Primitive chronic myeloid leukemia cells display a unique autocrine interleukin 3 (IL-3)/granulocyte–colony-stimluating factor (G-CSF) mechanism that may explain their abnormal proliferation and differentiation control. Here we show that BCR-ABL transduction of primitive Sca-1+ lin− mouse bone marrow (BM) cells causes immediate activation of IL-3, G-CSF, and granulocyte macrophage–colony-stimulating factor (GM-CSF) expression in these cells. Their autocrine IL-3–mediated growth dependence is thus demonstrable only in clonal cultures where paracrine effects are reduced. Interestingly, upon continued culture, these cells produce large populations of rapidly proliferating mast cells in which only the IL-3 autocrine mechanism is consistently maintained, together with evidence of hyperphosphorylation of p210BCR-ABL and STAT5 and retention of a multilineage but attenuated in vivo leukemogenic potential characterized by a prolonged latency. BCR-ABL transduction of IL-3−/− Sca-1+ lin− BM cells initially activates GM-CSF and G-CSF production, factor independence, and the ability to generate phenotypically indistinguishable populations of mast cells. However, maintenance of factor independence, and p210BCR-ABL and STAT 5 activation beyond 4 to 6 weeks, requires rescue with an IL-3 transgene. The cultured BCR-ABL–transduced IL-3−/− cells also lack leukemogenic activity in vivo. These findings provide new evidence that IL-3 production is a rapid, sustained, and biologically relevant consequence of BCR-ABL expression in primitive hematopoietic cells with multilineage leukemogenic activity.
Introduction
Autocrine mechanisms have been well documented in many types of hematologic neoplasms, but whether or how they contribute to the initial development or subsequent maintenance of the malignant stem cell compartment is not known. Expression of BCR-ABL, the oncogene responsible for human chronic myeloid leukemia (CML; reviewed in Sawyers1 and Holyoake2) confers autonomous growth properties on a variety of growth factor–dependent hematopoietic cell lines.3-6 In several cases, acquisition of this growth factor independence has been shown to be accompanied by, and at least partially dependent on, activation of interleukin 3 (IL-3) production3,5,6 and activation of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway (reviewed in Sattler and Salgia7). Recently we showed that an autocrine IL-3/granulocyte–colony-stimulating factor (G-CSF) mechanism is activated in the primitive (CD34+) neoplastic cells from patients with CML and the associated activation of STAT5 in these cells is specifically dependent on their autocrine IL-3 response.8 An unusual feature of this autocrine mechanism is that it declines when the leukemic cells reach the terminal stages of differentiation. This explains the well-known growth factor dependence that human CML progenitors display when plated in semisolid assays to detect their ability to form colonies of mature cells,9 which contrasts with the extensive factor-independent proliferative activity seen when primitive CML cells are incubated in liquid suspension cultures,10 because in liquid suspension cultures, the growth factor requirement at the later stages of differentiation can be met by paracrine mechanisms.8
IL-3 is also produced by the rapidly fatal leukemic cell populations generated in mice injected with BCR-ABL–transduced mouse bone marrow (BM) cells11,12 and by transformed hematopoietic cell lines derived from BCR-ABL–transduced embryonic stem cells.13Interestingly, it was recently shown that BCR-ABL–transduced IL-3−/− or granulocyte macrophage–colony-stimulating factor (GM-CSF)−/− BM cells can, like their +/+ counterparts, produce rapidly lethal leukemias in vivo when assessed immediately after transduction.14 These findings have raised questions about the potential role of autocrine mechanisms in the process by which primitive BCR-ABL+ hematopoietic cells develop a transformed phenotype. However, the much slower and indolent course of the chronic phase of human CML suggests that the more aggressive disease models used to date to address the question of the role of autocrine mechanisms in this disease may have precluded detection of subtle but clinically relevant effects.
To address this issue, we have re-examined the role of IL-3 in the BCR-ABL–mediated transformation of very primitive hematopoietic cells in a model in which the generation of disease in vivo is more attenuated. The results of these studies show that BCR-ABL rapidly activates a stable but initially redundant autocrine IL-3 mechanism in primitive hematopoietic cells. However, after several weeks in culture, both autonomous growth in vitro and leukemogenic activity in vivo are lost by BCR-ABL+–transformed IL-3−/− cells.
Materials and methods
Retroviral vectors and virus production
An MSCV-BCR-ABL-IRES-GFP (BCR-ABL) retroviral vector (Figure1) was constructed by ligating the 7.3-kilobase (kb) EcoR1 fragment containing the full-length BCR-ABL cDNA (from J. Griffin, Dana Farber Cancer Institute, Boston, MA) to the upstream internal ribosomal entry site (IRES) of the MSCV-IRES-GFP (MIG) vector (modified by K. Humphries, Terry Fox Laboratory, Vancouver, BC, Canada, from a vector originally developed and provided by R. Hawley, American Red Cross, Derwood, MD). The murine IL-3 cDNA was released by EcoR1 and Sal1 digestion from a PAX142 vector (from R. Kay, Terry Fox Laboratory) and cloned into the EcoR1 and Xho1 sites of a MSCV-IRES-YFP (MIY) vector (also from K. Humphries). Constructs were verified by restriction enzyme digestion analysis.
Retroviral vectors.
The EcoR1 and Xba1 restriction sites used for determining proviral integration sites and the full-length provirus are indicated. LTR, long terminal repeat; IRES, internal ribosome entry site; X, Xba1; E, EcoR1; N, Nco1; S,Sal1; B, BamH1; H, Hind111.
Retroviral vectors.
The EcoR1 and Xba1 restriction sites used for determining proviral integration sites and the full-length provirus are indicated. LTR, long terminal repeat; IRES, internal ribosome entry site; X, Xba1; E, EcoR1; N, Nco1; S,Sal1; B, BamH1; H, Hind111.
Plasmid DNA was isolated by Qiagen plasmid purification (Qiagen, Hilden, Germany) and ecotropic Phoenix packaging cells15 then transfected with 10 to 30 μg retroviral plasmid DNA using calcium phosphate.16 The medium was changed 24 hours later and virus-containing supernatants (VCS) harvested after another 24 hours. Supernatants were passed through a 0.45 μ filter, aliquoted, and frozen at −80°C. Viral titers (2-5 × 105/mL for the BCR-ABL virus and 5-10 × 105/mL for the IL-3 and MIG virus) were determined by assessment of GFP+ or YFP+ BaF3 cells 2 days after a single infection.
Infection of mouse BM cells
BM cells from 6- to 10-week-old C57BL/6 and congenic IL-3−/− mice (breeding pairs obtained from G. Dranoff, Dana Farber Cancer Institute) were harvested 4 days after injection of 150 mg/kg 5-fluorouracil (5-FU). The cells were then washed with phosphate-buffered saline (PBS) containing 2% fetal calf serum (FCS) and low-density cells isolated by centrifugation on a 1.077 g/mL Ficoll-Hypaque solution.17 Low-density cells were then cultured for 48 hours at 3-5 × 105/mL in serum-free medium (SFM), that is, Iscove medium supplemented with BSA plus insulin plus transferrin (BIT; StemCell Technologies, Vancouver, BC, Canada) and 40 μg/mL low-density lipoproteins (Sigma Chemical, St Louis, MO), to which 100 ng/mL murine steel factor (SF, purified from cDNA-transfected COS cell supernatants) and 10 ng/mL human IL-6 (Cangene, Mississauga, ON, Canada) was also added. The cells were then collected and resuspended at 5 × 105 cells/mL in VCS diluted 1:3 in Dulbecco modified Eagle medium (DMEM) containing 15% FCS, protamine sulfate (5 μg/mL) and SF and IL-6 at the same concentrations as noted previously. The cells were then transferred to petri dishes that had been precoated with 3 μg/cm2fibronectin (Sigma Chemical) and preloaded with virus for 30 minutes at room temperature.18 This transduction was repeated once 4 hours later. The cells were then incubated at 37°C for a further 24 hours before being injected into mice, or for a further 48 hours prior to cell purification by fluorescence-activated cell sorting (FACS; described below) and additional studies in vitro.
Cell sorting and flow cytometry
After transduction, the cells were washed and suspended in Hanks solution with 2% FCS (HF) at 5-10 × 106 cells/mL and then incubated first with 6 μg/mL 2.4G2 (anti–Fc receptor antibody19), followed by a mixture of biotinylated lineage-specific rat monoclonal antibodies (anti-B220 [RA3-6B2], anti–Gr-1 [RB6-8C5], anti–Ly-1 [53-7.3], Ter119 [anti–Ly-76]), and finally with phycoerythrin (PE)–labeled anti–Sca-1 (anti–Ly-6A/E; E13-161.7), each time for 30 minutes at 4°C as described.20 Anti–Mac-1 was omitted from this cocktail of antilineage (lin) markers because expression of this antigen is up-regulated on activated stem cells.21 The antibody-labeled cells were then washed once, incubated with allophycocyanin (APC)–labeled streptavidin (SA-APC) for 30 minutes at 4°C, washed twice again, and finally resuspended in 1 μg/mL propidium iodide (PI; Sigma Chemical). A FACStar Plus (Becton Dickinson Immunocytometry Systems, San Jose, CA) was used to isolate populations of more than 99% pure, viable GFP+ Sca-1+lin− or GFP+ lin− or YFP+ lin− or GFP+YFP+lin− cells. Bulk cells were collected into eppendorf tubes containing DMEM with 10% FCS or PBS. Single cells were sorted directly into the wells of 96-well microtiter plates preloaded with 100 μL of DMEM plus 10% FCS using the FACS single-cell deposition unit. Subsequent surface expression on cultured cells of Gr-1, Mac-1, B220, CD4, CD8, Ter119, Sca-1, c-kit, IgE, Ly5.1, and Ly5-2 was evaluated using directly conjugated PE- or APC-conjugated monoclonal antibodies against these antigens after staining as described above. Data acquisition using the FACScan and FACScalibur and analysis was performed with Cell Quest software (Becton Dickinson). For cell cycle analyses, cells were washed with PBS containing 0.1% glucose, and the cells then fixed with 70% ethanol for 1 hour at 4°C. After washing the cells in 2% HF and incubation in a PI staining solution (PBS with 0.1% glucose, 200 μg/mL RNase A and 10 μg/mL PI) at 4°C for 1 hour, FACS profiles were collected on 2 × 104 cells per sample.
Cell culture
Sorted cells were cultured at 1-5 × 105cells/mL in DMEM plus 10% FCS with or without 100 ng/mL SF and 10 ng/mL IL-6 at 37°C. Viable cell numbers were determined from hematocytometer counts of trypan blue–excluding cells. Clone formation (presence of ≥ 2 refractile cells) and clone size (total number of refractile cells) in wells seeded with single cells (using the FACS cloning device) were assessed by direct visual inspection using an inverted microscope.
Progenitor assays
Colony-forming cells (CFCs) and long-term culture-initiating cells (LTC-ICs) were assayed using culture media from StemCell Technologies. No IL-3 was added to the CFC methylcellulose assay medium (which contained FCS plus 10 ng/mL human IL-6, 50 ng/mL murine SF and 3 U/mL human erythropoietin [EPO; StemCell Technologies] unless otherwise indicated) to ensure equal stimulation of IL-3−/− and +/+ CFCs. Colony counts were performed after 12 to 14 days of incubation at 37°C using standard scoring criteria.20 LTCs were set up using pre-established irradiated mouse marrow LTC feeder layers and weekly half medium changes of LTC medium containing fresh hydrocortisone as previously described.20 In some CFC assays the following antibodies were added alone or in combination: an anti–IL-3 antibody (SH15 purAB, from H. Ziltener, Biomedical Research Centre, University of British Columbia, Vancouver, BC, Canada), an anti–GM-CSF antibody (RAB 143, also from H. Ziltener), an anti–G-CSF antibody (R&D Systems, Minneapolis, MN), or control sheep or rabbit IgG (from H. Ziltener).
Expanded populations of BCR-ABL–transduced +/+ and IL-3−/− cells were generated from single colonies generated in methylcellulose cultures or clones initially generated in liquid cultures of single Sca-1+ lin−GFP+ cells deposited into 96-well plates using the FACS. Initial clones were then expanded in DMEM containing 10% FCS, 100 ng/mL SF and 20 ng/mL IL-6, or 5% pokeweed mitogen (PWM)–stimulated murine spleen cell conditioned medium (StemCell Technologies). Morphologic assessment of cells was performed on Wright-Giemsa–stained cytospin preparations.
Reverse transcriptase–polymerase chain reaction analyses
A semiquantitative procedure to assess cytokine mRNA expression was performed as previously described.8,22 23 Briefly, total RNA was extracted from aliquots of 1-2 × 104cells and reverse transcriptase–polymerase chain reaction (RT-PCR) was performed using an oligo (dT)–based primer and poly (A) tailing procedure. For specific RT-PCR amplification, the RT reaction was performed in 20 μL with superscript II reverse transcriptase (Gibco/BRL, Rockville, MD) using random hexamer oligonucleotides in 50 μL 1X PCR buffer (Amersham Pharmacia) containing 20 mM Tris-HCL (pH 8.4), 50 mM KCl, 2mM MgCl2, 200 μM of each dNTP (Gibco/BRL), 1 U Taq polymerase, and 10 pM of specific primers for BCR-ABL (5′-CAGGGTGCACACCGCAACGGCAA-3′ and 5′-GTCCAGCGAGAAGGTTTTCCTTGGA-3′), mouse IL-3 (5′-CTCCAAGCTTCAATCAGTGGCC-3′ and 5′-CGGTTCCACGGTTAGGAGAGAC-3′), mouse GM-CSF (5′-CTGCAGAATTTACTTTTCCTGGGC-3′ and 5′-GCATTCAAAGGGGATATCAGTCAG-3′), mouse G-CSF (5′-CAAGTGAGGAAGATCCAGGCCA-3′ and 5′-AGCCCTGCAGGTACGAAATGGC-3′), glyceraldehyde phosphate dehydrogenase (GAPDH) (5′-GTCTTCACCACCATGGAGAAGG-3′ and 5′-GCCTGCTTCACCACCTTCTTGA-3′) to give DNA fragments of 374 base pair (bp) (BCR-ABL), 455 bp (IL-3), 309 bp (GM-CSF), 409 bp (G-CSF), and 493 bp (GAPDH). We then performed 35 cycles of amplification (94°C for 30 seconds, 62°C for 60 seconds, 72°C for 60 seconds). Aliquots (10 μL) of the total amplified cDNA or specific amplified PCR products were then electrophoresed in a 1% agarose gel stained with ethidium bromide for quantification using a Kodak Digital Science ID scanner and image analysis software (Eastman Kodak Company, Rochester, NY). The total amplified cDNA was then transferred onto a nylon membrane (Zeta-probe; BioRad) and hybridized. cDNA probes for murine IL-3, murine GM-CSF, and murine G-CSF were provided by R. Kay and K. Humphries. Blots were exposed to Kodak XAR film (Eastman-Kodak) at −70°C for periods of 3 to 24 hours.
Southern blotting
High-molecular-weight DNA was isolated using DNAZol (Gibco/BRL). DNA (15 μg) was digested with EcoR1, electrophoresed, and transferred to a nylon membrane.8 The blot was then hybridized with a GFP probe derived from the retroviral vector, and a BCR-ABL cDNA used to confirm the presence of an intactBCR-ABL gene.
Western blotting
Cells (105) were lysed in phosphorylation solubilization buffer (PSB; 50 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] buffer, 100 mM NaF, 10 nM sodium pyrophosphate, 2 mM Na3VO4, 4 mM ethylenediaminetetraacetic acid [EDTA]) containing 0.5% Nonidet P-4010 and Western analyses performed using a monoclonal anti–ABL antibody (8E9; Pharmingen, Mississauga, ON, Canada), an anti–phosphotyrosine antibody (4G10; Upstate Biotechnology, Lake Placid, NY) and rabbit polyclonal anti–phospho-STAT5A/B and anti–STAT5A antibodies (Upstate Biotechnology).
Animals
C57BL/6 mice with a homozygous null mutation in theIL-3 gene24 (IL-3−/−mice, provided by G. Dranoff, Dana Farber Cancer Institute), C57BL/6+/+ mice, nonobese diabetic-scid(NOD/SCID) mice25 and NOD/SCID-β2microglobulin (β2m)−/− mice26 were all bred and maintained in the animal facility of the British Columbia Cancer Research Centre (Vancouver, BC, Canada) in microisolator cages and were provided with autoclaved food and water. NOD/SCID and NOD/SCID-β2m−/− mice were irradiated at 8 to 12 weeks of age with 350 cGy 137Cs γ-rays and thereafter received acidified water containing 100 mg/mL Ciprofloxacin (Bayer AG, Leverkusen, Germany). Cells were injected into mice intravenously a few hours after they had been irradiated. Mice were monitored daily for signs of weight loss or lethargy. Peripheral blood was collected every 1 to 2 weeks and leukocytes analyzed by FACS as described above after lysis of the red cells with ammonium chloride.
Results
BCR-ABL rapidly activates an autocrine mechanism mediated by IL-3, GM-CSF, and G-CSF in primitive adult mouse BM cells
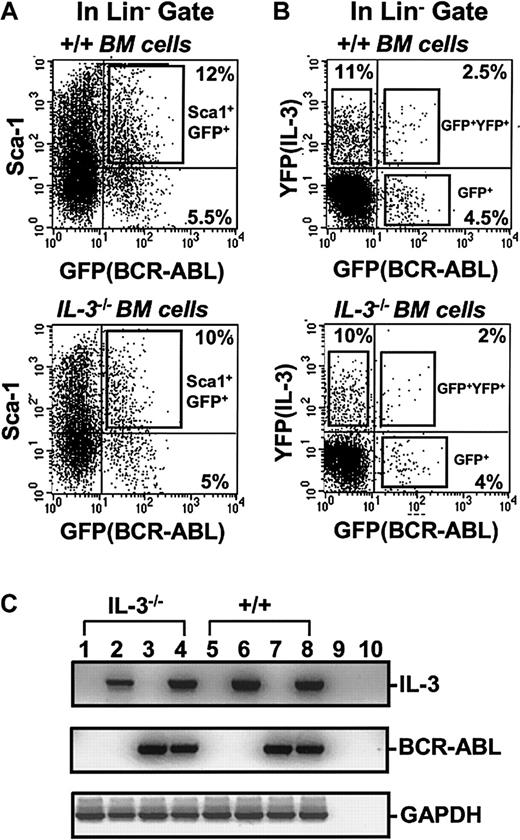
To obtain a highly stem cell–enriched population of BCR-ABL (or control) virus-transduced cells, BM cells from 5-FU–treated +/+ mice were first exposed to a BCR-ABL or control MIG virus (Figure 1). The transduced (GFP+) fraction of Sca-1+lin− cells (11% ± 4% of the total cells present immediately after transduction, n = 6) was then isolated by FACS (Figure 2A). Already by this time, that is, 4 days after the first exposure of the cells to BCR-ABL virus (labeled as day 0 in the representative experiment shown in Figure3), BCR-ABL transcripts were at maximum levels, as shown by semiquantitative gene expression analysis. Continued culture of both BCR-ABL–transduced and control (MIG-transduced) cells in the presence of growth factors supported their expansion at an indistinguishable rate (data not shown) and sequential gene expression analyses of the BCR-ABL–transduced cells demonstrated that IL-3 and GM-CSF transcripts consistently appeared in them 3 days later (Figure 3). G-CSF transcripts were also detected in them after another 3 days. No transcripts for any of these growth factors were detected at any time in MIG-transduced cells. The activated expression of IL-3, GM-CSF, and G-CSF in the BCR-ABL–transduced cells initiated a multifactorial autocrine growth mechanism. This was demonstrated by the inhibitory effects of adding neutralizing IL-3, GM-CSF, or G-CSF antibodies to CFC assays of the initially BCR-ABL–transduced (GFP+) Sca-1+lin− cells. As shown in Table1, addition of antibody to a single growth factor gave a 15% to 35% reduction in colony formation and, when antibodies to all 3 growth factors were combined, up to 50% inhibition of factor-independent colony formation was seen.
FACS profiles of transduced +/+ and IL-3−/− BM cells.
(A) Dot plot showing gates used to isolate the Sca-1+lin− subset of GFP+ cells after exposure of +/+ and IL-3−/− BM cells to BCR-ABL-GFP. (B) Dot plot showing gates used to isolate the GFP+(BCR-ABL+, lower right), YFP+(IL-3+, upper left), and GFP+ and YFP+ (BCR-ABL+andIL-3+, upper right) cells within the lin−subset after simultaneous exposure of +/+ and IL-3−/− BM cells to BCR-ABL-GFP and IL-3-YFP virus. (C) RT-PCR analysis of RNA isolated from FACS purified GFP+, YFP+, and GFP+ and YFP+ fractions of cells shown in Figure 1B. Lanes 1 and 5 show results for MIG-transduced GFP+ IL-3−/− and +/+ cells; lanes 2 and 6 for cells cotransduced with BCR-ABL and IL-3 and purified for YFP (IL-3) expression only; lanes 3 and 7 for cells cotransduced with BCR-ABL and IL-3 and purified for GFP (BCR-ABL) expression only; and lanes 4 and 8 for cells cotransduced with BCR-ABL and IL-3 and purified for dual GFP and YFP (BCR-ABL and IL-3) expression. Lanes 9 and 10 were water controls (negative controls) for the RT and PCR reactions.
FACS profiles of transduced +/+ and IL-3−/− BM cells.
(A) Dot plot showing gates used to isolate the Sca-1+lin− subset of GFP+ cells after exposure of +/+ and IL-3−/− BM cells to BCR-ABL-GFP. (B) Dot plot showing gates used to isolate the GFP+(BCR-ABL+, lower right), YFP+(IL-3+, upper left), and GFP+ and YFP+ (BCR-ABL+andIL-3+, upper right) cells within the lin−subset after simultaneous exposure of +/+ and IL-3−/− BM cells to BCR-ABL-GFP and IL-3-YFP virus. (C) RT-PCR analysis of RNA isolated from FACS purified GFP+, YFP+, and GFP+ and YFP+ fractions of cells shown in Figure 1B. Lanes 1 and 5 show results for MIG-transduced GFP+ IL-3−/− and +/+ cells; lanes 2 and 6 for cells cotransduced with BCR-ABL and IL-3 and purified for YFP (IL-3) expression only; lanes 3 and 7 for cells cotransduced with BCR-ABL and IL-3 and purified for GFP (BCR-ABL) expression only; and lanes 4 and 8 for cells cotransduced with BCR-ABL and IL-3 and purified for dual GFP and YFP (BCR-ABL and IL-3) expression. Lanes 9 and 10 were water controls (negative controls) for the RT and PCR reactions.
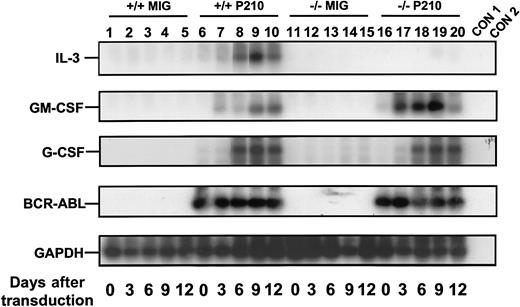
Rapid activation of growth factor gene expression in primitive BCR-ABL–transduced +/+ and IL-3−/− BM cells.
Sca-1+ lin− BCR-ABL–transduced cells were sorted immediately after transduction and the cells analyzed after varying periods of culture in the presence of SF and IL-6. A semiquantitative RT-PCR was performed followed by Southern blot analysis using gene-specific cDNA probes. Each filter was then stripped and reprobed with a GAPDH probe. Lanes 1 to 5 and 6 to 10 are for MIG- and BCR-ABL–transduced +/+ cells, and lanes 11 to 15 and 16 to 20 are for the MIG- and BCR-ABL–transduced IL-3−/− cells. Controls (CON) 1 and 2 are negative controls with no RT and water only, respectively. Results shown here are from a representative experiment.
Rapid activation of growth factor gene expression in primitive BCR-ABL–transduced +/+ and IL-3−/− BM cells.
Sca-1+ lin− BCR-ABL–transduced cells were sorted immediately after transduction and the cells analyzed after varying periods of culture in the presence of SF and IL-6. A semiquantitative RT-PCR was performed followed by Southern blot analysis using gene-specific cDNA probes. Each filter was then stripped and reprobed with a GAPDH probe. Lanes 1 to 5 and 6 to 10 are for MIG- and BCR-ABL–transduced +/+ cells, and lanes 11 to 15 and 16 to 20 are for the MIG- and BCR-ABL–transduced IL-3−/− cells. Controls (CON) 1 and 2 are negative controls with no RT and water only, respectively. Results shown here are from a representative experiment.
Partial inhibition of colony formation by BCR-ABL-transduced +/+ CFCs and LTC-IC-derived CFCs in the presence of antibodies to IL-3, GM-CSF, and G-CSF
| Antibodies added to CFC assays . | Origin of CFCs . | No. of colonies* . | % inhibition† . |
|---|---|---|---|
| Control‡ | Sca-1+lin−GFP+cells | 150 ± 30 | — |
| Anti–IL-3 | 97 ± 22 | 35 ± 5 | |
| Anti–GM-CSF | 114 ± 26 | 25 ± 4 | |
| Anti–G-CSF | 120 ± 26 | 20 ± 3 | |
| Anti–IL-3 + anti–GM-CSF | 81 ± 14 | 46 ± 3 | |
| Anti–IL-3 + anti–GM-CSF + anti–G-CSF | 80 ± 20 | 48 ± 4 | |
| Control‡ | LTC-IC | 2000 ± 200 | — |
| Anti–IL-3 | 1290 ± 200 | 36 ± 4 | |
| Anti–GM-CSF | 1350 ± 180 | 31 ± 3 | |
| Anti–G-CSF | 1688 ± 142 | 16 ± 2 | |
| Anti–IL-3 + anti–GM-CSF | 1000 ± 50 | 49 ± 3 | |
| Anti–IL-3 + anti–GM-CSF + anti–G-CSF | 994 ± 90 | 50 ± 4 |
| Antibodies added to CFC assays . | Origin of CFCs . | No. of colonies* . | % inhibition† . |
|---|---|---|---|
| Control‡ | Sca-1+lin−GFP+cells | 150 ± 30 | — |
| Anti–IL-3 | 97 ± 22 | 35 ± 5 | |
| Anti–GM-CSF | 114 ± 26 | 25 ± 4 | |
| Anti–G-CSF | 120 ± 26 | 20 ± 3 | |
| Anti–IL-3 + anti–GM-CSF | 81 ± 14 | 46 ± 3 | |
| Anti–IL-3 + anti–GM-CSF + anti–G-CSF | 80 ± 20 | 48 ± 4 | |
| Control‡ | LTC-IC | 2000 ± 200 | — |
| Anti–IL-3 | 1290 ± 200 | 36 ± 4 | |
| Anti–GM-CSF | 1350 ± 180 | 31 ± 3 | |
| Anti–G-CSF | 1688 ± 142 | 16 ± 2 | |
| Anti–IL-3 + anti–GM-CSF | 1000 ± 50 | 49 ± 3 | |
| Anti–IL-3 + anti–GM-CSF + anti–G-CSF | 994 ± 90 | 50 ± 4 |
From assays of 103 Sca-1+lin− GFP+ cells assayed directly or after their maintenance in LTCs for 4 weeks. CFC assays were performed in FCS-containing methylcellulose medium without added growth factors. Antibodies were added as shown at 10 μg/mL.
Calculated as a percent of the value obtained in the control assays in the same experiment.
Control antibodies were sheep or rabbit IgG as appropriate.
Data shown are the mean ± SEM of values obtained from 3 independent experiments.
CFC indicates colony-forming cell; LTC-IC, long-term culture-initiating cell; IL-3, interleukin 3; GM-CSF, granulocyte macrophage–colony-stimulating factor; G-CSF, granulocyte–colony-stimulating factor; FCS, fetal calf serum.
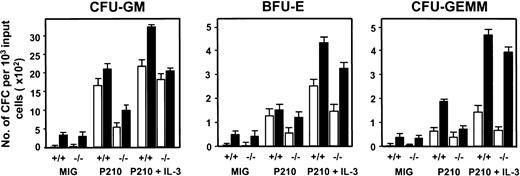
To assess effects on a more primitive, functionally defined subset of the initially transduced cells, LTC-IC assays were also performed on freshly isolated GFP+ Sca-1+ lin−cells. The 4-week output of all types of CFCs from BCR-ABL–transduced cells in these LTC-IC assays was increased (∼6-fold, Figure4) relative to results for MIG-transduced control cells. Moreover, more than 70% of the BCR-ABL–transduced LTC-IC–derived CFCs were factor-independent and antibody studies again showed that this was due at least partially to the continued activity of an autocrine mechanism involving all 3 growth factors (Table 1).
Reduced numbers of total and factor-independent LTC-IC–derived CFCs from BCR-ABL–transduced IL-3−/− vs +/+ lin−GFP+ cells.
The numbers of LTC-IC–derived CFU-GMs (left panel), erythroid burst-forming units (BFU-Es; middle panel), and CFU-granulocyte/erythroid/macrophage/megakaryocytes (GEMMs) (right panel) able to make colonies in the presence of SF, IL-6, and EPO (▪) or without any added growth factors (□) are shown for the various combinations of target cells and viruses indicated. Values shown are the means ± SEM from 3 experiments each with MIG-, BCR-ABL– and BCR-ABL plus IL-3–transduced +/+ and IL-3−/− cells.
Reduced numbers of total and factor-independent LTC-IC–derived CFCs from BCR-ABL–transduced IL-3−/− vs +/+ lin−GFP+ cells.
The numbers of LTC-IC–derived CFU-GMs (left panel), erythroid burst-forming units (BFU-Es; middle panel), and CFU-granulocyte/erythroid/macrophage/megakaryocytes (GEMMs) (right panel) able to make colonies in the presence of SF, IL-6, and EPO (▪) or without any added growth factors (□) are shown for the various combinations of target cells and viruses indicated. Values shown are the means ± SEM from 3 experiments each with MIG-, BCR-ABL– and BCR-ABL plus IL-3–transduced +/+ and IL-3−/− cells.
Reduced growth autonomy of BCR-ABL–transduced IL-3−/− BM cells
A similar series of experiments was then carried out with BM cells from congenic IL-3−/− mice. As expected, the Sca-1+ lin− BCR-ABL–transduced IL-3−/− cells produced no IL-3 transcripts. However, both GM-CSF and G-CSF transcripts appeared with the same rapid kinetics as seen in the +/+ cells (Figure 3). Consistent with this finding was the observation of similar growth rates of the BCR-ABL–transduced IL-3−/− and +/+ cells during the first 2 weeks both in liquid suspension culture and in semisolid medium either with or without added growth factors (data not shown). However, as shown in Figure 4, the output of all types of CFCs generated in 4-week LTC-IC assays of BCR-ABL–transduced IL-3−/− cells was consistently lower than in the corresponding +/+ assays (∼2-fold less, P < .002, although still increased above MIG-transduced control values, P < .05). In addition, the BCR-ABL–transduced IL-3−/− LTC-ICs generated very few factor-independent CFCs. To determine whether a unique role of BCR-ABL–activated autocrine IL-3 production might be demonstrable sooner, we assessed the growth potential of single BCR-ABL–transduced Sca-1+ lin− GFP+ +/+ and IL-3−/− cells in serum-free liquid medium without added growth factors. Under these very stringent conditions, which preclude any initial paracrine effects, individual BCR-ABL–transduced IL-3−/− cells showed a significantly reduced ability to proliferate (P < .05, Figure5).
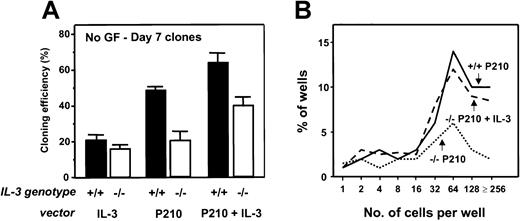
Rescue of reduced factor-independent growth of single BCR-ABL-transduced IL-3−/−BM cells by restoration of IL-3 production.
(A) Percentage of transduced cells generating clones of more than or equal to 2 refractile cells after 7 days in the absence of added growth factors. Values shown are the means ± SEM from 3 experiments with each genotype of BM cells. (B) Clone size distributions obtained from single BCR-ABL ± IL-3–transduced IL-3−/− cells by comparison to BCR-ABL–transduced +/+ cells after being cultured for 7 days in the absence of growth factors. Data for a representative experiment of 3 performed are shown.
Rescue of reduced factor-independent growth of single BCR-ABL-transduced IL-3−/−BM cells by restoration of IL-3 production.
(A) Percentage of transduced cells generating clones of more than or equal to 2 refractile cells after 7 days in the absence of added growth factors. Values shown are the means ± SEM from 3 experiments with each genotype of BM cells. (B) Clone size distributions obtained from single BCR-ABL ± IL-3–transduced IL-3−/− cells by comparison to BCR-ABL–transduced +/+ cells after being cultured for 7 days in the absence of growth factors. Data for a representative experiment of 3 performed are shown.
To further examine the role of IL-3 in contributing to this reduced growth factor autonomy, we evaluated the behavior of primitive BCR-ABL–transduced IL-3−/− BM cells that were coinfected with an MSCV murine IL-3-IRES-YFP virus (Figure 1). The specificity of the FACS gates used to select cells that had been successfully cotransduced with both BCR-ABL+ (GFP+) and IL-3+ (YFP+) cDNAs (illustrated in Figure 2B) was established by RT-PCR analysis of the various populations isolated (Figure 2C). As shown in Table 2, restored expression of IL-3 in IL-3−/− cells enabled them to produce as many factor-independent CFCs in 4-week LTC-IC assays as were produced by IL-3–transduced +/+ cells. Moreover, when the IL-3−/− cells were transduced with both IL-3 and BCR-ABL, their output of both total and factor-independent CFCs in 4-week LTC-IC assays became equivalent to that of BCR-ABL–transduced +/+ cells (Figure 4 and Table 2).
Reduced factor-independent colony formation by BCR-ABL-transduced IL-3−/− LTC-IC-derived CFCs is restored when the cells are cotransduced with an IL-3 cDNA
| Cells . | Virus . | No. of LTC-IC–derived CFCs detected (per 103 input cells) . | |||
|---|---|---|---|---|---|
| BFU-E . | CFU-GEMM . | CFU-GM . | Total . | ||
| −/− | IL-3 | 75 ± 15 | 37 ± 12 | 690 ± 30 | 800 ± 60 |
| +/+ | IL-3 | 90 ± 12 | 45 ± 30 | 800 ± 57 | 830 ± 70 |
| −/− | BCR-ABL | 60 ± 12 | 40 ± 15 | 650 ± 60 | 760 ± 60 |
| +/+ | BCR-ABL | 120 ± 20 | 70 ± 30 | 1750 ± 150 | 1940 ± 200 |
| −/− | IL-3 + BCR-ABL | 140 ± 20 | 70 ± 30 | 1830 ± 150 | 2040 ± 200 |
| +/+ | IL-3 + BCR-ABL | 250 ± 30 | 140 ± 20 | 2235 ± 115 | 2625 ± 110 |
| Cells . | Virus . | No. of LTC-IC–derived CFCs detected (per 103 input cells) . | |||
|---|---|---|---|---|---|
| BFU-E . | CFU-GEMM . | CFU-GM . | Total . | ||
| −/− | IL-3 | 75 ± 15 | 37 ± 12 | 690 ± 30 | 800 ± 60 |
| +/+ | IL-3 | 90 ± 12 | 45 ± 30 | 800 ± 57 | 830 ± 70 |
| −/− | BCR-ABL | 60 ± 12 | 40 ± 15 | 650 ± 60 | 760 ± 60 |
| +/+ | BCR-ABL | 120 ± 20 | 70 ± 30 | 1750 ± 150 | 1940 ± 200 |
| −/− | IL-3 + BCR-ABL | 140 ± 20 | 70 ± 30 | 1830 ± 150 | 2040 ± 200 |
| +/+ | IL-3 + BCR-ABL | 250 ± 30 | 140 ± 20 | 2235 ± 115 | 2625 ± 110 |
LTC-IC assays were initiated with 103lin− cells from each population. The LTCs were harvested after 4 weeks and the cells in them were then assayed for CFCs in FCS-containing methylcellulose medium in the absence of added growth factors. Data shown are the mean ± SEM of values obtained from 3 independent experiments. LTC-IC indicates long-term culture-initiating cell; CFC, colony-forming cell; IL-3, interleukin 3; BFU-E, erythroid-burst-forming unit; CFU-GEMM, colony-forming unit–granulocyte/erythroid/macrophage/megakaryocyte; CFU-GM, granulocyte macrophage-colony-forming unit; FCS, fetal calf serum.
Similar results were obtained when the effects of forced expression of IL-3 were examined in single cell cultures of Sca-1+lin− +/+ and IL-3−/− cells. Notably, coinfection of the IL-3−/− cells with BCR-ABL and IL-3 increased the frequency of factor-independent clones to levels similar to those obtained by BCR-ABL–transduced +/+ cells (Figure 5). Taken together, these results demonstrate the ability of restored IL-3 expression to immediately reverse the in vitro growth deficiencies exhibited by BCR-ABL–-transduced IL-3−/−cells.
Characterization of clonal isolates generated from primitive BCR-ABL–transduced +/+ and IL-3−/− mouse BM cells
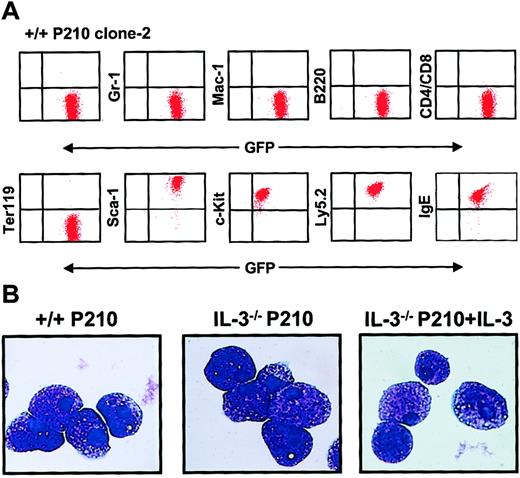
Continued culture of single BCR-ABL–transduced (GFP+) +/+ and IL-3−/− cells for several weeks in medium containing growth factors yielded rapidly growing populations of cells, more than 95% of which showed a Sca-1+ c-kit+lin− IgE receptor+ (IgER+) phenotype (Figure 6A) and mast cell morphology (Figure 6B). Southern blot analysis and hybridization with a GFP probe of EcoR1-digested extracts of DNA from these expanded clones showed that most contained a single copy of the retroviral cDNA with occasional examples of 2 integration sites (Figure7). Rehybridization with a BCR-ABL cDNA probe confirmed the presence of the full-length 7.3-kbBCR-ABL gene in each case.
Homogeneous mast cell phenotype of populations generated from single BCR-ABL–transduced +/+ and IL-3−/− cells.
(A) FACS profiles from a representative clonal population of expanded BCR-ABL–transduced +/+ cells are shown. All cells express Ly 5.2, GFP, Sca-1, and c-kit, and bind IgE, but do not express Gr-1, Mac-1, B220, CD4, CD8, or the antigen recognized by Ter119. Similarly derived expanded clones of BCR-ABL–transduced IL-3−/− cells showed an indistinguishable phenotype. (B) Wright-Giemsa–stained cytospin preparations from representative clones of BCR-ABL–transduced +/+ cells (left panel), BCR-ABL–transduced IL-3−/− cells (middle panel), and BCR-ABL– and IL-3–cotransduced IL-3−/− cells. All of the cells in all cultures examined exhibited this typical mature mast cell morphology.
Homogeneous mast cell phenotype of populations generated from single BCR-ABL–transduced +/+ and IL-3−/− cells.
(A) FACS profiles from a representative clonal population of expanded BCR-ABL–transduced +/+ cells are shown. All cells express Ly 5.2, GFP, Sca-1, and c-kit, and bind IgE, but do not express Gr-1, Mac-1, B220, CD4, CD8, or the antigen recognized by Ter119. Similarly derived expanded clones of BCR-ABL–transduced IL-3−/− cells showed an indistinguishable phenotype. (B) Wright-Giemsa–stained cytospin preparations from representative clones of BCR-ABL–transduced +/+ cells (left panel), BCR-ABL–transduced IL-3−/− cells (middle panel), and BCR-ABL– and IL-3–cotransduced IL-3−/− cells. All of the cells in all cultures examined exhibited this typical mature mast cell morphology.
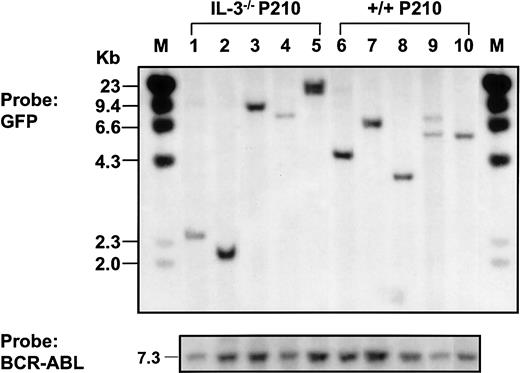
Southern blot analysis of populations derived from single BCR-ABL–transduced +/+ and IL-3−/− cells.
Genomic DNA was digested with EcoR1 and probed for GFP.EcoR1 cleaves once within the retroviral sequence (Figure1). Therefore, each fragment represents a unique site of retroviral integration. Lanes 1-5: 5 BCR-ABL–transduced IL-3−/−clones; lanes 6-10: 5 BCR-ABL–transduced +/+ clones. Molecular size markers are also indicated. The same filter was stripped and reprobed with a BCR-ABL cDNA probe showing the full-length BCR-ABL cDNA in each line isolated (at bottom).
Southern blot analysis of populations derived from single BCR-ABL–transduced +/+ and IL-3−/− cells.
Genomic DNA was digested with EcoR1 and probed for GFP.EcoR1 cleaves once within the retroviral sequence (Figure1). Therefore, each fragment represents a unique site of retroviral integration. Lanes 1-5: 5 BCR-ABL–transduced IL-3−/−clones; lanes 6-10: 5 BCR-ABL–transduced +/+ clones. Molecular size markers are also indicated. The same filter was stripped and reprobed with a BCR-ABL cDNA probe showing the full-length BCR-ABL cDNA in each line isolated (at bottom).
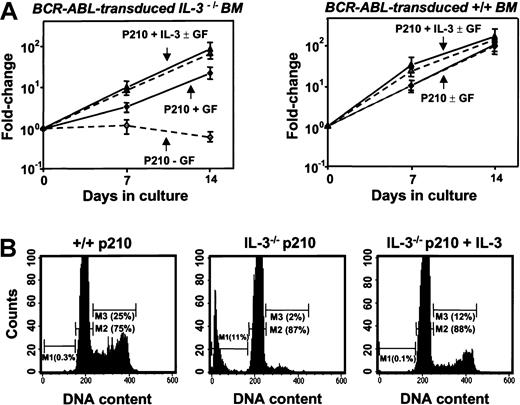
Even in the presence of SF and IL-6, the cells in all of the clones derived from BCR-ABL–transduced IL-3−/− cells (n = 6) grew more slowly (∼2-fold) than their BCR-ABL–transduced +/+ counterparts (n = 8). Moreover, in the absence of added growth factors, the numbers of BCR-ABL–transduced IL-3−/− cells could not be maintained (Figure 8A). In contrast, their +/+ counterparts continued to grow as well as when growth factors were present and the same was true for expanded clones of BCR-ABL–transduced IL-3−/− cells that had been cotransduced with IL-3. Assessment of the cell cycle distribution of the expanded clones 7 days after removal of exogenous growth factors showed that a relatively high proportion (25%) of the BCR-ABL–transduced +/+ cells remained in S phase. In contrast, the S-phase fraction of similarly treated BCR-ABL–transduced IL-3−/− cells was only 2% with another 11% of these cells showing the reduced DNA content of apoptotic cells (Figure 8B). Again, both of these parameters were normalized (by comparison to the BCR-ABL–transduced +/+ cells) in the clones expanded from BCR-ABL– and IL-3–cotransduced IL-3−/− cells.
Loss of growth factor independence by cultured BCR-ABL–transduced IL-3−/− cells.
(A) Growth of cells from expanded clones of BCR-ABL–transduced (diamonds), IL-3−/− (left panel) and +/+ BM cell cells (right panel) in the presence (solid symbols) and absence (open symbols) of exogenous factors (SF and IL-6) compared with data for clones derived from IL-3−/− or +/+ cells cotransduced with BCR-ABL and IL-3 (triangles). Viable cell numbers were determined by hematocytometer counts of trypan blue–excluding cells. Results are expressed as the mean fold-changes compared with input values (± SEM) from 3 independent experiments. (B) DNA profiles of PI-stained cells from representative expanded clones of BCR-ABL–transduced +/+ (left), IL-3−/− (middle), and BCR-ABL– and IL-3–cotransduced IL-3−/− cells (right) assessed after their maintenance for 7 days in the absence of growth factors. Percentage of apoptotic cells (M1) in the G0/G1 (M2) and S/G2/M (M3) phases of the cell cycle were determined using the Cell Quest software (Becton Dickinson).
Loss of growth factor independence by cultured BCR-ABL–transduced IL-3−/− cells.
(A) Growth of cells from expanded clones of BCR-ABL–transduced (diamonds), IL-3−/− (left panel) and +/+ BM cell cells (right panel) in the presence (solid symbols) and absence (open symbols) of exogenous factors (SF and IL-6) compared with data for clones derived from IL-3−/− or +/+ cells cotransduced with BCR-ABL and IL-3 (triangles). Viable cell numbers were determined by hematocytometer counts of trypan blue–excluding cells. Results are expressed as the mean fold-changes compared with input values (± SEM) from 3 independent experiments. (B) DNA profiles of PI-stained cells from representative expanded clones of BCR-ABL–transduced +/+ (left), IL-3−/− (middle), and BCR-ABL– and IL-3–cotransduced IL-3−/− cells (right) assessed after their maintenance for 7 days in the absence of growth factors. Percentage of apoptotic cells (M1) in the G0/G1 (M2) and S/G2/M (M3) phases of the cell cycle were determined using the Cell Quest software (Becton Dickinson).
Cultures expanded from single BCR-ABL–transduced IL-3−/− cells lack an autocrine mechanism and show reduced tyrosine phosphorylation of BCR-ABL and STAT5
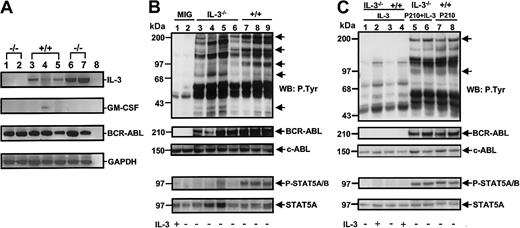
As a first step toward defining the molecular basis of the deficient growth exhibited by the expanded clones of BCR-ABL–transduced IL-3−/− cells, RT-PCR analyses were undertaken to examine their growth factor gene expression profile. All 8 of the expanded clones of BCR-ABL–transduced +/+ cells still contained IL-3 transcripts and 2 of these also still contained GM-CSF transcripts (see Figure 9A versus Figure3). However, none contained detectable levels of G-CSF mRNA (data not shown). Interestingly, no transcripts for any of these 3 growth factors were detected in any of the 6 expanded clones of BCR-ABL–transduced IL-3−/− BM cells, although their expression of BCR-ABL appeared undiminished (Figure 9A).
Loss of growth factor gene expression and reduced tyrosine phosphorylation of BCR-ABL and STAT5 in cultured BCR-ABL–transduced IL-3−/− cells.
(A) RT-PCR analysis of RNA isolated from representative clones generated from IL-3−/− and +/+ cells transduced with BCR-ABL only (lanes 1-5) or BCR-ABL and IL-3 (lanes 6 and 7). Lanes 1 and 2 show results for 2 BCR-ABL–transduced IL-3−/−clones; lanes 3-5 for 3 BCR-ABL–transduced +/+ clones; and lanes 6-7 for 2 clones of IL-3−/− cells cotransduced by IL-3 and BCR-ABL. Lane 8 was a negative control (water control). (B) Western blot analyses of cell lysates from control (MIG-transduced) cells with or without IL-3 (lanes 1 and 2); 4 clones of BCR-ABL–transduced IL-3−/− cells (lanes 3-6); and 3 clones of BCR-ABL–transduced +/+ cells (lanes 7-9). The cells were incubated in the presence or absence of IL-3 (as indicated below each lane) for 16 hours and lysates from equal numbers of cells separated on 8% polyacrylamide gradient gels. Filters were first probed with antiphosphotyrosine (4G10) and anti–phospho-STAT5 (P-STAT 5A/B) antibodies, then stripped and reprobed with anti–ABL antibodies and anti-STAT5 (STAT5A). The positions of prestained molecular weight markers are indicated on the left side of each blot. (C) Western blot analysis of IL-3–transduced IL-3−/− (lanes 1 and 2) and +/+ cells (lanes 3 and 4); BCR-ABL– and IL-3–transduced IL-3−/− cells (lanes 5 and 6); and BCR-ABL–transduced +/+ cells (lanes 7 and 8). Culture conditions and antibodies were the same as in panel B.
Loss of growth factor gene expression and reduced tyrosine phosphorylation of BCR-ABL and STAT5 in cultured BCR-ABL–transduced IL-3−/− cells.
(A) RT-PCR analysis of RNA isolated from representative clones generated from IL-3−/− and +/+ cells transduced with BCR-ABL only (lanes 1-5) or BCR-ABL and IL-3 (lanes 6 and 7). Lanes 1 and 2 show results for 2 BCR-ABL–transduced IL-3−/−clones; lanes 3-5 for 3 BCR-ABL–transduced +/+ clones; and lanes 6-7 for 2 clones of IL-3−/− cells cotransduced by IL-3 and BCR-ABL. Lane 8 was a negative control (water control). (B) Western blot analyses of cell lysates from control (MIG-transduced) cells with or without IL-3 (lanes 1 and 2); 4 clones of BCR-ABL–transduced IL-3−/− cells (lanes 3-6); and 3 clones of BCR-ABL–transduced +/+ cells (lanes 7-9). The cells were incubated in the presence or absence of IL-3 (as indicated below each lane) for 16 hours and lysates from equal numbers of cells separated on 8% polyacrylamide gradient gels. Filters were first probed with antiphosphotyrosine (4G10) and anti–phospho-STAT5 (P-STAT 5A/B) antibodies, then stripped and reprobed with anti–ABL antibodies and anti-STAT5 (STAT5A). The positions of prestained molecular weight markers are indicated on the left side of each blot. (C) Western blot analysis of IL-3–transduced IL-3−/− (lanes 1 and 2) and +/+ cells (lanes 3 and 4); BCR-ABL– and IL-3–transduced IL-3−/− cells (lanes 5 and 6); and BCR-ABL–transduced +/+ cells (lanes 7 and 8). Culture conditions and antibodies were the same as in panel B.
Western analyses were also performed on extracts of the BCR-ABL–transduced +/+ and IL-3−/− clones. As illustrated in Figure 9B, these showed bands of the size expected for p210BCR-ABL, STAT5, and another, as-yet-unidentified 150-kd protein to be consistently less tyrosine phosphorylated in the BCR-ABL–transduced IL-3−/− cells (n = 4) by comparison to the BCR-ABL–transduced +/+ cells (n = 3). Conversely, examples of phosphorylated 75-kd and 35-kd proteins were found in the IL-3−/− cells but not in the +/+ cells. Importantly, all of these differences in protein tyrosine-phosphorylation in the IL-3−/− cells disappeared in the BCR-ABL–transduced IL-3−/− cells that had been cotransduced with IL-3 (Figure 9C). The use of phosphospecific antibodies for STAT5 confirmed the increased phosphorylation of STAT5 in the IL-3 expressing (BCR-ABL–transduced) +/+ cells and IL-3−/− cells (cotransduced with BCR-ABL and IL-3).
Expanded clonal populations of primitive, BCR-ABL–transduced +/+ cells produce a delayed but multilineage and ultimately fatal myeloproliferative disease in vivo whereas IL-3−/−cells do not
Preliminary experiments with BCR-ABL–transduced +/+ and IL-3−/− BM cells confirmed the previously reported ability of these cells to generate rapidly lethal leukemias (within 20 days) in irradiated (900 cGy) syngeneic recipients injected with large numbers of unselected cells (3 to 5 × 105cells/mouse) immediately after transduction.14 Even when limiting numbers of transplantable transduced cells were injected by decreasing the transplant 30- to 50-fold, death from the transduced +/+ cells occurred within 30 days and from the IL-3−/− cells within 45 days. Because the growth factor dependence of the in vitro–expanded clones of BCR-ABL–transduced IL-3−/− BM cells appeared more profoundly compromised than the initial cells from which they were derived, we also tested their in vivo leukemogenic potential. In these experiments, we used sublethally irradiated (350 cGy) NOD/SCID and NOD/SCID-β2m−/− mice as hosts. This was based on the need for strategies to avoid the lethal consequences of a myeloablative preparative regimen and at the same time minimize immune reactivity in a nonmyeloablated host to potential neoantigens acquired by the cultured BCR-ABL–transduced cells. Cells from 3 different expanded clones of BCR-ABL–transduced +/+ BM cells (2 producing only IL-3 and one producing IL-3 and GM-CSF) and from 3 independently isolated clones of IL-3−/− cells were injected into both types of mice after a total period in vitro of 4 to 6 weeks. As shown in Figure 10A, no mice injected with clonally expanded populations of BCR-ABL–transduced +/+ cells died during the first 3 weeks and the first deaths in the NOD/SCID mice did not occur until another 5 weeks later. Nevertheless, all mice injected with BCR-ABL–transduced +/+ cells eventually developed a GFP+ leukemia (Figure 10B) and were dead within 100 days, with the more immunodeficient NOD/SCID-β2m−/−mice dying approximately 2 weeks faster than the NOD/SCID hosts. However, there was no significant difference between the rate of death in mice injected with BCR-ABL–transduced +/+ cells producing IL-3 only and those producing GM-CSF as well as IL-3 (data not shown).
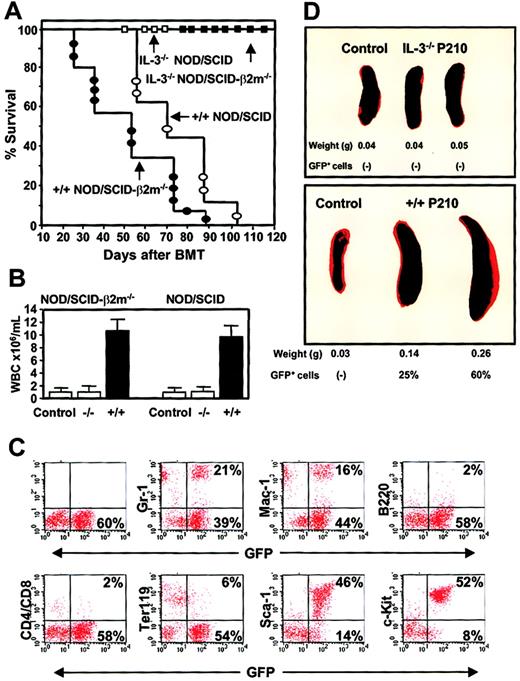
Differences in the leukemogenic activity of cultured BCR-ABL–transduced +/+ and IL-3−/− cells.
(A) Survival curves of NOD/SCID (open symbols) and NOD/SCID-β2m−/− (solid symbols) mice injected with 5 × 106 cells from expanded clones of BCR-ABL–transduced +/+ (circles) and IL-3−/− (squares) cells. (B) WBC counts of the blood from mice injected with BCR-ABL–transduced IL-3−/− and +/+ cells. Controls were normal NOD/SCID-β2m−/− and NOD/SCID mice. (C) FACS profiles of GFP+ bone marrow cells isolated from a representative moribund NOD/SCID mouse with leukemia, killed 60 days after injection of cells from a BCR-ABL+ +/+ culture and showing expression by the GFP+ cells of Gr-1, Mac-1, B220, CD4/CD8, Ter119, Sca-1, and c-kit. (D) Spleen weight of untreated mice or mice injected with BCR-ABL–transduced IL-3−/− (top) or +/+ (bottom) cells.
Differences in the leukemogenic activity of cultured BCR-ABL–transduced +/+ and IL-3−/− cells.
(A) Survival curves of NOD/SCID (open symbols) and NOD/SCID-β2m−/− (solid symbols) mice injected with 5 × 106 cells from expanded clones of BCR-ABL–transduced +/+ (circles) and IL-3−/− (squares) cells. (B) WBC counts of the blood from mice injected with BCR-ABL–transduced IL-3−/− and +/+ cells. Controls were normal NOD/SCID-β2m−/− and NOD/SCID mice. (C) FACS profiles of GFP+ bone marrow cells isolated from a representative moribund NOD/SCID mouse with leukemia, killed 60 days after injection of cells from a BCR-ABL+ +/+ culture and showing expression by the GFP+ cells of Gr-1, Mac-1, B220, CD4/CD8, Ter119, Sca-1, and c-kit. (D) Spleen weight of untreated mice or mice injected with BCR-ABL–transduced IL-3−/− (top) or +/+ (bottom) cells.
Remarkably, in spite of the apparently homogeneous mast cell phenotype of the cells transplanted (Figure 6), the GFP+ leukemia generated from the +/+ cells in vivo had multilineage features that included the production of GFP+ B220+(B-lineage) cells, GFP+ CD4+ or CD8+ (T-lineage) cells, and GFP+Ter119+ (erythroid) cells as well as large numbers of GFP+ Gr-1+ or Mac-1+ (myeloid) cells (Figure 10C). Although double-staining studies were not performed, the fact that more than 80% of all the circulating leukocytes appeared as morphologically recognizeable neutrophils and monocytes means that many of these must have also phenotypically abnormal in their expression of Sca-1 and/or c-kit. Conversely, only a minority of the cells produced in vivo (< 2%) exhibited a mast cell/basophil morphology (Table 3). The leukemic mice also developed splenomegaly (Figure 10D), hepatomegaly, and multilineage GFP+ hematopoiesis in all hematopoietic tissues (Figure 10C) and BM and spleen cells from primary diseased mice produced GFP+ leukemia with the same prolonged latent period when transplanted into secondary recipients (n = 4, data not shown). In marked contrast, none of the clonally expanded populations of BCR-ABL–transduced IL-3−/− cells produced detectable GFP+ cells in either the primary NOD/SCID or NOD/SCID-β2m−/− hosts and all of these mice remained free of any evidence of disease for more than 4 months. Bulk cultures of similarly expanded MIG-transduced +/+ cells also failed to engraft either type of host when followed for a similar observation period (n = 4 data not shown).
Pathologic parameters of leukemic mice injected with clonally expanded BCR-ABL transduced cells.
| Donor genotype . | Recipient genotype . | Disease3-150 latency . | WBC3-151(× 106/mL) . | Differential counts (%) . | Spleen weight (g) . | GFP+ cells in PB, BM . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gr . | Ly . | Mo . | Baso . | Eo . | ||||||
| +/+ | β2m | 57 ± 7.5 | 9.5 ± 3 | 88 ± 4 | 8 ± 4 | 3 ± 1 | 2 ± 1 | 0 | 0.25 ± 0.03 | (+) |
| +/+ | NOD/SCID | 71 ± 7.3 | 8.5 ± 2 | 87 ± 2 | 11 ± 2 | 4 ± 2 | 1 ± 0.5 | 0 | 0.18 ± 0.05 | (+) |
| IL-3−/− | β2m | >120 | 1.1 ± 0.2 | 74 ± 2 | 24 ± 4 | 5 ± 2 | 0.5 ± 0.3 | 0 | 0.04 ± 0.003 | (−) |
| IL-3−/− | NOD/SCID | >120 | 1.0 ± 0.1 | 76 ± 4 | 19 ± 4 | 7 ± 1 | 0.5 ± 0.3 | 0 | 0.04 ± 0.005 | (−) |
| β2m control | — | 1.0 ± 0.3 | 67 ± 5 | 26 ± 5 | 9 ± 2 | 0.5 ± 0.3 | 0 | 0.05 ± 0.005 | ||
| NOD/SCID control | — | 1.2 ± 0.2 | 71 ± 3 | 24 ± 3 | 7 ± 2 | 0 | 0.5 ± 0.3 | 0.04 ± 0.005 | ||
| Donor genotype . | Recipient genotype . | Disease3-150 latency . | WBC3-151(× 106/mL) . | Differential counts (%) . | Spleen weight (g) . | GFP+ cells in PB, BM . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gr . | Ly . | Mo . | Baso . | Eo . | ||||||
| +/+ | β2m | 57 ± 7.5 | 9.5 ± 3 | 88 ± 4 | 8 ± 4 | 3 ± 1 | 2 ± 1 | 0 | 0.25 ± 0.03 | (+) |
| +/+ | NOD/SCID | 71 ± 7.3 | 8.5 ± 2 | 87 ± 2 | 11 ± 2 | 4 ± 2 | 1 ± 0.5 | 0 | 0.18 ± 0.05 | (+) |
| IL-3−/− | β2m | >120 | 1.1 ± 0.2 | 74 ± 2 | 24 ± 4 | 5 ± 2 | 0.5 ± 0.3 | 0 | 0.04 ± 0.003 | (−) |
| IL-3−/− | NOD/SCID | >120 | 1.0 ± 0.1 | 76 ± 4 | 19 ± 4 | 7 ± 1 | 0.5 ± 0.3 | 0 | 0.04 ± 0.005 | (−) |
| β2m control | — | 1.0 ± 0.3 | 67 ± 5 | 26 ± 5 | 9 ± 2 | 0.5 ± 0.3 | 0 | 0.05 ± 0.005 | ||
| NOD/SCID control | — | 1.2 ± 0.2 | 71 ± 3 | 24 ± 3 | 7 ± 2 | 0 | 0.5 ± 0.3 | 0.04 ± 0.005 | ||
Gr, indicates granulocytes; Ly, lymphocytes; Mo, monocytes; Baso, basophils; Eo, eosinophils; PB, peripheral blood; BM, bone marrow.
Time (in days) to death due to leukemia.
Maximum WBC count prior to death.
Discussion
In the generation of human CML, expression of theBCR-ABL oncogene is initiated in a hematopoietic stem cell and this leads to the generation of an amplified clone of multiple lineages of differentiating myeloid and lymphoid progeny and a deregulated overproduction of granulocytes. These biologic features of the chronic phase of the human disease are explained by multiple abnormalities in the complex biologic mechanisms that control normal hematopoiesis. However, a clear understanding of molecular changes important at particular stages in the expansion of the chronic phase clone has remained elusive. Recently, we demonstrated the consistent activation in primitive CML cells of a novel IL-3– and G-CSF–mediated autocrine growth-stimulatory mechanism that appears to explain many of the altered biologic properties of the earliest neoplastic elements.8
In the present study we provide new evidence that IL-3–mediated growth factor autonomy is important for the maintenance of leukemogenic activity in BCR-ABL–transduced cells able to display hematopoietic stem cell activity in vivo. First, phenotypically enriched (Sca-1+ lin−) and functionally defined (LTC-ICs) populations of BCR-ABL–transduced murine BM cells showed a rapid activation of a multifactorial autocrine mechanism involving IL-3, GM-CSF, and G-CSF. Thus, the acquisition of an autocrine phenotype by BCR-ABL–induced leukemic populations is not a late secondary event nor a feature restricted to terminally differentiating BCR-ABL+ cells. Second, each of the 3 growth factors induced was shown to contribute to the immediate factor-independence exhibited by primitive BCR-ABL–transduced cells, as indicated by their reduced autonomous growth in the presence of neutralizing anti–growth factor antibodies and by the decreased ability of BCR-ABL–transduced IL-3−/− cells to proliferate in single cell cultures. Third, after 4 to 6 weeks of growth in vitro, the clonal progeny of primitive BCR-ABL–transduced +/+ cells remained leukemogenic in vivo but only after a prolonged latency of up to 15 weeks. This leukemogenic activity was dependent on the ability of the expanded transduced cells to maintain an autocrine IL-3 mechanism because morphologically indistinguishable expanded clones of BCR-ABL–transduced IL-3−/− BM cells consistently reverted to a factor-dependent phenotype and were not leukemogenic in immunodeficient mice. Taken together, these findings demonstrate the importance of a persisting autocrine IL-3 phenotype to the sustained in vivo leukemic activity of BCR-ABL–expressing cells.
Previous investigations have described more modest effects on the induction of leukemia by BCR-ABL–transduced murine IL-3−/− BM cells using the same vector and target cells14 and these warrant discussion in light of the above conclusions. In fact, there are no substantive differences in the results obtained in the present studies, which confirmed the ability of BCR-ABL–transduced IL-3−/− BM cells assayed immediately after transduction to generate rapidly lethal leukemias in vivo, particularly after the injection of large numbers of transduced cells. However, we now show that the IL-3−/− cells, like the +/+ cells, initially produce sufficient GM-CSF and G-CSF to support their survival and rapid amplification in the absence of any exogenously supplied factor except insulin. Thus, their capacity immediately after transduction for rapid, uncontrolled growth in vivo leading to a fatal outcome should not be viewed as surprising. It is important to note that such a model of leukemogenesis differs markedly from the slow emergence over many years of the clonal disease that characterizes spontaneously arising BCR-ABL–associated CML in humans,27where IL-3 and G-CSF are the only growth factors whose expression has been consistently detected in the neoplastic cells.8 It may therefore be significant that the initial phase of autocrine GM-CSF and G-CSF production characteristic of primitive BCR-ABL–transduced murine hematopoietic cells proved to be a transient response, with the leukemogenic properties of derivative generations of cells more closely approximating those important to the genesis of BCR-ABL+human disease. Future investigations will be required to determine whether the explanation for this discrepancy lies in species differences, or the likely higher level of LTR-regulated BCR-ABL expression in virus-transduced cells as compared with human CML cells (where expression is controlled by endogenous BCR sequences).
The ability of BCR-ABL expression in primitive murine +/+ and IL-3−/− BM cells to facilitate the ready generation in vitro of rapidly expanding clonal populations of mast cells made it possible to undertake a preliminary comparison of their tyrosine phosphorylated protein profiles. Interestingly, evidence that both the BCR-ABL oncoprotein and STAT5 were more highly phosphorylated in the +/+ cells was obtained. This confirms our earlier observations of a dependence of STAT5 activation in primitive human CML cells on their IL-3–mediated autostimulation.8 It has been reported that targeted disruption of the STAT5 A/B gene reduces myeloid progenitor numbers, suggesting a nonredundant role of STAT5 in primitive normal hematopoiesis.28 In addition, BCR-ABL has been found to activate transcription of the Bcl-Xgene through STAT529,30 and mice given transplants of BCR-ABL–transduced STAT5 A/B−/− BM cells developed myeloid leukemia at a reduced frequency.31 These latter results suggest that IL-3–dependent STAT5 activation may contribute to disease aggressiveness.
An important and unexpected outcome of the present study was the observation that apparently homogeneous populations of proliferating mast cells generated in vitro from single primitive BCR-ABL–transduced +/+ mouse BM cells consistently produced an eventually fatal clone of leukemic cells in vivo that showed features of multilineage differentiation. This was shown by the appearance several months after the injection of the cultured cells of large numbers of GFP+ cells in many hematopoietic tissues including the blood, marrow, spleen, and liver that expressed antigens characteristic of mature myeloid, erythroid, and lymphoid cells. Although no attempt was made to explain how these phenotypes might have been generated in vivo, BCR-ABL–induced lineage switching in hematopoietic cells is an emerging theme.32,33 Thus, one possibility would be that certain cells within the proliferating clones of BCR-ABL+ mast cells generated in this study were able to dedifferentiate or transdifferentiate in vivo to produce a spectrum of myeloid and lymphoid cell types.34,35 However, evidence that BCR-ABL can transform primary adult mouse BM cells with multilineage potential is also well established,11 and recently, clonal analysis of differentiating murine embryonic stem cells expressing high levels of BCR-ABL revealed some that could produce primitive erythroid as well as adult lymphoid and myeloid progeny.36 A second possibility would therefore be that the clonally expanded cells transplanted in this study contained a rare subpopulation of BCR-ABL–transduced stem cells with multilineage potential. Although the majority of cells in the cultured BCR-ABL+ populations studied had a lin− Sca-1+ c-kit+IgER+ mast cell phenotype, occasional lin−cells lacking surface IgER and/or Sca-1 or c-kit were detected (Figure6A), suggestive of a phenotype described for a subset of stem cells in normal BM.37 38 Further studies will be required to characterize the leukemogenic cells persisting in these cultured populations of BCR-ABL–transduced +/+ cells and determine the origin of the different cell types produced in vivo.
In summary, we describe here an experimental protocol for obtaining BCR-ABL–transduced adult mouse BM cells that give rise to multilineage leukemias after a prolonged latency in nonmyeloablated immunodeficient hosts. We also show that a rapidly activated and persistent autocrine IL-3 mechanism is essential to the sustained leukemogenicity of these cells in this model. These findings reinforce the likely importance of autocrine IL-3 in the early stages of development of human CML and provide a new approach for further investigations of BCR-ABL–induced myeloid leukemia.
The authors thank Gayle Thornbury, Giovanna Cameron, and Rick Zapf for FACS operation; Dianne Reid, Maya Sinclair, Margaret Hale, and Karen Lambie for technical assistance; and Amy Ahamed for assistance in preparing the manuscript. We also thank G. Dranoff (Dana Farber Cancer Institute, Boston, MA) for providing the IL-3−/− mice; K. Humphries, R. Kay, P. Lansdorp, and G Krystal (Terry Fox Laboratory, Vancouver, BC) for various growth factor cDNAs, retroviral vectors and antibodies; J. Griffin (Dana Farber Cancer Institute, Boston, MA) for the BCR-ABL cDNA; and H. Ziltener (Biomedical Research Centre, University of British Columbia, Vancouver, BC) for anti–murine IL-3 and anti–murine GM-CSF antibodies.
C.J.E. was a Terry Fox Cancer Research Scientist of the NCIC.
Prepublished online as Blood First Edition Paper, July 5, 2002; DOI 10.1182/blood-2002-05-1324.
Supported by grants from the Leukemia Research Fund of Canada and the National Cancer Institute of Canada (NCIC) with funds from the Canadian Cancer Society and from the Terry Fox Run.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
C. J. Eaves, Terry Fox Laboratory, 601 West 10th Ave, Vancouver, BC, Canada V5Z 1L3; e-mail:ceaves@bccancer.bc.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal