding or transactivation compared to wild-type PU.1. fs indicates frame shift; X, new stop codon due to frame shift mutation; del, deleted sequences. ins: inserted sequences.
Schematic representation of the PU.1 mutations found in AML patients.
PU.1 wild-type consists of 2 transactivation domains (TAD), a PEST domain, and a DNA-binding domain (DBD); the numbers refer to the location of the amino acids of the human PU.1 protein. Mutated sequences or frame shift sequences downstream of the mutation are depicted with hatched bars. The FAB subtypes are shown in the second column. 9P-10PinsQ represents a 3 base pair insertion splice variant found equally in healthy volunteers as well as in AML patients, which demonstrated no difference in DNA binding or transactivation compared to wild-type PU.1. fs indicates frame shift; X, new stop codon due to frame shift mutation; del, deleted sequences. ins: inserted sequences.
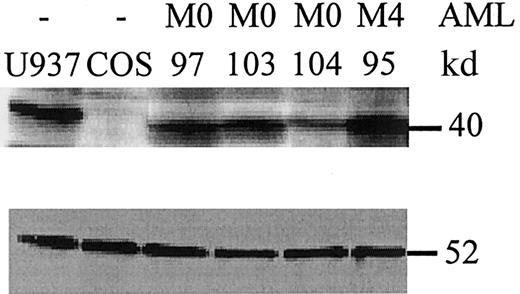
PU.1 mutant proteins are expressed in leukemic cells.
Whole cell lysates from leukemic cells at diagnosis were analyzed by Western blot for PU.1 expression (upper panel). U937 cells served as a positive control, whereas COS cells were negative for PU.1 expression. AML patients #97 and #103 had an AML-M0 subtype and lacked PU.1 mutations. In contrast, AML-M0 patient #104 carries the heterozygous V105-H264del mutation that encodes a mutant peptide lacking the PEST and Ets domains. This peptide is detected by an amino-terminal PU.1 antibody19 (data not shown), not by the antibody used in this blot, which is raised against a carboxyl terminal epitope deleted in the mutant allele. The peptide detected in patient #104 is that encoded by the wild-type allele, and approximately one-half as much protein is detected as compared to the other samples. Patient #95 is an AML-M4 with no PU.1 mutation. (Lower panel) The same blot was stained for β-tubulin as a loading control. The comparative amount of PU.1 protein was assessed by quantitation on a phosphorimager (Molecular Dynamics) and normalized to β-tubulin.
PU.1 mutant proteins are expressed in leukemic cells.
Whole cell lysates from leukemic cells at diagnosis were analyzed by Western blot for PU.1 expression (upper panel). U937 cells served as a positive control, whereas COS cells were negative for PU.1 expression. AML patients #97 and #103 had an AML-M0 subtype and lacked PU.1 mutations. In contrast, AML-M0 patient #104 carries the heterozygous V105-H264del mutation that encodes a mutant peptide lacking the PEST and Ets domains. This peptide is detected by an amino-terminal PU.1 antibody19 (data not shown), not by the antibody used in this blot, which is raised against a carboxyl terminal epitope deleted in the mutant allele. The peptide detected in patient #104 is that encoded by the wild-type allele, and approximately one-half as much protein is detected as compared to the other samples. Patient #95 is an AML-M4 with no PU.1 mutation. (Lower panel) The same blot was stained for β-tubulin as a loading control. The comparative amount of PU.1 protein was assessed by quantitation on a phosphorimager (Molecular Dynamics) and normalized to β-tubulin.
Mutations in the Ets domain of PU.1
PU.1 acts as a transactivator that requires coactivators to achieve potent activation function through physical interactions.35,38-40 The carboxyl terminus of the PU.1 Ets-homology domain is a winged helix-turn-helix (wHTH) motif that serves as a DNA-binding domain.41 The Ets domain of PU.1 has also been found to physically interact with many proteins, including the negative regulator GATA-1.39,42-44 We recently demonstrated that it is the β3/β4 region (amino acids 243-254) of PU.1, downstream of the wHTH motif, that mediates the interaction with a number of myeloid regulators including c-Jun, AML1B, and C/EBPα.28,35 39
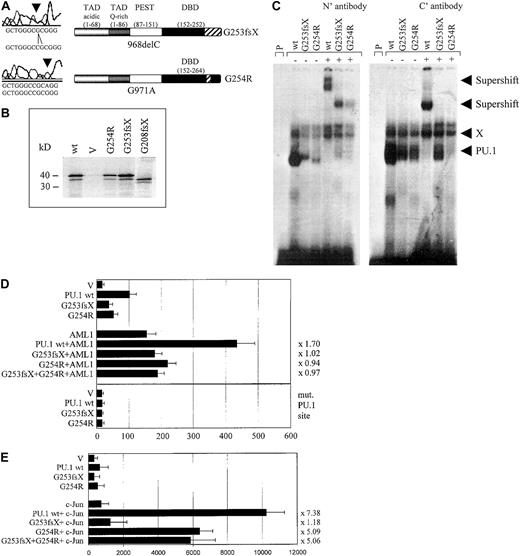
We identified 3 patients with heterozygous mutations in the Ets domain of PU.1. One patient (#70) had a different mutation in each allele, both of which affected the β3/β4 region of the Ets domain (Figures 1, 3A). One allele consisted of a point mutation, which selectively caused a G254R substitution in the β3/β4 region. The other mutation in patient #70 represented a one base-pair deletion causing a frame shift deletion downstream of amino acid 253 in the β3/β4 region of the Ets domain. Both of these Ets domain mutations in patient #70 encoded stable proteins (Figure3B). We asked whether these mutant PU.1 proteins of AML patient #70 might still be able to bind to target gene promoter sequences and retain the ability to activate target genes, such as the M-CSF receptor promoter.20,23 We found that the DNA-binding potential of the mutant PU.1 peptide, encoded by the point mutation G254R in the DNA-binding domain, was significantly reduced (Figure 3C). This DNA-binding complex could be supershifted using antiserum directed against both the amino and carboxyl terminus of PU.1. The deletion mutant G253fsX of patient #70 showed an equal decrease in its DNA-binding potential, which could be supershifted with amino- but not carboxyl-terminal PU.1 antibody (Figure 3C). Altogether, these findings suggest that PU.1 DNA-binding activity is dramatically reduced in leukemic cells from this patient, which contains in both alleles a PU.1 mutation involving the β3/β4 region. In addition, we found that both #70 mutants have a decrease of their activating potential20 to 53% for the G254R mutant and to 39% for the G253fsX mutant (Figure 3D). We therefore conclude that leukemic cells of this particular AML patient have a decisively reduced ability to activate PU.1 target genes.
The PU.1 mutations G253fsX and G254R identified in AML M4 patient #70 demonstrate decreased DNA binding, transactivation, and synergism with AML1 and c-Jun.
(A) The top left panel represents the one base pair deletion G253fsX; the lower left panel depicts the point mutation G254R. Sequences are shown below the left panels for the mutation (above) and the wild-type (below). The panels on the right are schematic representations of these 2 mutations in the DNA-binding domain (DBD) with the frame shift sequences depicted with hatched bars. (B) Western blot using a FLAG antibody. FLAG-tagged PU.1 wild-type (wt), vector only, which lacks a FLAG-tag (V), or FLAG-tagged G254R, G253fsX, or G208fsX mutants were in vitro translated and run on the SDS gel. Molecular weight markers are shown on the left. (C) Electrophoretic mobility shift assay (EMSA) analyzing DNA binding to the PU.1 site in the M-CSF receptor promoter of in vitro translated proteins encoded by PU.1 wild-type and the PU.1 mutants G253fsX and G254R. The input protein for wild-type and mutant PU.1 proteins is shown in Figure 3B. (Left panel) Binding was supershifted using an antiserum directed against the amino terminus of the PU.1 protein. Consistently, we observed that the complex obtained for PU.1 wild-type with the amino terminal antibody migrated more slowly than the complex containing one of the mutant proteins. (Right panel) Supershift was achieved with an antibody directed against amino acids 251 to 271 of the murine PU.1 protein. X indicates nonspecific binding activity (does not compete with self oligonucleotide); P, labeled probe alone. In both panels, the complex migrating more slowly than wild-type PU.1, which does not react with either antibody, has been observed previously in EMSA using this probe.20 (D, upper panel) COS7 cells were transfected with PU.1 wild-type (wt) or one of the 2 PU.1 mutants identified in AML patient #70 (G253fsX and G254R) together with either AML1 or pcDNA3 vector alone (V). Either 500 ng of a single PU.1 allele or 250 ng each of 2 PU.1 alleles were transfected. The AML1 cofactor CBFβ was present in all transfections in equimolar amounts. The reporter consisted of a luciferase construct with a wild-type PU.1 site.35 The ability to activate the PU.1 site derived from the M-CSF receptor promoter is indicated in luciferase units normalized to wild-type PU.1 ( = 100). Synergy was calculated by the ratio of the activity observed with cotransfected AML1 and PU.1 wild-type divided by the arithmetic addition of AML1 activation alone and PU.1 wild-type activation alone. The same ratio was determined for the PU.1 mutant G254R and indicated to the right of each bar. (Lower panel) The same assay as above, except the reporter consisted of a luciferase construct with the PU.1 site mutated (mut. PU.1 site).35(E) F9 cells that are c-Jun deficient were transfected with PU.1 wild-type (wt) or the 2 PU.1 mutants identified in AML patient #70 (G253fsX and G254R) together with c-Jun or the empty expression vector (V). Empty expression vector was added in all transfections to ensure that equal amounts of DNA were transfected. The ability to activate the PU.1 site in the M-CSF receptor promoter and synergism with c-Jun was determined as described for panel D.
The PU.1 mutations G253fsX and G254R identified in AML M4 patient #70 demonstrate decreased DNA binding, transactivation, and synergism with AML1 and c-Jun.
(A) The top left panel represents the one base pair deletion G253fsX; the lower left panel depicts the point mutation G254R. Sequences are shown below the left panels for the mutation (above) and the wild-type (below). The panels on the right are schematic representations of these 2 mutations in the DNA-binding domain (DBD) with the frame shift sequences depicted with hatched bars. (B) Western blot using a FLAG antibody. FLAG-tagged PU.1 wild-type (wt), vector only, which lacks a FLAG-tag (V), or FLAG-tagged G254R, G253fsX, or G208fsX mutants were in vitro translated and run on the SDS gel. Molecular weight markers are shown on the left. (C) Electrophoretic mobility shift assay (EMSA) analyzing DNA binding to the PU.1 site in the M-CSF receptor promoter of in vitro translated proteins encoded by PU.1 wild-type and the PU.1 mutants G253fsX and G254R. The input protein for wild-type and mutant PU.1 proteins is shown in Figure 3B. (Left panel) Binding was supershifted using an antiserum directed against the amino terminus of the PU.1 protein. Consistently, we observed that the complex obtained for PU.1 wild-type with the amino terminal antibody migrated more slowly than the complex containing one of the mutant proteins. (Right panel) Supershift was achieved with an antibody directed against amino acids 251 to 271 of the murine PU.1 protein. X indicates nonspecific binding activity (does not compete with self oligonucleotide); P, labeled probe alone. In both panels, the complex migrating more slowly than wild-type PU.1, which does not react with either antibody, has been observed previously in EMSA using this probe.20 (D, upper panel) COS7 cells were transfected with PU.1 wild-type (wt) or one of the 2 PU.1 mutants identified in AML patient #70 (G253fsX and G254R) together with either AML1 or pcDNA3 vector alone (V). Either 500 ng of a single PU.1 allele or 250 ng each of 2 PU.1 alleles were transfected. The AML1 cofactor CBFβ was present in all transfections in equimolar amounts. The reporter consisted of a luciferase construct with a wild-type PU.1 site.35 The ability to activate the PU.1 site derived from the M-CSF receptor promoter is indicated in luciferase units normalized to wild-type PU.1 ( = 100). Synergy was calculated by the ratio of the activity observed with cotransfected AML1 and PU.1 wild-type divided by the arithmetic addition of AML1 activation alone and PU.1 wild-type activation alone. The same ratio was determined for the PU.1 mutant G254R and indicated to the right of each bar. (Lower panel) The same assay as above, except the reporter consisted of a luciferase construct with the PU.1 site mutated (mut. PU.1 site).35(E) F9 cells that are c-Jun deficient were transfected with PU.1 wild-type (wt) or the 2 PU.1 mutants identified in AML patient #70 (G253fsX and G254R) together with c-Jun or the empty expression vector (V). Empty expression vector was added in all transfections to ensure that equal amounts of DNA were transfected. The ability to activate the PU.1 site in the M-CSF receptor promoter and synergism with c-Jun was determined as described for panel D.
We next tested the ability of these 2 mutants to synergistically activate the M-CSF receptor together with AML1,28 a function that has been attributed to the β3/β4 region that is affected by both mutations.28,39 Indeed, we observed a complete lack of synergy for both #70 mutants (Figure 3D). We therefore conclude that these 2 PU.1 mutations have lost the ability to activate crucial PU.1 target genes alone or in synergy with factors such as AML1. In addition, PU.1 uses c-Jun as a coactivator to activate target genes such as the M-CSF receptor.35 Again, this function is mediated by the β3/β4 region.35 We observed that the G253fsX mutant peptide has completely lost its synergistic potential if cotransfected with c-Jun, whereas the G254R mutant retained some synergy with c-Jun (64% of the activation when c-Jun is cotransfected with wild-type PU.1) (Figure 3E). One possible explanation for the retention of some synergism between G254R and c-Jun is that the 2 proteins can still physically interact (Figure5). In summary, both PU.1 mutant alleles of patient #70 involving the β3/β4 region have significant defects in their ability to coactivate target genes with AML1, and one mutant allele, G253fsX, is also defective in coactivation with c-Jun.
The #68 mutant G208fsX
Whereas the deletion mutation of #70 (G253fsX) caused a loss of parts of the β3/β4 region in the Ets domain, the frame shift mutation of #68 (G208fsX) disrupted the entire wHTH motif and the β3/β4 region (Figure 4A). Despite this deletion, the G208fsX mutant encoded a stable protein (Figure 3B). We predicted that DNA binding would be affected, and indeed no DNA-binding activity to the M-CSF receptor promoter oligonucleotide was observed for the G208fsX mutant (Figure 4C). Consequently, this mutant also failed to activate a M-CSF receptor promoter construct in transient transfection assays (Figure4D), consistent with its lack in DNA binding (Figure 4B). We next tested the ability of this mutant to synergistically activate the M-CSF receptor together with AML1 or c-Jun. Because G208fsX fails to physically interact with AML1, and binds very weakly to c-Jun compared to wild-type PU.1 (Figure 5), we predicted that it might be defective in synergism with both factors. The G208fsX mutant not only failed to synergize with AML1 (Figure 4C), but it appeared to block AML1 function. Cotransfection of this mutant together with AML1 results in luciferase activity that is only 36% of what is observed with AML1 alone (Figure 4C). In addition, we observed almost no synergy between the G208fsX mutant and c-Jun (Figure 4D). We therefore conclude that this PU.1 mutation involving the β3/β4 region negatively affects the coactivation of PU.1 by c-Jun.
Functional consequences of the G208fsX mutation.
(A) Schematic representation of the G208fsX mutation in the DNA-binding domain (DBD) with the frame shift sequences depicted with hatched bars. (B) EMSA analysis of the binding of nuclear extracts from COS7 cells transfected with PU.1 wild-type (wt; lane 2) or G208fsX (mut; lane 4) to the PU.1 binding site in the M-CSF receptor. ss indicates that PU.1 wild-type binding was supershifted with carboxyl terminal-specific PU.1 antiserum (lane 3); C, competition of wild-type binding with 100-fold excess of unlabeled oligonucleotide (lane 5); and p, labeled probe alone (lane 1). As in Figure 3C, the complex migrating more slowly than wild-type PU.1, and which does not react with the anti-PU.1 antibody, has been observed previously in EMSA using this probe.20 (C) COS7 cells were transfected with PU.1 wild-type (wt) or the PU.1 mutant G208fsX (mut) together with either AML1 or the pcDNA3 vector (V). The ability to activate the PU.1 site in the M-CSF receptor promoter was measured. Synergy represents the ratio of the activity seen with cotransfected AML1 and PU.1 wild-type divided by the arithmetic addition of AML1 activation alone and PU.1 wild-type activation alone. The same ratio was determined for the PU.1 mutant and indicated to the right of each bar. (D) c-Jun–deficient F9 cells were transfected with PU.1 wild-type (wt) or the G208fsX mutant (mut) together with c-Jun or the empty expression vector (V). Empty expression vector was added in all transfections to ensure that equal amounts of DNA were transfected. The ability to activate the PU.1 site in the M-CSF receptor promoter and synergy was determined as described for panel C.
Functional consequences of the G208fsX mutation.
(A) Schematic representation of the G208fsX mutation in the DNA-binding domain (DBD) with the frame shift sequences depicted with hatched bars. (B) EMSA analysis of the binding of nuclear extracts from COS7 cells transfected with PU.1 wild-type (wt; lane 2) or G208fsX (mut; lane 4) to the PU.1 binding site in the M-CSF receptor. ss indicates that PU.1 wild-type binding was supershifted with carboxyl terminal-specific PU.1 antiserum (lane 3); C, competition of wild-type binding with 100-fold excess of unlabeled oligonucleotide (lane 5); and p, labeled probe alone (lane 1). As in Figure 3C, the complex migrating more slowly than wild-type PU.1, and which does not react with the anti-PU.1 antibody, has been observed previously in EMSA using this probe.20 (C) COS7 cells were transfected with PU.1 wild-type (wt) or the PU.1 mutant G208fsX (mut) together with either AML1 or the pcDNA3 vector (V). The ability to activate the PU.1 site in the M-CSF receptor promoter was measured. Synergy represents the ratio of the activity seen with cotransfected AML1 and PU.1 wild-type divided by the arithmetic addition of AML1 activation alone and PU.1 wild-type activation alone. The same ratio was determined for the PU.1 mutant and indicated to the right of each bar. (D) c-Jun–deficient F9 cells were transfected with PU.1 wild-type (wt) or the G208fsX mutant (mut) together with c-Jun or the empty expression vector (V). Empty expression vector was added in all transfections to ensure that equal amounts of DNA were transfected. The ability to activate the PU.1 site in the M-CSF receptor promoter and synergy was determined as described for panel C.
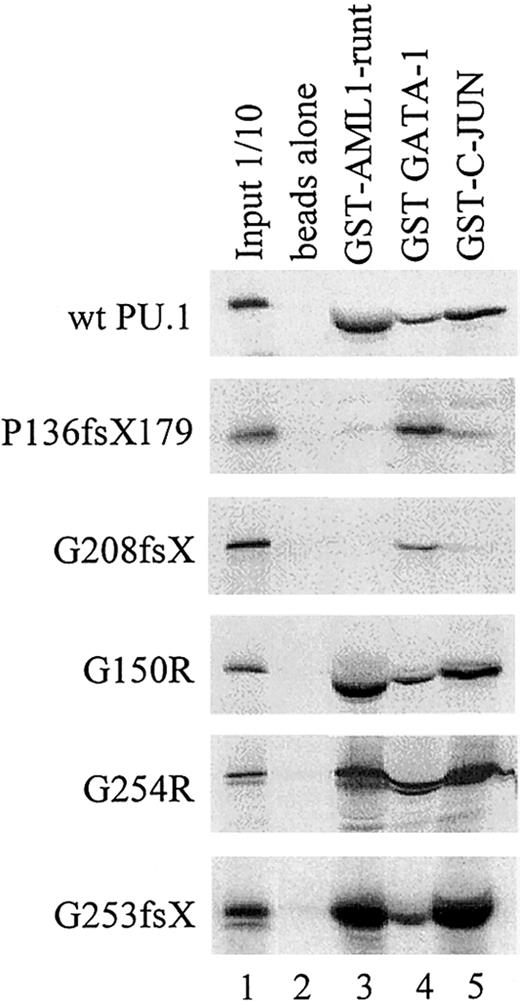
Physical interaction of wild-type and mutant PU.1 peptides with AML1, GATA-1, and c-Jun.
GST-fusion proteins for AML1 (runt domain), GATA-1, or c-Jun were incubated with in vitro translated PU.1 wild-type or one of the PU.1 mutant peptides. The interaction with GATA-1 served as a positive control, since GATA-1 interacts with both the amino and carboxyl terminus of PU.1.39 43 Input: 1/10: 10% of in vitro translated protein used for binding reaction was loaded as a control.
Physical interaction of wild-type and mutant PU.1 peptides with AML1, GATA-1, and c-Jun.
GST-fusion proteins for AML1 (runt domain), GATA-1, or c-Jun were incubated with in vitro translated PU.1 wild-type or one of the PU.1 mutant peptides. The interaction with GATA-1 served as a positive control, since GATA-1 interacts with both the amino and carboxyl terminus of PU.1.39 43 Input: 1/10: 10% of in vitro translated protein used for binding reaction was loaded as a control.
PU.1 mutants involving the β3/β4 region fail to physically interact with AML1 and/or c-Jun. We previously have shown that PU.1 and AML1 physically interact via the runt domain of AML1 and the DNA-binding Ets domain of PU.1, resulting in synergistic activation of the M-CSF receptor promoter.20,28 45 We consequently observed that PU.1 mutations involving the Ets domain (such as the G253fsX and G254R mutations in patient #70 or the G208fsX mutation in patient #68) have lost their ability to synergistically activate with AML1. We asked whether the lack of physical interaction of these mutants with AML1 could be demonstrated using GST pull-down assays. Figure 5 demonstrates that the runt domain of AML1 strongly interacted with wild-type PU.1 protein, mutants G150R (a point mutation in the PEST domain in patients #63 and #38), and G254R (a single point mutation in the Ets domain in patient #70). In contrast, the 2 mutants P136fsX179 (frame shift deletion with loss of the entire Ets domain in patient #57) and G208fsX (frame shift deletion destroying most of the Ets domain in patient #68) showed no interaction with AML1. Therefore, the lack of synergy observed between AML1 and PU.1 mutants P136fsX179 and G208fsX is likely due to the inability of these PU.1 mutants to physically interact via their Ets domain with AML1. Interestingly, the 2 mutants in patient #70 (G254R and G253fsX) retained their ability to interact with AML1, whereas transcriptional synergy was lost if each of the mutants was cotransfected with AML1 (Figure 3D). This suggests that the structural alterations caused by these mutations in the β3/β4 region were sufficient to affect DNA binding to target genes (Figure 3C) but did not affect interaction with additional coactivators such as AML1. This was also the case for the interaction of G253fsX and c-Jun (Figures3C,E; Figure 5).
Many of the patients in this study had been previously analyzed for AML1 mutations.5 An H58N point mutation in the AML1 gene was detected in AML-M0 patient #57, which also harbored a heterozygous P136fsX179 mutation in PU.1. This AML1 mutation showed an increased transactivation potential of the M-CSF receptor.5 It is thus an interesting finding that this single patient harbored a “super-activating” mutation in AML1 and a mutation in PU.1 that abrogates interaction with wild-type AML1 (Figure 5) and most likely cannot synergize in activating target genes.
As described above, we also studied synergistic effects between PU.1 and c-Jun.35 c-Jun does not directly bind to the M-CSF receptor promoter but associates via its basic domain with the Ets domain of PU.1.35 Again, we used GST pull-down assays to test whether our mutants in the Ets domain, which do not synergistically activate with c-Jun, have also lost the ability to physically interact with c-Jun. As a control, we also tested the ability of our mutant PU.1 proteins to interact with GATA-1, which interacts with both the amino terminus and β3/β4 carboxyl region of PU.1.39 43 Figure 5 indeed indicates that the mutants G150R (a point mutation in the PEST domain in patients #63 and 38) and G254R (point mutation in the Ets domain in patient #70) interact with c-Jun. In contrast, the 2 mutants P136fsX179 (frame shift deletion involving PEST and Ets domains in patient #57) and G208fsX (frame shift deletion of parts of the Ets domain in patient #68) showed no interaction with c-Jun. This evidence supports the hypothesis that PU.1 mutations in the Ets domain have lost the ability to synergistically activate PU.1 target gene promoters because of loss of physical interaction between PU.1 and c-Jun.
The G150R point mutation in the PEST domain in AML-M4/M5 patients
We observed a G150R point mutation in the PEST domain of 2 AML patients. We did not detect this abnormality in DNA from 43 healthy volunteers. Unfortunately, no remission or nonleukemic material was available from these patients. Because this domain has been shown to mediate interaction with members of the interferon responsive factor (IRF) family, including IRF-4 and ICSBP,46-48 we hypothesized that the point mutation of amino acid 150 in these 2 AML patients (Figure 1) might affect IRF recruitment. However, the G150R mutant was still capable of properly binding to DNA, and both transactivation of the M-CSF receptor promoter and synergy with IRF family members in activating the interleukin (IL)-1β promoter was not significantly affected (data not shown). Furthermore, the G150R mutant protein was still capable of physically interacting with the interferon consensus sequence binding protein (ICSBP) in a manner similar to that of PU.1 wild-type in a GST pull-down assay (data not shown). Therefore, the nature of the defect of the G150R mutant, if any, remains unknown.
Loss of exons 3 to 5 in AML patients
In one patient (#54), only a shortened splice variant and not the full-length PU.1 sequence could be identified using cDNA as a template (Figure 6). This variant deletes exons 3, 4, and much of exon 5. In this deletion, the sequence immediately following exon 2 derives from sequences 50 bp downstream of the translation stop codon in exon 5. Using genomic DNA from leukemic cells of this patient and exon-specific primers, we failed to amplify exons 3 to 5 by PCR (data not shown). We therefore believe that both alleles in this patient lack a large part of the wild-type sequence involving at least exons 3 to 5. Unfortunately, no material was available for confirmation by FISH analysis. No DNA binding to the M-CSF receptor oligonucleotide could be observed, and the potential to activate the M-CSF receptor promoter was completely abolished in transient transfection assays (data not shown). Loss of both alleles for the AML1 transcription factor has been previously described in patients with AML.7
Inability to detect exons 3, 4, and 5 from patient #54.
Shown is an ethidium-bromide–stained agarose gel demonstrating PCRs from cDNA of 5 AML patients amplifying the full-length wild-type sequence of PU.1 (1051 bp). In patient #54 (lane 3), only the splice variant involving exons 1 and 2 is detectable (335 bp). C indicates PU.1 wild-type plasmid serves as a positive control. H, water as a negative control.
Inability to detect exons 3, 4, and 5 from patient #54.
Shown is an ethidium-bromide–stained agarose gel demonstrating PCRs from cDNA of 5 AML patients amplifying the full-length wild-type sequence of PU.1 (1051 bp). In patient #54 (lane 3), only the splice variant involving exons 1 and 2 is detectable (335 bp). C indicates PU.1 wild-type plasmid serves as a positive control. H, water as a negative control.
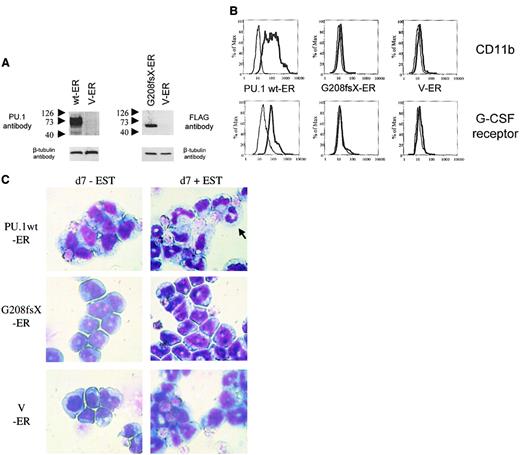
Conditional expression of mutant G208fsX in PU.1−/−cells fails to induce granulocytic differentiation
PU.1−/− cells represent early myeloid precursors that can be induced to differentiate following transduction with a retrovirus expressing the wild-type PU.1 protein.26,27 Therefore, we asked whether a PU.1 mutation identified in AML patients had lost this potential. We transduced PU.1−/− cells with a retrovirus expressing either the wild-type human PU.1-estrogen receptor fusion protein or the PU.1 mutant G208fsX estrogen receptor fusion protein (Figure7A). Both constructs contained a FLAG sequence at the amino terminus of PU.1. In the absence of estradiol, the estrogen receptor (and thus the PU.1 protein fused to it) was localized to the cytoplasm. Treatment of the cells with 1 mM estradiol induced translocation of the PU.1-ER protein into the nucleus (data not shown). Expression of wild-type PU.1 induced differentiation of PU.1−/− cells. CD11b is a PU.1 target gene19that is up-regulated during myeloid differentiation. PU.1−/− cells expressing the PU.1-ER fusion showed a dramatic increase in CD11b expression after 4 days of treatment with estradiol (Figure 7B). In contrast, CD11b levels were unchanged after treatment with estradiol in the parental PU.1−/− cells. In addition, we determined expression of the G-CSF receptor as a marker for neutrophil differentiation.33 Again, PU.1-ER expressing PU.1−/− cells demonstrated a marked increase in G-CSF receptor expression, whereas the levels remained unchanged in the parental line. Finally, Wright-Giemsa staining of PU.1−/− cells expressing the PU.1-ER fusion protein before and 7 days after treatment with 1 mM β-estradiol demonstrated neutrophilic differentiation of immature myeloid blasts (Figure 7C). We therefore confirmed that expression of PU.1 protein in PU.1−/− cells is sufficient to induce terminal granulocytic differentiation in this system as described previously.27 We also tested the G150R mutant using this system and found that this mutant retained the ability to induce neutrophil differentiation and activate PU.1 target genes (data not shown).
The G208fsX mutant is defective in induction of myeloid differentiation of PU.1−/− cells.
Conditional expression of the estrogen receptor alone, or fused to wild-type PU.1 or the G208fsX mutant in PU.1−/− cells. (A) Left panel: Western blot using carboxyl terminal PU.1 antiserum (1:500; Santa Cruz, catalog #sc-352) of whole cell lysates from PU.1−/− cells transfected with wild-type human PU.1-estrogen receptor fusion plasmid (wt-ER) or the estrogen receptor (V-ER) alone. The V-ER estrogen receptor alone contains no PU.1 or FLAG sequences. The migration of molecular weight markers is shown to the left of each panel. The blot was reprobed for β-tubulin as a loading control (lower panel). Right panel: Western blot using a FLAG antibody detecting the G208fsX PU.1 mutant fused to the estrogen receptor. Shown below is the β-tubulin control. (B) Flow cytometric analysis for CD11b expression (upper panel) and G-CSF receptor expression (lower panel). PU.1−/− cells expressing the PU.1 wt-ER, the PU.1 mutant G208fsX-ER, or the estrogen receptor alone (V-ER) were untreated (fine lines) or treated (thick lines) with 1 mM β-estradiol for 7 days. CD11b and G-CSF receptor expression were determined by flow cytometry. (C) Wright-Giemsa staining of PU.1−/−cells expressing PU.1 wild-type or mutant-ER fusion proteins. Cells were untreated (d7-EST) or treated with (d7 + EST) 1 mM β-estradiol for 7 days. The arrow in the upper right panel indicates a mature neutrophil in cells expressing PU.1 wild-type protein. Magnification × 100.
The G208fsX mutant is defective in induction of myeloid differentiation of PU.1−/− cells.
Conditional expression of the estrogen receptor alone, or fused to wild-type PU.1 or the G208fsX mutant in PU.1−/− cells. (A) Left panel: Western blot using carboxyl terminal PU.1 antiserum (1:500; Santa Cruz, catalog #sc-352) of whole cell lysates from PU.1−/− cells transfected with wild-type human PU.1-estrogen receptor fusion plasmid (wt-ER) or the estrogen receptor (V-ER) alone. The V-ER estrogen receptor alone contains no PU.1 or FLAG sequences. The migration of molecular weight markers is shown to the left of each panel. The blot was reprobed for β-tubulin as a loading control (lower panel). Right panel: Western blot using a FLAG antibody detecting the G208fsX PU.1 mutant fused to the estrogen receptor. Shown below is the β-tubulin control. (B) Flow cytometric analysis for CD11b expression (upper panel) and G-CSF receptor expression (lower panel). PU.1−/− cells expressing the PU.1 wt-ER, the PU.1 mutant G208fsX-ER, or the estrogen receptor alone (V-ER) were untreated (fine lines) or treated (thick lines) with 1 mM β-estradiol for 7 days. CD11b and G-CSF receptor expression were determined by flow cytometry. (C) Wright-Giemsa staining of PU.1−/−cells expressing PU.1 wild-type or mutant-ER fusion proteins. Cells were untreated (d7-EST) or treated with (d7 + EST) 1 mM β-estradiol for 7 days. The arrow in the upper right panel indicates a mature neutrophil in cells expressing PU.1 wild-type protein. Magnification × 100.
Consequently, we tested the PU.1 mutant G208fsX, which deletes the last 56 amino acids of the PU.1 gene, including the β3/β4 region. The G208fsX protein has lost the potential to activate PU.1 target genes (Figure 4D), and no up-regulation of CD11b as well as of G-CSF receptor expression was detectable following estradiol treatment in G208fsX-ER transduced cells (Figure 7B). In addition, induction of the PU.1 mutant G208fsX-ER fusion with estradiol failed to induce the marked granulocytic morphologic changes in PU.1−/− cells compared to wild-type PU.1-ER (Figure 7C). We therefore conclude that the PU.1 G208fsX mutant has lost the potential to activate important myeloid target genes, such as the M-CSF receptor,23 and induce terminal differentiation.
Discussion
We report here for the first time mutations in the PU.1 gene in malignant cells isolated from patients with cancer. Screening 126 AML patients, we identified 9 with mutations in the coding region of the PU.1 gene. As assessed by conventional karyotype analysis, 5 of these patients had an otherwise normal karyotype. Thus, PU.1 mutations represent the only genomic abnormalities detected so far in these particular patients. Comprehensive clinical information was available for 6 of the 9 AML patients with PU.1 mutations and for 66 of the 117 AML patients with wild-type PU.1. Based on the relatively small numbers of patients, we found that patients with PU.1 mutations fared worse than patients without PU.1 mutations (median survival, 80 days and 364 days, respectively; with a complete remission achieved in 33% and 57%). These results suggest that the presence of PU.1 mutations carries a worse prognosis, but clearly additional studies with more patients will be required to answer this question in a definitive fashion. In addition, no mutations were observed in a collection of 24 patients with a good risk karyotype involving either the t(8;21) or the t(15;17) translocation, or inv(16). These results suggest that mutations in PU.1 might define a distinct subgroup of AML patients, and therefore detection of PU.1 mutations may be of possible prognostic importance in the future.
What is the significance of these PU.1 mutations? Since we identified mutant and wild-type alleles in cells from AML patients with PU.1 mutations, one hypothesis is that haploinsufficiency contributes to leukemogenesis, as has been described for the AML1 transcription factor.6 Support for this idea comes from a recent report demonstrating that PU.1+/− mice showed increased hematopoiesis.49 Spleens from PU.1+/− mice were not only enlarged, but also contained increased numbers of hematopoietic progenitors.49 These findings point to a dosage effect as a potential pathogenetic mechanism underlying PU.1 mutations. Since PU.1 binds DNA as a monomer, we anticipated that mutants defective in DNA binding alone might not affect the PU.1 wild-type protein in activation of target genes. Indeed, we did not observe that 5 mutants in the DNA-binding domain affected the ability of the PU.1 wild-type to activate the M-CSF receptor promoter.
An emerging concept from the role of transcription factors in hematopoiesis is that not only are single factors of importance, but rather combination of factors are needed.3,4,28,29,35,39,50 We previously reported that PU.1 synergizes with its coactivator c-Jun, as well as with AML1, in activating target genes such as the M-CSF receptor.28 35In both instances, it is the β3/β4 region in the Ets domain of PU.1 that mediates this interaction. Consequently, we tested the transactivation potential of PU.1 mutants involving the β3/β4 region in competitive cotransfection studies in these assay. We observed loss of transactivation synergy for these mutants with AML1 and with c-Jun. Surprisingly, one of these, G208fsX, which did not bind DNA (Figure 4B), also inhibited the function of AML1 (Figure 4C), even though we could not detect a physical interaction between them (Figure5), suggesting that effects on other factors, such as AML1, rather than the wild-type PU.1 allele, might mediate some of the adverse effects on myeloid differentiation.
Of note is the fact that we were able to identify mutations in PU.1 predominantly in very immature (M0) or monocytic (M4/M5) AML subgroups (as described above), while we previously found such mutations in the myeloid transcription factor C/EBPα to be limited to the myeloblastic subtypes M1 and M2.8 Together, these results are consistent with gene targeting studies, showing that disruption of C/EBPα function results in a block in granulocyte differentiation at an early stage,51 while the hematopoietic system of the PU.1 knockouts is blocked at a very early stage of myeloid development and affects monocytic development to a greater degree than granulocytic.18,24,26 Thus, mutations in PU.1 and C/EBPα are observed in human diseases with phenotypes similar to, or predictable from, the murine knockout phenotypes. Furthermore, we predict that small cytogenetically undetectable mutations in other myeloid transcription factors will play a role in the pathogenesis of other AMLs. Indeed, recent studies reported mutations in the runt domain of the AML1 gene predominantly in AML with an M0 subtype,5,7 9 which underlines the importance of this gene at a very early stage in hematopoiesis, and parallels our findings for the PU.1 gene. Interestingly, while mutant G208fsX was unable to induce markers such as CD11b and the G-CSF receptor, there were slight morphologic changes suggestive of some partial differentiation (Figure7), consistent with detection of this mutation in a patient with M4 rather than M0 AML. These results suggest that the different PU.1 mutant peptides might have distinct functions in differentiation. Additional studies with more mutants found in AML will be required to attempt to correlate structure-function studies of the PU.1 mutants with differentiation of PU.1−/− cells in culture and the AML phenotype observed in patients.
The human PU.1 gene is located at chromosome 11p11.22, which is not a site of known chromosome translocations in leukemia.1,3The PU.1 gene itself has never been reported to be a partner gene of a chromosomal translocation. This is similar to other transcription factors such as C/EBPα, in which only cytogenetically undetectable small mutations are reported so far in AML patients.8 10In contrast, the AML1 gene can be either mutated or be a partner in chromosomal translocations in AML patients. If interruption of PU.1 function is an important step in induction of myeloid leukemia, then we predict that in other AML subtypes, other mutant gene products will adversely affect PU.1 function. Finally, we anticipate finding alterations in PU.1 in other human neoplasms involving cell types in which PU.1 is expressed, including B-cell cancers.
We thank Christian Busse and Chris Hetherington for assistance with the mutational analysis; Pu Zhang for biotinylated G-CSF; Bruce Torbett for PU.1−/− cells; Richard Maki for the amino terminal PU.1 antibody; Alan Friedman for the pBabePuro-vector; Eric Johannsen and Elliot Kieff for assistance with EBV immortalization of bone marrow cells; Gary Gilliland, Linda Clayton, and the members of the Tenen laboratory for suggestions; and M. Singleton and A. Lugay for assistance with the preparation of the manuscript.
Supported by grants from the Swiss National Science Foundation (81BS-52825) (B.U.M.) and the National Institutes of Health (grants CA41456 and CA72009) (D.G.T.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Daniel G. Tenen, Harvard Institutes of Medicine, 77 Avenue Louis Pasteur, Rm 954, Boston, MA 02115; e-mail:dtenen@caregroup.harvard.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal