Abstract
In this report we demonstrate a role for Runx1 (AML1)at the hemangioblast stage of hematopoietic and endothelial development in embryonic stem (ES) cell–derived embryoid bodies (EBs).Runx1 is expressed in EBs during the appearance of precursors with hemangioblast properties, the blast colony-forming cells (BL-CFCs). Cell sorting studies revealed that all BL-CFCs within EBs express Runx1. Runx1-deficient EBs consistently generate 10- to 20-fold fewer blast colonies than wild-type controls and display a complete block in definitive hematopoiesis. Despite this defect, Runx1−/−EBs and yolk sacs from mutant embryos generate normal numbers of primitive erythroid precursors. These observations clearly demonstrate that Runx1 functions early in hematopoietic development, and they support the interpretation that the primitive erythroid lineage is established early by a subset of BL-CFCs that develop in aRunx1-independent fashion.
Introduction
Hematopoiesis in the mouse embryo begins in the yolk sac, where blood islands of mesodermal origin develop at approximately day 8 of gestation (E8).1,2 These blood islands consist of 2 distinct lineages, a population of erythroblasts and a surrounding layer of angioblasts that will form the first vascular structures.3 The parallel temporal development of these lineages in physical proximity provided the basis for the hypothesis that they arise from a common precursor, a cell called the hemangioblast.4,5 Erythroid cells within the blood islands, known as embryonic or primitive erythrocytes, are large and nucleated, and they produce the embryonic forms of globin.1,6,7 Generation of the primitive erythroid lineage is known as primitive hematopoiesis, and it represents a transient developmental program that is restricted to the yolk sac between the primitive streak and 20 somite pair (sp) stages of development.8-10 Definitive hematopoiesis encompasses the development of all lineages other than primitive erythroid and includes definitive erythroid, myeloid, and lymphoid. As with primitive hematopoiesis, the first definitive hematopoietic precursors also develop in the yolk sac and can be detected as early as the primitive streak stage of development.10 Although initiated in this extra-embryonic region, definitive hematopoiesis is most often associated with intra-embryonic sites such as the para-aortic splanchnopleura (P-Sp), the aorta–gonad–mesonephros (AGM), and the fetal liver, where long-term repopulating stem cells and precursor populations from different lineages undergo significant expansion and maturation.11-15
Although yolk sac blood islands were identified as the earliest site of hematopoietic and endothelial development almost 100 years ago, attempts to identify, isolate, and characterize the precursors representing these initial stages of lineage commitment, including the elusive hemangioblast, have been largely hampered by the inaccessibility of the early mammalian embryo. One promising alternative approach to study early hematopoietic development is the model system based on the differentiation potential of embryonic stem (ES) cells in culture.16-21 Most evidence suggests that the events leading to the establishment of the hematopoietic and endothelial lineages in embryoid bodies (EBs) generated from ES cells in culture are similar, if not identical, to those in the early yolk sac.19,22-29 Using the ES/EB differentiation model, cells with hemangioblast potential have been identified.30,31 In the presence of vascular endothelial growth factor (VEGF) in methylcellulose cultures, these EB-derived precursors generate blast cell colonies that display hematopoietic and endothelial potential.30 32 The cells that give rise to these blast colonies, the blast colony-forming cells (BL-CFCs), represent a transient population that appears in EBs before the establishment of any other hematopoietic lineages. The developmental potential of the BL-CFC strongly suggests that it represents the in vitro equivalent of the hemangioblast and, as such, the earliest stage of hematopoietic and endothelial commitment.
Although a precursor with hemangioblast properties remains to be identified in the early embryo, insights into the molecular events involved in the establishment of the earliest hematopoietic and endothelial lineages have been provided by gene-targeting experiments. Such studies have uncovered the role of specific genes at distinct stages in this process and, in doing so, have been instrumental in defining key developmental steps in the commitment, growth, and maturation of these lineages. Flk-1, a gene that encodes a receptor tyrosine kinase, is required early in ontogeny and is essential for the development of the hematopoietic and the endothelial components of the blood island.33 Cell tracking studies have indicated that the Flk-1 receptor is initially involved in migration of the mesodermal precursor cells to the extraembryonic region of the embryo, the site of yolk sac development.34These migrating precursors may be similar to the EB-derived BL-CFCs that have been shown to express Flk-135 and to grow in response to its ligand, VEGF.32
Scl, a member of the helix-loop-helix family of transcription factors,36 appears to function at a slightly later developmental stage than Flk-1. The yolk sacs ofScl−/− embryos develop a primary vascular network but contain no primitive or definitive hematopoietic cells, indicating a pivotal role for Scl in hematopoietic commitment but not in the early stages of vasculogenesis.36-38 Although not essential for establishment of the endothelial lineage, Scl does appear to be required for remodeling and maturation of the vascular system.39 In vitro differentiation studies withScl−/− ES cells showed a complete block in primitive and definitive hematopoietic potential, confirming the developmental defect observed in vivo.24 Further analysis of the Scl−/− EBs revealed a defect as early as the BL-CFC because the mutant ES cells were unable to generate blast cell colonies.29,35Scl−/−ES cells were, however, able to generate precursors that gave rise to transitional colonies that represent a stage of development slightly earlier than the blast cell colony.29
The AML1 gene (recently renamed Runx1), which encodes the DNA-binding subunit of a transcription factor of the core binding factor (CBF) family,40-42 is required for the establishment of definitive but not primitive hematopoiesis.Runx1−/− embryos develop normal blood islands and progress through the yolk sac phase of hematopoiesis but die between E11 and E12.5.26,27 Before death, the liver rudiment contains primitive nucleated erythrocytes but lacks all definitive erythroid, myeloid, and megakaryocytic cells, indicating a complete block in the development of the definitive hematopoietic program. Analysis of Runx1−/− EBs revealed a similar block in definitive hematopoietic potential.26 43In addition to the hematopoietic abnormalities,Runx1-deficient animals show extensive central nervous system hemorrhage and necrosis, suggesting an additional vascular defect.
Although these targeting studies position Runx1 at the establishment and expansion of the definitive hematopoietic program, expression analysis suggest that it may function at earlier stages of development. Using mice with the LacZ gene targeted to theRunx1 locus (Runx1+/Z mice), North et al44 demonstrated expression of Runx1 (LacZ) in subpopulations of endothelial cells in the yolk sac, in the vitelline and umbilical arteries, and in the ventral wall of the dorsal aorta. Expression was also detected in emerging primitive erythrocytes early in the yolk sac. Expression in these cells was transient, declined significantly by E8.5, and was undetectable by E10.5. The presence ofRunx1 transcripts in primitive erythrocytes and in subpopulations of endothelial cells suggests that this gene may be expressed and may function at the level of a cell with hemangioblast properties.
To investigate the role of Runx1 at the earliest stage of hematopoietic commitment, we analyzed its expression pattern and function during ES/EB differentiation and in early yolk sac development. Our results indicate that Runx1 is expressed in yolk sac mesodermal cells before the establishment of the blood islands and within the BL-CFCs in EBs. Analysis of Runx1-deficient ES cells demonstrated that this gene is essential for the generation of normal numbers of blast colonies and, as such, provides evidence that it does function at the equivalent of the hemangioblast stage of development. BL-CFCs that develop in the deficient EBs appear to be primitive erythroid restricted, suggesting that the functional requirement for Runx1 may define subpopulations of these precursors.
Materials and methods
In situ hybridization
In situ hybridization of outbred ICR (Taconic, Germantown, NY) murine embryos was performed as previously described9 with the following modifications. Single-stranded 33P-labeled antisense Runx1 and sense control probes (accession number W29200) were prepared at specific activities of 4 × 109 dpm/μg. After hybridization and high-stringency washes, tissues were counterstained with hematoxylin. Bright-field and dark-field images were captured with a Polaroid digital camera and were processed, including pseudocolorizing grains, using Adobe Photoshop (Adobe Systems). No signal above background was observed in the negative controls.
Embryonic stem cell growth and differentiation
The generation of Runx1+/−,Runx1−/−, and Runx1+/zJ1 ES cells has been previously described.27 44 ES cells were maintained on irradiated embryonic feeder cells in Dulbecco modified Eagle medium supplemented with 15% fetal calf serum (FCS), penicillin, streptomycin, leukemia inhibitory factor (LIF) (1% conditioned medium), and 1.5 × 10−4 M monothioglycerol (MTG; Sigma, St Louis, MO). Two days before the onset of differentiation, cells were transferred to gelatinized plates in the same media. For the generation of EBs, ES cells were trypsinized and plated at various densities in differentiation cultures. Differentiation of EBs was carried out in 60-mm Petri-grade dishes in Iscoves modified Dulbecco medium (IMDM) supplemented with 15% FCS, 2 mM L-glutamine (Gibco/BRL, Grand Island, NY), transferrin (200 μg/mL), 0.5 mM ascorbic acid (Sigma), and 4.5 × 10−4M MTG. Cultures were maintained in a humidified chamber in a 5% CO2–air mixture at 37°C.
Colony assays
For the generation of blast cell colonies (BL-CFC assay), EB cells were plated in 1% methylcellulose supplemented with 10% FCS (Summit, Fort Collins, CO), VEGF (5 ng/mL), c-kit ligand (KL; 1% conditioned medium), interleukin-6 (IL-6; 5 ng/mL), and 25% D4T endothelial cell-conditioned medium.32 Transitional colonies were generated in the same basic conditions in the absence of VEGF. Colonies were scored after 4 days of culture. For the growth of hematopoietic precursors, cells were plated in 1% methylcellulose containing 10% plasma-derived serum (Antech, Tyler, TX), 5% protein-free hybridoma medium (PFHM-II; Gibco-BRL), and cytokines KL (1% conditioned medium), thrombopoietin (5 ng/mL), Erythropoietin (2 U/mL), IL-11 (25 ng/mL), IL-3 (1% conditioned medium), granulocyte-macrophage colony-stimulating factor (GM-CSF; 3 ng/mL), G-CSF (30 ng/mL), M-CSF (5 ng/mL), and IL-6 (5 ng/mL). Cultures were maintained at 37°C, 5% CO2. Primitive erythroid colonies were scored at day 5 to 6 of culture, whereas definitive erythroid, macrophage, and multilineage colonies were counted after 7 to 10 days of culture.
For expansion of blast cell colonies, individual colonies were transferred to Matrigel-coated (Collaborative Research, San Jose, CA) microtiter wells containing IMDM with 10% FCS (Hyclone, Logan, UT), 10% horse serum (Biocell, Rancho Dominguez, CA), VEGF (5 ng/mL), insulinlike growth factor 1 (IGF-1) (10 ng/mL), erythropoietin (2 U/mL), basic fibroblast growth factor (bFGF) (10 ng/mL), IL-11 (50 ng/mL), KL (1% conditioned medium), IL-3 (1% conditioned medium), L-glutamine (1%), and 4.5 × 10−4 M MTG. After 4 days of growth, the nonadherent cells of each well were harvested and cultured in 1 mL methylcellulose containing the above mixture of cytokines used for the growth of hematopoietic precursors. LIF and c-kit ligand were derived from media conditioned by Chinese hamster ovary cells transfected with LIF and KL expression vectors, respectively (kindly provided by Genetics Institute, Cambridge, MA). IL-3 was obtained from medium conditioned by X63 AG8-653 myeloma cells transfected with a vector expressing IL-3.45 VEGF, GM-CSF, M-CSF, G-CSF, thrombopoietin, IL-6, and IL-11 were purchased from R&D Systems (Minneapolis, MN).
X-gal staining
Undifferentiated ES cells, differentiated EBs, and hematopoietic colonies were fixed in 1× phosphate-buffered saline (PBS) containing 0.5% glutaraldehyde (Sigma) and 1 mM MgCl2 for 10 to 15 minutes. After fixation, the cells were rinsed in 1× PBS and stained with 1 mM MgCl2, 3.3 mM K4Fe(CN)6, 3.3 mM K3Fe(CN)6, 0.02% NP-40, and 0.1 vol 2% X-gal (Sigma) overnight at 37°C. Positive cells were detected by the presence of blue staining visualized under light microscopy.
FACS-gal analysis
Fluorescein di-β-D-galactopyrosanide (FDG; Sigma), hydrolyzed to fluorescein by intracellular β-galactosidase, was used to detect LacZ activity. EB-derived cells were washed and resuspended in 1× PBS–20% FCS to a final maximum concentration of 107cells/mL. Hypotonic loading was achieved by a 2-minute incubation at 37°C with an equal volume of prewarmed 2 mM FDG (in water). After the loading procedure, 10 vol cold IMDM–15% FCS were added, and the mixture was incubated 20 minutes on ice. Stained suspensions were analyzed on a FACScan (Becton Dickinson, San Jose, CA) or sorted on a MoFlo (Cytomation Systems, Fort Collins, CO) cell sorter.
Gene expression analysis
For polyA+ global amplification polymerase chain reaction (PCR), reverse transcription (RT), poly-A tailing, and PCR procedures were performed as described by Brady et al,46with the exception that the X-dT oligonucleotide was shortened to 5′-GTTAACTCGAGAATTC(T)24-3′. Amplified products from PCR were separated on agarose gels and transferred to a Zeta-probe GT membrane (Bio-Rad, Hercules, CA). Resultant blots were hybridized with32P randomly primed cDNA fragments (Ready-to-Go Labeling; Pharmacia, Piscataway, NJ) corresponding to the 3′ region of the genes (for all except β-H1). A β H1-specific probe was prepared by annealing 2 oligonucleotides, TGGAGTCAAAGAGGGCATCATAGACACATGGG and CAGTACACTGGCAATCCCATGTG, that share an 8-base homology at their 3′ termini. This β H1-specific probe was labeled with 32P using a Klenow fill-in reaction. For gene-specific PCR, total RNA was extracted from each sample with the RNeasy mini-kit and treated with Rnase-free DNase (Qiagen, Valencia, CA). Two micrograms total RNA were reverse-transcribed into cDNA with random hexamer using the Omniscript RT kit (Qiagen). PCR was carried out using the following oligonucleotides: β-actin, 5′ATGAAGATCCTGACCGAGCG3′ (sense), 5′TACTTGCGCTCAGGAGGAGC3′ (antisense); Brachyury, 5′CTAGTACTCTTTCTTGCTGG3′ (sense), 5′GGTCTCGGGAAAGCAGTGGC3′ (antisense);Runx1, 5′CCAGCAAGCTGAGGAGCGGCG3′ (sense), 5′CGGATTTGTAAAGACGGTGA3′ (antisense); Flk1,5′CACCTGGCACTCTCCACCTTC3′(sense), 5′GATTTCATCCCACTACCGAAAG3′ (antisense); and Scl, 5′ATGGAGATTTCTGATGGTCCTCAC3′ (sense), 5′AAGTGTGCTTGGGTGTTGGCTC3 (antisense).
PCR was performed with 2.5 U Taq polymerase (Promega, Madison, WI), PCR buffer, 2.5 mM MgCl2, 0.2 μM each primer, and 0.2 mM dNTP. Cycling conditions were as follows; 94°C for 5 minutes followed by 35 cycles of amplification (94°C denaturation for 1 minute, 60°C annealing for 1 minute, 72°C elongation for 1 minute), with a final incubation at 72°C for 7 minutes.
Results
Runx1 is expressed during gastrulation in extra-embryonic mesoderm before formation of blood islands
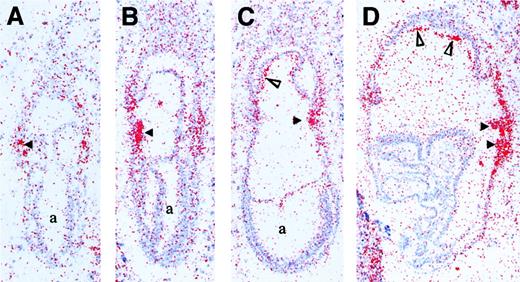
To further assess the role of Runx1 in the establishment of the hematopoietic system, we mapped its expression before and during the development of blood islands in E7.25 to E8.25 embryos. Runx1 transcripts were detected at the mid-to-late primitive streak stage (E7.25), specifically in extraembryonic mesoderm cells adjacent to visceral endoderm (Figure1A). This pattern of expression in extraembryonic mesoderm persisted in early neural plate embryos (E7.5, Figure 1B). At mid-to-late neural plate stages, Runx1 mRNA was present predominantly in nascent yolk sac blood islands (Figure 1C and data not shown). In addition, there was a low level of expression in the developing chorion that increased by early somite pair stages (E8.25, Figure 1D). At E8.25, the predominant accumulation ofRunx1 mRNA was in the developing yolk sac blood islands (Figure 1D). The early expression of Runx1 described here is similar to that observed for Scl47 and is consistent with a role in the commitment of mesoderm to hematopoietic–endothelial fates.
Expression ofRunx1 mRNA during gastrulation in the mouse embryo (E7.25-E8.25).
(A) Mid-to-late primitive streak stage embryo (E7.25) withRunx1 mRNA accumulation in mesoderm cells adjacent to visceral endoderm in the forming yolk sac (arrowhead). (B) Early neural plate embryo (E7.5) with increased Runx1 transcripts in the yolk sac mesoderm (arrowhead). (C) Late neural plate embryo (E7.75) reveals Runx1 expression in nascent yolk sac blood islands (arrowhead) and in the mesoderm component of the chorion (open triangle). (D) Early somite-stage embryo (E8.25) displaysRunx1 mRNA accumulation in expanding blood islands of the yolk sac (arrowheads) and in the chorion (open triangles). a indicates amniotic cavity.
Expression ofRunx1 mRNA during gastrulation in the mouse embryo (E7.25-E8.25).
(A) Mid-to-late primitive streak stage embryo (E7.25) withRunx1 mRNA accumulation in mesoderm cells adjacent to visceral endoderm in the forming yolk sac (arrowhead). (B) Early neural plate embryo (E7.5) with increased Runx1 transcripts in the yolk sac mesoderm (arrowhead). (C) Late neural plate embryo (E7.75) reveals Runx1 expression in nascent yolk sac blood islands (arrowhead) and in the mesoderm component of the chorion (open triangle). (D) Early somite-stage embryo (E8.25) displaysRunx1 mRNA accumulation in expanding blood islands of the yolk sac (arrowheads) and in the chorion (open triangles). a indicates amniotic cavity.
Runx1 expression is up-regulated at the hemangioblast stage of EB differentiation
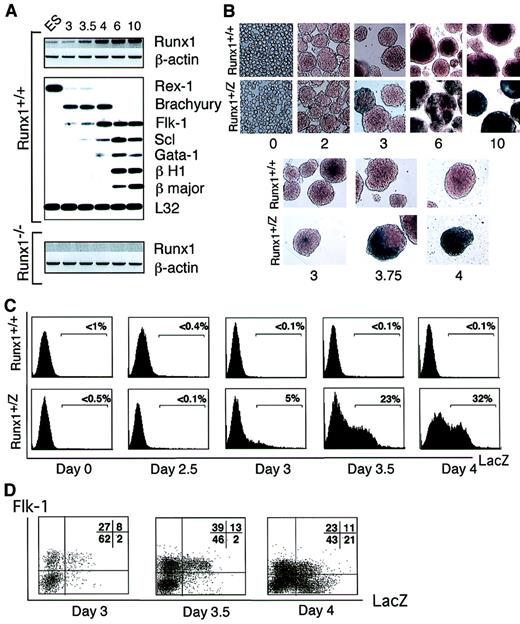
The above analyses indicate that Runx1 is expressed at the earliest stage of blood island development, suggesting a potential role at the level of the putative hemangioblast. To further investigate the function of Runx1 at the onset of hematopoietic development, we analyzed its expression pattern in EBs over a 10-day differentiation period. A low level of Runx1expression was detected in undifferentiated ES cells and in EBs after the first few days of differentiation (Figure2A). Runx1 expression increased significantly between days 3 and 4 of differentiation and remained elevated thereafter. Based on real-time PCR analysis, the magnitude of the increase in Runx1 expression over this 24-hour period was found to be approximately 10-fold (not shown). NoRunx1 cDNA was detected in EBs generated fromRunx1−/− ES cells. Comparative analysis ofRunx1 expression with other genes demonstrated a striking similarity to the temporal expression pattern of Flk-1. The up-regulation of both Runx1 and Flk-1 was preceded by the expression of Brachyury, a marker of early mesoderm development.48Rex-1, a zinc finger transcription factor expressed in ES cells but not in their differentiated progeny,49 was readily detected in undifferentiated ES cells but not significantly in day 3 EBs.Scl was expressed at low levels as early as day 3.5 of differentiation. The levels increased over the next few days and then remained relatively constant. Gata-1, a transcription factor expressed in hematopoietic but not endothelial cells50 and in the embryonic β H1 and adult β majorglobins, followed the onset of Scl expression. These findings suggest that Runx1 expression is up-regulated at the hemangioblast stage of development (as defined by Flk-1and Scl), following the establishment of the earliest mesodermal population (Brachyury) but preceding the commitment to the hematopoietic lineage (Gata-1, β H1, β major globin).
Gene expression analysis of EBs.
(A) Gene expression in Runx1+/+ andRunx1−/− EBs. Numbers on top of the figure indicate day of EB differentiation. ES represents undifferentiated ES cells. Runx1 and β-actin expression was determined using specific oligonucleotides. Expression of other genes was evaluated using a polyA+ global amplification PCR method.46 Hybridization with a 3′ probe from the L32 ribosomal protein gene was included to control for amounts of cDNA loaded. (B) X-gal staining of Runx1+/+ andRunx1+/Z EBs. Numbers indicate day of EB differentiation. Original magnification day 0, × 400; days 2 to 4, × 200; and days 6 to 10, × 40. (C) Flow cytometry analysis of FDG stained Runx1+/+ andRunx1+/Z EBs. (D) FACS analysis ofFlk-1 and Runx1 expression in days 3, 3.5, and 4Runx1+/Z EBs. Numbers in quadrant represent the percentage of total population in each fraction.
Gene expression analysis of EBs.
(A) Gene expression in Runx1+/+ andRunx1−/− EBs. Numbers on top of the figure indicate day of EB differentiation. ES represents undifferentiated ES cells. Runx1 and β-actin expression was determined using specific oligonucleotides. Expression of other genes was evaluated using a polyA+ global amplification PCR method.46 Hybridization with a 3′ probe from the L32 ribosomal protein gene was included to control for amounts of cDNA loaded. (B) X-gal staining of Runx1+/+ andRunx1+/Z EBs. Numbers indicate day of EB differentiation. Original magnification day 0, × 400; days 2 to 4, × 200; and days 6 to 10, × 40. (C) Flow cytometry analysis of FDG stained Runx1+/+ andRunx1+/Z EBs. (D) FACS analysis ofFlk-1 and Runx1 expression in days 3, 3.5, and 4Runx1+/Z EBs. Numbers in quadrant represent the percentage of total population in each fraction.
The up-regulation of Runx1 expression detected during EB differentiation could reflect an increase in the level of expression within a subset of cells or an increase in the number of cells expressing this gene. To distinguish between these 2 possibilities, we analyzed EBs generated from Runx1+/Z ES cells that contain the LacZ gene targeted to the Runx1gene.44LacZ expression was evaluated either by direct X-gal staining or by FDG staining followed by flow cytometry analyses. Using both methods of detection, no significant staining was observed in undifferentiated ES cells (day 0) or in early EBs (up to 2.5 days) (Figure 2B-C). LacZ+ cells were first detected within the EBs by day 3 of differentiation, at which time approximately 5% of cells expressed this marker as determined by fluorescence-activated cell sorter (FACS) analysis. The number of positive cells increased dramatically over the next 24 hours, reaching levels greater than 30% of the total EB population by day 4 of differentiation. The frequency of LacZ+ cells remained elevated (from 30% to 50%) in EBs between days 5 and 10 of differentiation (Figure 2B and not shown). X-gal staining revealed that a large portion of individual EBs at day 4 to 6 of differentiation expressed LacZ. No significant level of β-galactosidase activity was detected in control wild-type (Runx1+/+) EBs. These findings demonstrate an early and rapid expansion of cells expressing Runx1, with kinetics almost identical to the expansion of the Flk-1+population51 and the development of the BL-CFC.30 To determine whether the Runx1+ and Flk-1+ cells represent distinct or overlapping populations, we analyzed developing EBs for the presence of both markers. At days 3.0 and 3.5 of differentiation, most LacZ+cells also expressed Flk-1 (Figure 2D). By day 4 of differentiation, however, a significant population of Flk1−/LacZ+ cells was detected. These patterns suggest that the expression of Runx1 within a subpopulation of Flk-1+ cells at these early stages of differentiation could define the emergence of the BL-CFC.
Runx1 is expressed in blast colonies and BL-CFCs
Given the early expression pattern observed in EBs, we next investigated Runx1 expression in blast colonies and populations derived from them. As shown in Figure3A, blast colonies generated fromRunx1+/Z EBs uniformly stained positive for β-galactosidase activity. In contrast, no staining was observed in control Runx1+/+ blast colonies. This indicates that the cells within these colonies, which have previously been shown to represent hematopoietic and endothelial precursors,30express Runx1. When individual blast colonies are transferred to microtiter wells under appropriate growth conditions, these precursors grow and mature into adherent endothelial cells and a nonadherent population of hematopoietic cells after 4 days of culture. Analysis of these subpopulations demonstrated extensive LacZexpression in the hematopoietic cells (Figure 3B). Expression was also detected in the adherent cells, though the levels were significantly lower than in the hematopoietic population (Figure 3B, arrowheads). Expression in the adherent population is consistent with the observation that Runx1 is expressed in subsets of endothelial cells in the yolk sac and embryo proper.44
Runx1 gene expression in blast colonies and BL-CFC.
(A) X-gal staining of Runx1+/+ andRunx1+/Z blast colonies. Colonies were generated from day 3.5 EB-derived cells and were stained after 4 days of growth. Original magnification × 200. (B) X-gal staining of nonadherent and adherent cells derived from Runx1+/+ andRunx1+/Z blast colonies. Arrowheads indicate LacZ+ adherent cells. Original magnification × 400. (C) Upper portion of figure shows FACS profile of FDG-stained day 3.75Runx1+/Z EBs. LacZ-negative (Neg) and -positive (Pos) fractions isolated by cell sorting are indicated. Sorting gates were set by comparing the staining of the +/Z cells to the wild-type (+/+) cells. Middle portion shows number of blast colonies generated by the different sorted fractions. Colonies were scored at day 4 of culture. Bars represent standard error of the mean number of colonies from at least 3 cultures. RT-PCR gene expression analysis of LacZ-positive and -negative fractions of day 3.75 EBs is depicted in the lower portion. PCR products for Runx1 andβ-actin were detected by hybridization with specific32P-labeled probes. Products for Brachyury,Flk-1, Scl, Gata-1, and β H1 were visualized following ethidium bromide staining and are presented as a negative image. (D) Cell sorting of FDG-stained day 3Runx1+/Z EBs. Secondary EBs (2°EBs), transitional colonies (Trans), and blast colonies (Blast) were scored 4 days following replating of the isolated fractions. Bars represent standard error of the mean number of colonies from at least 3 cultures.
Runx1 gene expression in blast colonies and BL-CFC.
(A) X-gal staining of Runx1+/+ andRunx1+/Z blast colonies. Colonies were generated from day 3.5 EB-derived cells and were stained after 4 days of growth. Original magnification × 200. (B) X-gal staining of nonadherent and adherent cells derived from Runx1+/+ andRunx1+/Z blast colonies. Arrowheads indicate LacZ+ adherent cells. Original magnification × 400. (C) Upper portion of figure shows FACS profile of FDG-stained day 3.75Runx1+/Z EBs. LacZ-negative (Neg) and -positive (Pos) fractions isolated by cell sorting are indicated. Sorting gates were set by comparing the staining of the +/Z cells to the wild-type (+/+) cells. Middle portion shows number of blast colonies generated by the different sorted fractions. Colonies were scored at day 4 of culture. Bars represent standard error of the mean number of colonies from at least 3 cultures. RT-PCR gene expression analysis of LacZ-positive and -negative fractions of day 3.75 EBs is depicted in the lower portion. PCR products for Runx1 andβ-actin were detected by hybridization with specific32P-labeled probes. Products for Brachyury,Flk-1, Scl, Gata-1, and β H1 were visualized following ethidium bromide staining and are presented as a negative image. (D) Cell sorting of FDG-stained day 3Runx1+/Z EBs. Secondary EBs (2°EBs), transitional colonies (Trans), and blast colonies (Blast) were scored 4 days following replating of the isolated fractions. Bars represent standard error of the mean number of colonies from at least 3 cultures.
To determine whether the BL-CFC also expresses Runx1, day 3.75 Runx1+/Z EB cells were fractionated for β-galactosidase activity by cell sorting (Figure 3C). Higher levels of Runx1 expression were detected in cells from the LacZ-positive fraction than in those from the negative fraction, confirming that separation of cells based on β-galactosidase activity resulted in an enrichment of Runx1-expressing cells. Analysis of BL-CFC content revealed that most precursors were present in the LacZ+ population, demonstrating that most BL-CFCs isolated from day 3.75 EBs express Runx1. To further define the developmental status of the day 3.75 LacZ+(Runx1+) cells, we analyzed this fraction for the expression of other genes known to play a role in early hematopoietic commitment and maturation. Runx1+ cells were found to contain lower levels of Brachyury than those in the negative fraction, indicating that this population is exiting the mesodermal stage of development. Flk-1 was expressed in both the positive and the negative fractions, a finding consistent with the previous FACS analyses that demonstrated the presence of Flk-1+/Runx1+ and Flk-1+/Runx1− populations in comparably staged EBs (Figure 2D). The higher levels of Scl andGata-1 in the positive fraction are consistent with its elevated BL-CFC content and further support the interpretation thatRunx1 expression defines one of the earliest stages of hematopoietic commitment. Low levels of βH1 globin were present in the LacZ+ (Runx1+) fraction, indicating the onset of differentiation to the primitive erythroid lineage.
To evaluate whether earlier developing BL-CFCs also expressRunx1, day 3 Runx1+/Z EBs were fractionated for β-galactosidase activity. In addition to BL-CFCs, EBs at this stage of development contain some residual ES cells and precursors (Trans-CFC) that generate transitional colonies. Transitional colonies represent an earlier stage of development than blast colonies and contain cell populations undergoing commitment of mesoderm to the hematopoietic and endothelial lineages.29As observed for day 3.75 BL-CFCs, almost all day 3.0 BL-CFCs were present in the LacZ+ fraction (Figure 3D). In contrast, the Trans-CFC and the ES cells that generate secondary EBs segregated to the LacZ− fraction, indicating that they do not expressRunx1 (Figure 3D). Taken together, the findings from the cell-sorting studies clearly demonstrate that the BL-CFC, a precursor with hemangioblast characteristics, expresses Runx1.
Runx1−/−ES cells display a defect in BL-CFC developmental potential
To examine the requirement for the Runx1 gene in BL-CFC development, day 3.5 and 4 EBs generated fromRunx1−/−, Runx1+/−, and Runx1+/+ ES cells were analyzed for BL-CFC content. At both time points, the Runx1−/− EB cells displayed a profound defect in BL-CFC potential.Runx1-deficient EBs consistently generated 10- to 20-fold fewer blast colonies than wild-type controls (Figure4A). The blast colonies that did develop from the Runx1−/− EB cells were similar in morphology to those generated from wild-type cells (Figure5A, top). To confirm thatRunx1 was indeed critical for blast colony development, we attempted to rescue the defect by retroviral-mediated expression of this gene in deficient cells. As shown in Figure 4B, day 3 and 3.75 EBs generated from Runx1−/− ES cells infected with a retroviral vector encoding Runx1, and selected for puromycin resistance, produced higher numbers of blast colonies than EBs from ES cells infected with the empty retrovirus. These findings strongly suggest that Runx1 plays a pivotal role at the stage of BL-CFC development.
Analysis of BL-CFC potential ofRunx1+/+, Runx1+/−, and Runx1−/− EBs.
(A) Number of blast colonies generated by day 3.5 and 4Runx1+/+ (+/+), Runx1+/−(+/−), and Runx1−/− (−/−) EB-derived cells. Two Runx1−/− clones (−/−A, −/−B) were analyzed. Bars represent standard error of the mean number of colonies from at least 3 cultures. (B) Rescue of the BL-CFC potential ofRunx1−/− ES cells.Runx1−/− ES cells infected with either a retrovirus expressing the Runx1b isoform of the gene (MSCVRunx1) or with an empty virus control (MSCV) and selected with puromycin were differentiated for the indicated periods of time. EBs were harvested and analyzed for BL-CFCs. Bars represent standard error of the mean number of colonies from at least 3 cultures.
Analysis of BL-CFC potential ofRunx1+/+, Runx1+/−, and Runx1−/− EBs.
(A) Number of blast colonies generated by day 3.5 and 4Runx1+/+ (+/+), Runx1+/−(+/−), and Runx1−/− (−/−) EB-derived cells. Two Runx1−/− clones (−/−A, −/−B) were analyzed. Bars represent standard error of the mean number of colonies from at least 3 cultures. (B) Rescue of the BL-CFC potential ofRunx1−/− ES cells.Runx1−/− ES cells infected with either a retrovirus expressing the Runx1b isoform of the gene (MSCVRunx1) or with an empty virus control (MSCV) and selected with puromycin were differentiated for the indicated periods of time. EBs were harvested and analyzed for BL-CFCs. Bars represent standard error of the mean number of colonies from at least 3 cultures.
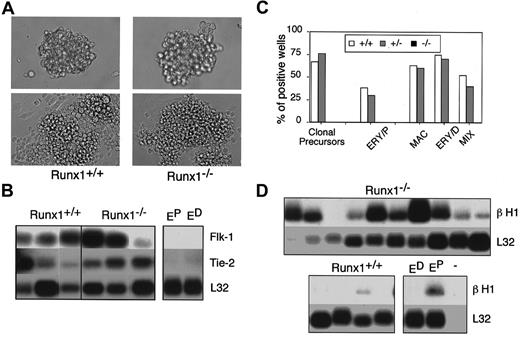
Developmental potential of Runx1−/− blast colonies.
(A) Morphology of Runx1+/+ andRunx1−/− blast cell colonies (top, original magnification × 200) and the adherent and nonadherent cells generated from them following 2 days of culture (bottom, original magnification × 400). (B) Gene expression analysis of adherent cells generated by individual blast colonies. Analysis was performed by the polyA+ global amplification method. Each lane represents adherent cells derived from one blast colony. Control primitive erythroid (EP) and definitive erythroid (ED) samples are indicated. (C) Hematopoietic potential of the nonadherent population derived from expandedRunx1+/+ (+/+), Runx1+/−(+/−), and Runx1−/− (−/−) blast colonies. Nonadherent cells derived from individual colonies were harvested, and the cells were plated in conditions that support the growth of primitive and definitive colonies. The frequency of wells giving rise to total secondary colonies (clonal precursors), primitive erythroid colonies (ERY/P), or definitive hematopoietic colonies (MAC, macrophage; ERY/D, definitive erythroid; and MIX, multilineage) is represented. (D) β H1 globin expression in nonadherent cells generated by individual colonies. Each lane corresponds to the nonadherent cells from one expanded blast colony.
Developmental potential of Runx1−/− blast colonies.
(A) Morphology of Runx1+/+ andRunx1−/− blast cell colonies (top, original magnification × 200) and the adherent and nonadherent cells generated from them following 2 days of culture (bottom, original magnification × 400). (B) Gene expression analysis of adherent cells generated by individual blast colonies. Analysis was performed by the polyA+ global amplification method. Each lane represents adherent cells derived from one blast colony. Control primitive erythroid (EP) and definitive erythroid (ED) samples are indicated. (C) Hematopoietic potential of the nonadherent population derived from expandedRunx1+/+ (+/+), Runx1+/−(+/−), and Runx1−/− (−/−) blast colonies. Nonadherent cells derived from individual colonies were harvested, and the cells were plated in conditions that support the growth of primitive and definitive colonies. The frequency of wells giving rise to total secondary colonies (clonal precursors), primitive erythroid colonies (ERY/P), or definitive hematopoietic colonies (MAC, macrophage; ERY/D, definitive erythroid; and MIX, multilineage) is represented. (D) β H1 globin expression in nonadherent cells generated by individual colonies. Each lane corresponds to the nonadherent cells from one expanded blast colony.
In the next set of experiments, we assessed the developmental potential of the blast colonies generated from theRunx1−/− and the rescuedRunx1−/− ES cells and compared it to that of colonies from heterozygous and wild-type cells. Two different assays were used in this analysis. As a first approach, colonies were picked, transferred to microtiter wells, and cultured for 4 days to determine their potential to generate adherent (endothelial) and nonadherent (hematopoietic) cells. Colonies from each of the groups generated both types of cells (Runx1+/+ andRunx1−/−, shown in Figure 5A, bottom). After expansion, a fraction of the nonadherent cells of each well was replated in methylcellulose to assay hematopoietic potential. The remainder of the nonadherent population was analyzed forβH1 globin gene expression as a measure of primitive erythroid potential. Adherent cells from the wild-type andRunx1−/− colonies were cultured for an additional week and then harvested and analyzed for the expression of genes associated with the endothelial lineage. This expression analysis demonstrated the presence of Flk-1 andTie-252 transcripts in the adherent populations derived from Runx1−/− and theRunx1+/+ individual blast colonies, indicating that these cells are of the endothelial lineage (Figure 5B).
Replating studies revealed that the nonadherent populations from mostRunx1+/+ and Runx1+/−blast colonies contained precursors with colony-forming potential (Figure 5C). All expanded populations that replated were found to contain definitive hematopoietic precursors. Approximately 30% of the cultures with definitive precursors also contained primitive erythroid precursors. Cultures with only primitive erythroid precursors were not detected in this set of experiments. In contrast to the wild-type cells, nonadherent populations from Runx1−/−blast colonies contained few, if any, precursors. No precursors were detected in the experiment shown in Figure 5C; however, primitive erythroid precursors were occasionally found in other studies. The nonadherent fraction of the rescued Runx1−/−blast colonies did contain precursors of the definitive hematopoietic lineages (macrophage and mix), demonstrating that this potential was rescued in the BL-CFCs. Although the precursor content was low in the nonrescued Runx1−/− cultures, βH1globin expression was readily detectable in the nonadherent cells, indicating the presence of primitive erythroid cells (Figure 5D). The presence of βH1, together with the lack of clonable precursors in the Runx1-deficient cultures, suggests that the primitive erythroid cells had matured beyond the precursor stage during the expansion phase. In the second analysis, we assayed the hematopoietic potential of the Runx1−/− blast colonies before expansion in culture to determine if they did contain primitive erythroid precursors. For this assay, individualRunx1−/− blast colonies were picked, and the cells were dispersed and replated directly in secondary methylcellulose cultures. Under these conditions, approximately 40% of the replated colonies gave rise to secondary primitive erythroid colonies. Colonies of definitive hematopoietic cells were never observed in these secondary cultures. Collectively, these findings demonstrate thatRunx1 is essential for the generation of normal numbers of blast colonies. Blast colonies that develop fromRunx1−/− EBs appear to be restricted to the primitive erythroid and endothelial lineages, a finding consistent with the interpretation that Runx1 is essential for definitive hematopoietic development.
Runx1 deficiency does not affect primitive hematopoietic development
The defect in blast colony development inRunx1−/− EBs prompted us to investigate the primitive hematopoietic potential of these cells and those of the yolk sac of Runx1−/− embryos. Previous analysis of the primitive erythroid lineage, by estimation of circulating red cell number in Runx1−/− embryos, suggested that this population was relatively unaffected by the deletion of this gene.26 27 However, because quantitative analysis of primitive erythroid precursors was not performed, it is possible that the Runx1 mutation did cause subtle defects in the development of this lineage.
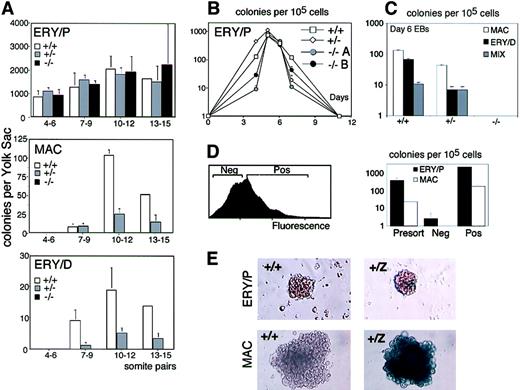
Analyses of the yolk sac of Runx1−/− embryos, ranging in stage from 4 to 6 sp (E8.25) to 13 to 15sp (E8.5), revealed the presence of normal numbers of primitive erythroid precursors (Figure 6A). This tissue was devoid of definitive hematopoietic precursors, including those of the macrophage and definitive erythroid lineages. In contrast, definitive precursors could be detected in the yolk sac of Runx1+/+and Runx1+/− embryos as early as the 7 to 9sp stage of development. Yolk sacs from Runx1+/−embryos contained fewer definitive precursors than those from wild-type embryos, indicating a hemizygous affect of Runx1 at these early stages of definitive hematopoiesis. These findings clearly demonstrate that the primitive lineage, at the level of the precursor stage, is intact in the Runx1−/− embryos and that the definitive hematopoietic program is affected at the earliest stages of its development.
Analysis of primitive and definitive hematopoietic potential ofRunx1+/+, Runx1+/−, and Runx1−/− embryos and ES cells.
(A) Primitive erythroid, definitive erythroid, and macrophage colonies generated by yolk sac cells fromRunx1+/+ (+/+), Runx1+/−(+/−), and Runx1−/− (−/−) embryos. The stage of development (number of somite pairs) is indicated. The number of embryos analyzed at each stage is as follows: 4-6sp: 3 +/+, 2 +/−, 2−/−; 7-9sp: 3 +/+, 3 +/−, 4−/−; 10-12sp: 3 +/+, 12 +/−, 3−/−; and 13-15sp: 1 +/+, 4 +/−, 2−/−. (B) Primitive erythroid precursors generated by Runx1+/+ (+/+),Runx1+/− (+/−), andRunx1−/− (−/−) EBs. Numbers are days of EB differentiation.−/−A and −/−B represent 2 independent clones ofRunx1−/− ES cells. Bars, where visible, represent standard error of the mean number of colonies from at least 3 cultures. (C) Definitive hematopoietic potential ofRunx1+/+ (+/+),Runx1+/− (+/−), andRunx1−/− (−/−) EBs at day 6 of differentiation. Macrophage (MAC), definitive erythroid (ERY/D), and multilineage (MIX) colonies were scored. Bars represent standard error of the mean number of colonies from at least 3 cultures. (D) FACS profile of day 4.75 Runx1+/Z EB cells stained with FDG (left panel). Negative (Neg) and positive (Pos) fractions were isolated and assayed for precursor potential. Number of primitive erythroid (ERY/P) and macrophage (MAC) colonies generated by the sorted fractions is shown in right panel. Bars represent standard error of the mean number of colonies from at least 3 cultures. (E) X-gal staining of Runx1+/+ (+/+) andRunx1+/Z (+/Z) primitive erythroid (ERY/P) and macrophage (MAC) colonies following 4 days of culture.
Analysis of primitive and definitive hematopoietic potential ofRunx1+/+, Runx1+/−, and Runx1−/− embryos and ES cells.
(A) Primitive erythroid, definitive erythroid, and macrophage colonies generated by yolk sac cells fromRunx1+/+ (+/+), Runx1+/−(+/−), and Runx1−/− (−/−) embryos. The stage of development (number of somite pairs) is indicated. The number of embryos analyzed at each stage is as follows: 4-6sp: 3 +/+, 2 +/−, 2−/−; 7-9sp: 3 +/+, 3 +/−, 4−/−; 10-12sp: 3 +/+, 12 +/−, 3−/−; and 13-15sp: 1 +/+, 4 +/−, 2−/−. (B) Primitive erythroid precursors generated by Runx1+/+ (+/+),Runx1+/− (+/−), andRunx1−/− (−/−) EBs. Numbers are days of EB differentiation.−/−A and −/−B represent 2 independent clones ofRunx1−/− ES cells. Bars, where visible, represent standard error of the mean number of colonies from at least 3 cultures. (C) Definitive hematopoietic potential ofRunx1+/+ (+/+),Runx1+/− (+/−), andRunx1−/− (−/−) EBs at day 6 of differentiation. Macrophage (MAC), definitive erythroid (ERY/D), and multilineage (MIX) colonies were scored. Bars represent standard error of the mean number of colonies from at least 3 cultures. (D) FACS profile of day 4.75 Runx1+/Z EB cells stained with FDG (left panel). Negative (Neg) and positive (Pos) fractions were isolated and assayed for precursor potential. Number of primitive erythroid (ERY/P) and macrophage (MAC) colonies generated by the sorted fractions is shown in right panel. Bars represent standard error of the mean number of colonies from at least 3 cultures. (E) X-gal staining of Runx1+/+ (+/+) andRunx1+/Z (+/Z) primitive erythroid (ERY/P) and macrophage (MAC) colonies following 4 days of culture.
As observed in the yolk sac, EBs generated fromRunx1−/− ES cells showed little or no defect in primitive erythroid potential (Figure 6B). The only discernible difference was that primitive erythroid development in theRunx1−/− EBs appeared to be restricted to a narrower window of differentiation than found in wild-type and heterozygous EBs. The reason for this restriction is unknown. No definitive precursors were found in theRunx1−/− EBs, confirming the findings in the mouse embryo that this gene is absolutely essential for the development of these lineages (Figure 6C). As with the yolk sacs,Runx1+/− EBs generated fewer definitive precursors than those differentiated from wild-type ES cells (Figure6C).
The previous observation of Runx1 expression in yolk sac primitive erythrocytes44 and our sorting studies indicating that all BL-CFCs are Runx1+ suggested that ES-derived committed primitive erythroid precursors are also likely to express this gene. To address this issue, we sorted day 4.75Runx1+/Z EBs for LacZ expression and analyzed the fractions for primitive erythroid potential (Figure 6D). Replating studies indicated that almost all primitive erythroid precursors were found in the Runx1+ fraction (Figure 6D). As expected, the earliest macrophage precursors developing in these EBs were also Runx1+. Additional studies demonstrated that all definitive precursors found in day 6 EBs were Runx1+ (data not shown). These findings extend our BL-CFC analysis and demonstrate that Runx1 expression defines the earliest stages of primitive and definitive hematopoiesis.
Although primitive erythroid precursors express Runx1, mature cells within colonies generated from them appear to have down-regulated expression as indicated by the low levels of β-galactosidase staining (Figure 6E). In contrast, colonies of definitive hematopoietic cells maintain high levels of expression (Figure 6E and data not shown). These results indicate that all hematopoietic precursors express Runx1. As the primitive erythroid lineage matures, the cells appear to rapidly loseRunx1 expression, a pattern similar to that observed in vivo in the yolk sac blood islands.
Discussion
Previous studies have established Runx1 as a pivotal player in the development of the definitive hematopoietic system in the mouse embryo.26 27 Given this association with definitive hematopoiesis, Runx1 is generally considered to exert its primary function within the embryo proper, following the primitive erythroid stage of development in the yolk sac. In this report, we investigated the role of Runx1 in hematopoietic development in ES cell–derived EBs and demonstrated that it is essential for the establishment or differentiation of the BL-CFC, a precursor that represents the earliest stage of hematopoietic and endothelial commitment in this model system. Our data clearly show thatRunx1 is expressed within BL-CFCs and is required for the development of normal numbers of blast colonies from these precursors. Several lines of evidence indicate that the observed effect on the number of blast colonies truly reflects a critical role for Runx1at this stage of development and is not simply due to reduced potential resulting from ES cell clonal variation. First, the defect in BL-CFC development was found in several independentRunx1−/− clones. Second,Runx1−/− ES cells were able to generate wild-type numbers of primitive erythroid precursors, indicating normal developmental potential for this lineage. Third, the defect at the level of BL-CFC development could be rescued by reintroduction and expression of a wild-type Runx1 gene inRunx1−/− ES cells.
The expression and cell separation studies presented here indicate that the up-regulation of Runx1 expression defines the earliest stage of hematopoietic and endothelial commitment. The low levels of expression found in ES cells and early EBs by RT-PCR were not detected by LacZ staining. This discrepancy may reflect the early expression of minor isoforms of Runx1 that do not use the exons containing the LacZ gene53 or differences in the sensitivity of the methods used in the analyses. Based on RT-PCR and LacZ expression, Runx1 expression was found to be up-regulated between days 3 and 4 of differentiation. The increase in Runx1 expression overlaps with the onset ofFlk-1 and Scl expression and with the development of BL-CFCs. Although previous analyses have demonstrated that all BL-CFCs express Flk-1,35 only a small fraction of the Flk-1+ population appears to represent the BL-CFC precursor. Our results clearly demonstrate that the BL-CFC expressesRunx1 and that Runx1 expression at days 3 and 3.5 of differentiation is limited to a subpopulation of Flk-1+cells. These observations suggest that the up-regulation ofRunx1 expression may mark the developmental progression from the prehemangioblast (Flk-1+/Runx1−) to the hemangioblast (Flk-1+/Runx1+) in EBs.
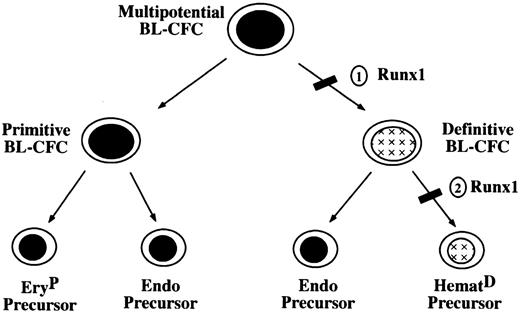
Although the deletion of Runx1 leads to a dramatic reduction in the number of blast colonies and a complete block in definitive hematopoiesis, it has little or no impact on the generation of the primitive erythroid lineage. These findings suggest that these hematopoietic programs may diverge at an early stage of development, possibly at the level of the BL-CFC as depicted in Figure7. In this model, 2 populations of BL-CFCs with distinct hematopoietic potential would be generated from a multipotential BL-CFC with primitive and definitive potential. Emergence of the primitive restricted BL-CFC would be aRunx1-independent event, whereas the generation of a functional definitive restricted precursor would requireRunx1. This model is supported by our previous studies in which we identified 3 different populations of BL-CFC, those with primitive and definitive hematopoietic potential, those that display definitive but little if any primitive potential, and those that appear to be restricted to the primitive erythroid lineage.30 32The primitive erythroid–restricted BL-CFCs were the least abundant and represented approximately 3% of the total population. This frequency is similar to the frequency of BL-CFC found inRunx1−/− EBs relative to wild-type controls. The BL-CFCs that develop in the Runx1−/− EBs may represent a combination of primitive erythroid– restricted precursors and multipotential precursors that are unable to generate definitive hematopoietic progeny. Although there is no direct evidence for the presence of hemangioblasts in the yolk sac, histologic studies demonstrating a close developmental association of the primitive erythroblasts and the endothelial cells within the blood islands would be consistent with the existence of a cell with properties of the primitive BL-CFC. The role of the proposed primitive restricted BL-CFC in vivo would be to generate the primitive erythroid lineage and the early yolk sac vasculature.
Model of early hematopoietic development.
Numbers indicate stages at which development could be blocked inRunx1-deficient cells.
Model of early hematopoietic development.
Numbers indicate stages at which development could be blocked inRunx1-deficient cells.
Most BL-CFCs within the developing EBs are Runx1 dependent and would belong to the definitive subpopulation of precursors in the above model. This finding is consistent with our previous studies demonstrating that most BL-CFCs had definitive but no detectable primitive potential.30 32 The significant reduction in blast cell colonies developing from Runx1−/−EBs could be interpreted as a block in the generation of definitive BL-CFC (Figure 7, block 1). Alternatively, it is possible that the developmental block is at the level of BL-CFC commitment to the definitive hematopoietic lineages (block 2) rather that in the generation of the BL-CFC. In this case, the definitive BL-CFC would be generated but only capable of giving rise to endothelial cells. The endothelial component alone may not grow well in methylcellulose or may generate colonies that are not recognized as typical blast colonies. We favor the second scenario (block 2) because a block at this level would not significantly impact endothelial development and be consistent with the observation that vascular development is not dramatically affected in the Runx1−/− embryos.
The existence of a definitive restricted hemangioblast in vivo is supported by a number of reports that indicate that hematopoietic cells “bud” from a subset of endothelial cells referred to as hemogenic endothelium54-56 found in the dorsal aorta of the developing embryo. This subpopulation of endothelial cells may represent definitive hemangioblasts. Evidence for a role forRunx1 at this stage of development has been provided by North et al,44 who demonstrated that a functional gene is essential for the development of these hematopoietic cell clusters but not for the underlying endothelial cells. Runx1-dependent hematopoietic clusters were also found associated with the endothelial lining of the vitelline and umbilical arteries and with vessels in the yolk sac capillaries.44 57 The function of the definitive hemangioblast in vivo would be first to generate the definitive precursors found early in the yolk sac and subsequently to establish intra-embryonic hematopoiesis in the P-Sp/AGM region of the embryo. These in vivo studies do support this model, but formal proof for the existence of a hemangioblast in the yolk sac or AGM will require clonal analysis.
In summary, the data presented in this report position Runx1functionally at the earliest stages of hematopoietic and endothelial commitment in developing EBs. Expression of Runx1 in early EBs defines a subset of the Flk-1+ population that contains most, if not all, BL-CFCs, the in vitro equivalent of the putative hemangioblast. Access to these early populations throughRunx1 expression provides a unique opportunity to define the molecular events involved in the establishment of the primitive and definitive hematopoietic programs.
We thank members of the Keller laboratory for critically reading the manuscript and Anne Trumble for expert technical assistance.
Prepublished online as Blood First Edition Paper, April 17, 2002; DOI 10.1182/blood-2001-12-0321.
Supported by National Institutes of Health grants R01 HL48834 and R01 HL65169.
G.L. and L.G. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Gordon Keller, Carl C. Icahn Institute for Gene Therapy and Molecular Medicine, Mount Sinai School of Medicine, New York, NY; e-mail: gordon.keller@mssm.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal