MLL (mixed lineage leukemia; alsoALL-1 or HRX) is a proto-oncogene that is mutated in a variety of acute leukemias. Its product is normally required for the maintenance of Hox gene expression during embryogenesis and hematopoiesis through molecular mechanisms that remain poorly defined. Here we demonstrate that MLL (mixed lineage leukemia) is proteolytically processed into 2 fragments (MLLN and MLLC) that display opposite transcriptional properties and form an intramolecular MLL complex in vivo. Proteolytic cleavage occurs at 2 amino acids (D2666 and D2718) within a consensus processing sequence (QXD/GZDD, where X is a hydrophobic amino acid and Z is an alanine or a valine) that is conserved in TRX, the Drosophila homolog of MLL, and in the MLL-related protein MLL2, suggesting that processing is important for MLL function. Processed MLLN and MLLC associate with each other via N-terminal (1253-2254 amino acids) and C-terminal (3602-3742 amino acids) intramolecular interaction domains. MLL processing occurs rapidly within a few hours after translation and is followed by the phosphorylation of MLLC. MLLNdisplays transcriptional repression activity, whereas MLLChas strong transcriptional activation properties. Leukemia-associated MLL fusion proteins lack the MLL processing sites, do not undergo cleavage, and are unable to interact with MLLC. These observations suggest that posttranslational modifications of MLL may participate in regulating its activity as a transcription factor and that this aspect of its function is perturbed by leukemogenic fusions.
Introduction
The MLL (mixed lineage leukemia) gene was originally identified as an oncogene that is often disrupted in various hematopoietic malignancies by rearrangements of chromosome band 11q23.1-4MLL gene abnormalities account for 5% to 10% of the acquired chromosomal rearrangements present in children and adults with ALL (acute lymphoblastic leukemia), acute myeloblastic leukemia (AML), poorly differentiated or biphenotypic leukemias, and myelodysplastic syndromes (MDS).5 About 30 genes have been identified that act as fusion partners with theMLL gene in leukemia-associated translocations. These translocations result in the production of characteristic chimeric proteins composed of the N-terminal region of MLL fused to a C-terminal region consisting of proteins encoded by the partner genes.5 It has been shown that the expression of MLL fusion proteins causes leukemia in mice.6-12
MLL is a predicted 431-kDa protein that appears to be a structural and functional homolog of the Drosophila trithorax (TRX) protein.1,2 Two domains that are highly conserved between MLL and TRX consist of a carboxy-terminal SET (Su(var)3-9, enhancer-of-zeste, and trithorax) domain and internal plant homeodomain (PHD) fingers. Both domains are found in many chromatin-associated transcriptional regulators and are thought to function either directly in chromatin modification or as protein-protein interaction surfaces for the recruitment of chromatin-modifying machinery.1,2,13-16 Genetic studies demonstrate that both MLL and TRX function as homeotic gene regulators.17-23 In MLL (−/−) embryos, expression of the homeobox genes Hoxa7 and Hoxc9is initiated but not maintained.22,23 Furthermore,MLL (+/−) heterozygotes have aberrations in segment identity and shifts in anterior Hox gene expression boundaries.22 23
MLL also has several motifs not found in TRX, including an AT hook region and a domain with homology to DNA methyltransferase (MT domain).24,25 The MLL AT hook region is reported to bind to a specific DNA sequence17 and interact with several proteins including SET (template-activating factor [TAF]-1α), TAF-1β, and growth arrest and DNA damage inducible protein 34 (GADD34).26,27 The MT domain represses transcription when tethered to DNA in the vicinity of a promoter28,29 and also confers DNA-binding activity onto MLL with a preference for unmethylated CpG sequences.30,31 Structure/function studies have revealed that both the AT hook region and the MT domain are critical for oncogenesis.30 In addition to transcriptional repression activity, MLL contains a strong transactivation domain28 that is reported to interact with the coactivator protein CBP.32
Despite the extensive analysis of MLL structure and identification of candidate interacting proteins, the mechanisms by which MLL functions as a homeotic gene regulator are not as yet understood. In this report, we show that MLL is proteolytically processed in vivo into 2 fragments that interact with each other and separately contain its transcriptional activator and repressor properties. We identified the processing sites and found that similar sequences are conserved in MLL/TRX family members, which suggests that processing of MLL is specific and evolutionally conserved. These observations provide additional insight into the mechanism of MLL function and suggest a model for regulating its bifunctional transcriptional activity.
Materials and methods
Cell culture
BOSC23 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). Raji, Jurkat, HL-60, and ML-1 cells were cultured in RPMI 1640 medium supplemented with 10% FCS.
cDNA cloning of full-length MLL
A size-fractionated cDNA library,33 which was made in pBlueScript from poly(A) RNA derived from human KG-1 cells, was screened by using the partial cDNA coding for 1-4218 base pairs (bp) ofMLL that was generously provided by Dr H. Hirai (University of Tokyo, Japan). The first screening yielded 8 clones spanning the N-terminal half of MLL cDNA. To obtain the C-terminal half of MLL cDNA, the same cDNA library was screened by using a DNA probe corresponding to 9599-10581 bp of MLL cDNA, which was amplified from a human bone marrow quick clone cDNA pool (Clontech, Palo Alto, CA) by using the primer pair 5′-GTCTCATCACCAGCACTTAC-3′ and 5′-TAAAGTTGGATACCTGGGGC-3′. We obtained 5 clones containing partial cDNA corresponding to the C-terminal half of MLL cDNA. Three different MLL cDNA clones obtained by these 2 screens were tethered together by restriction enzyme digestion and ligation to generate the full-lengthMLL cDNA.
Construction of expression vectors
pLNCX-MLL(H) was generated by cloning the XhoI/ClaI fragment of the full-length MLL cDNA tagged with HA at its C-terminal end into the SalI/ClaI sites of the pLNCX vector, whose HindIII site had been converted into a SalI site by using a SalI linker (NEB, Beverly, MA). MLL-p300 cDNA was created by ligation of 3 partial cDNAs. XhoI/BamHI fragment consisting the most of MLL portion and SpeI/ClaI fragment consisting the most of p300 portion were prepared by restriction enzyme digestion of each cDNA. BamHI/SpeI fragment consisting of the joint region of MLL and p300 was prepared from the polymerase chain reaction (PCR) products amplified from the cDNA pool of the acute myeloblastic leukemia (AML) patient associated with t(11;22)(q23;q13)34 using the primer pair 5′-AGTCAGAAACCTACCCCATC-3′ and 5′-GGCCTGAGAATTCACTTCTG-3′. XhoI/ClaI fragment of MLL-p300 cDNA was cloned into SalI/ClaI sites of pLNCX vector mentioned above to create pLNCX-MLL-p300 expression vector. pM-MLL(H) was generated by cloning the MluI/ClaI fragment of full-length MLL cDNA into the MluI/ClaI sites of the pM vector (Clontech), whose BglII site had been converted into a PacI site by using a PacI linker (NEB). pLNCX-GAL4-MLL(H) was generated by cloning the PacI/ClaI fragment of pM-MLL(H) into the PacI/ClaI site of the pLNCX vector, whose HindIII site had been converted into a PacI site by using a PacI linker. pLNCX-(HF)MLL(H) was generated by cloning the PacI/MluI fragment of the DNA fragment amplified from the pcDNA4/HISMAX vector (Invitrogen, Carlsbad, CA) by PCR with the primer pair 5′-TTAATTAATAATACGACTCACTATAGGG-3′ and 5′-GACGCGTTGCTTGTCATCGTCGTCCTTGTAGTCTGCCTTATCGTCATCGTCG- TACAGATC-3′ into PacI/MluI sites of the pLNCX-GAL4-MLL(H) vector. Substitution mutants were generated by PCR-mediated mutagenesis. Partial cDNA of MLLD2718A, in which D2718 was substituted by A2718, was generated by PCR with the primer pair 5′-AAACCTCAGGAGGATGGCTCTT-3′ and 5′-GGCAGTTATCCATCTTTGGC-3′ and 2 template DNA fragments containing the mutation, which had been separately amplified fromMLL cDNA by PCRs using the primer sets 5′-AAACCTCAGGAGGATGGCTCTT-3′/5′-ATCAACACCAGCCAACTGTGA-3′ and 5′-TCACAGTTGGCTGGTGTTGAT-3′/5′-GGCAGTTATCCATCTTTGGC-3′. pLNCX-MLL D2718A(H) was generated by cloning the AflII/BalI fragment of the partial cDNA of MLL D2718A into the AflII/BalI sites of pLNCX-MLL(H). pLNCX-MLL D2666A and pLNCX-MLL DD2666/2718AA were generated by cloning the StuI/AflII fragment amplified by PCR from MLL cDNA with the primer pair 5′-AGGCCTTTCTATGCCAGGAGT-3′ and 5′-GCTTAAGTCATCGGCCCCAGCCACCTGTCCTTC- AGCTGAT-3′ into the StuI/AflII sites of pLNCX-MLL(H) and pLNCX-MLL D2718A(H), respectively. The series of C-terminal pLNCX-MLL(H) deletion mutants were generated by cloning partial cDNA amplified by PCR into appropriate restriction enzyme sites. The series of N-terminal pM-MLL(H) deletion mutants were generated by cloning amplified GAL4 cDNA, in which appropriate restriction enzyme sites were added, into pM-MLL(H). The series of pLNCX-GAL4-MLL deletion mutants were generated by placing the PacI/ClaI fragment of the pM-MLL(H) C-terminal deletion mutants or the MluI/ClaI fragment of the pLNCX-MLL(H) N-terminal deletion mutants into the relevant sites of the pLNCX-MLL vector. The series of deletion mutants of GAL4-MLL C-terminal fragment tagged with His were created by replacing the PacI/XhoI fragment of pLNCX GAL4-MLL C-terminal fragment deletion mutants with the PacI/XhoI fragment of GAL4 DNA binding domain tagged with His (His-GAL4 cDNA). One of the partial DNA fragments of His-GAL4 cDNA was created by PCR from pcDNA4/HISMAX vector with the primer pair 5′-TTAATTAATACGACTCACTATAGGG-3′ and 5′-TCGATAGAAGACAGTAGCTTCTTATCGTCATCGTCGTACA-3′. The other one was created by PCR from pM vector with the primer pair 5′-TGTACGACGATGACGATAAGAAGCTACTGTCTTCTATCGA-3′ and 5′-TGCGGCCGCCCG- TTTGATCTCCGATACAGTCAACTGTCTTT-3′. Then His-GAL cDNA was created by PCR from the mixture of these partial DNA fragments with the primer pair 5′-TTAATTAATACGACTCACTATAGGG-3′ and 5′-TGCGGCCGCCCGTTTGATCTCCGATACAGTCAACTGTCTTT-3′.
Antibodies
Anti-MLL N-terminal polyclonal antibody (rpN1) was generated by immunizing rabbits with a glutathione-S–transferase (GST) fusion protein containing MLL residues 4-304. Anti-MLL N- and C-terminal monoclonal antibodies were raised against maltose-binding protein (MBP) fusion proteins containing MLL amino acids 3317 to 3604 (mmC2 and mmC3) or 161 to 356 (mmN3, mmN4, and mmN6).9Rabbit anti-MOZ antibody (N13-2) was described previously.35 Anti-HA (3F10; Roche, Indianapolis, IN), anti-FLAG (M2; Sigma), anti-His (D-8; Santa Cruz, CA), anti-GAL4 (RK5C1; Santa Cruz), and anti-p300 (RW128; Upstate Biotechnology, Lake Placid, NY) were obtained commercially. Horseradish-peroxidase (HRP)–conjugated goat anti–mouse Ig(H+L), HRP-conjugated goat anti–rat Ig(H+L), and HRP-conjugated goat anti–rabbit Ig(H+L) antibodies (Southern Biotechnology Associates) were used as secondary reagents for Western blotting.
Transfections
COS-7 or BOSC23 cells were seeded one day before transfection, and 60%-80% confluent cells were transfected with various expression plasmids by calcium phosphate precipitation methods used by the Profection mammalian transfection system (Promega, Madison, WI). The cells were washed and placed in fresh DMEM growth medium 12 hours after transfection. Cells were harvested 48 hours after transfection and used for experiments.
Protein sequencing
A total of 100 plates (100 mm diameter) of BOSC23 cells were transfected with pLNCX-MLL-HA, and MLL proteins were immunoprecipitated with anti-HA antibody 3F10. The immunoprecipitates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a membrane filter (polyvinylidenefluoride [PVDF] sequencing membrane; Millipore, Bedford, MA). The blotted 180-kDa polypeptides were subjected to N-terminal microsequencing using a G1005A protein sequencer (Hewlett Packard, Palo Alto, CA).
Protein analyses
Reporter gene assay
BOSC23 cells in 24-well culture plates were transfected with 1 μg of pLNCX-GAL4-MLL and 0.1 μg of pFR-luc (Stratagene, La Jolla, CA) together with 0.01 μg of pRL-tk (Promega), which acts as an internal control for the transfection efficiency as described by the manufacturer. The cells were harvested 48 hours after transfection and assayed for luciferase activity using the Dual-luciferase reporter assay system according to the manufacturer's instructions (Promega). The activation activity was set relative to that of the control expression vector pLNCX-GAL4.
Results
MLL is proteolytically processed in vivo
The expression of MLL protein was examined in various cell lines by immunoblot analysis with multiple anti-MLL antibodies. Although the predicted molecular mass of MLL is 431 kDa, a band corresponding to full-length MLL was only weakly detected by each of the antibodies used (indicated with a plus mark in Figure 1A) in whole cell extracts of several cell lines. Instead, a 300-kDa band (indicated with an asterisk in Figure 1A, lanes 1-6) was clearly detected by antibodies against the N-terminal portions of MLL (rpN1, mmN3). Conversely, antibodies against C-terminal portions of MLL (mmC2, mmC3) detected a broad 180-kDa band (indicated with an asterisk in Figure 1A, lanes 7-12). The observed immunoreactive proteins were not due to degradation during preparation of cell lysates because the cells were lysed by addition of SDS-PAGE sample buffer directly to the cell pellets. In fact, no degradation of high-molecular-weight proteins such as p300 and MOZ was observed (Figure 1A, lanes 13-18). These results suggested that MLL might be cleaved into 2 major fragments of 300 and 180 kDa in vivo.
MLL is proteolytically processed in vivo.
(A) Expression of MLL proteins in various cell lines. Cell lysates were analyzed by immunoblot with anti-MLL N-terminal (rpN1, mmN3) or C-terminal antibodies (mmC2, mmC3) or antibodies specific for p300 (RW128) or MOZ (N13-2), as denoted beneath the panels. Identities of cell lines are indicated above the gel lanes. The band corresponding to predicted 430-kDa full-length MLL is indicated by a plus mark. Bands corresponding to MLL N- and C-terminal fragments, p300 and MOZ, are indicated by asterisks. Migration of molecular weight standards is shown on the left. (B) BOSC23 cells were transfected with empty pLNCX vector (vector) or pLNCX expressing full-length MLL with an HA tag at its C-terminus [MLL(H)]. Cell lysates were analyzed by immunoblot with anti-MLL N-terminal antibodies (rpN1, mmN3, mmN4, mmN6), anti-HA antibody 3F10, or anti-MLL C-terminal antibodies (mmC2, mmC3). (C) Expression of MLL and MLL-AF6 in HL-60 and ML-1 cells. Cell lysates were analyzed by immunoblot with anti-MLL N-terminal antibody (rpN1). MLL N-terminal fragment and MLL-AF6 are indicated by arrows. (D) Expression of MLL and MLL-p300 in transiently transfected BOSC23 cells. Following transfection with the constructs indicated above the gel lanes, cell lysates were analyzed by immunoblot with anti-MLL N-terminal antibody (mmN3). MLL N-terminal fragment and MLL-p300 are indicated by arrows.
MLL is proteolytically processed in vivo.
(A) Expression of MLL proteins in various cell lines. Cell lysates were analyzed by immunoblot with anti-MLL N-terminal (rpN1, mmN3) or C-terminal antibodies (mmC2, mmC3) or antibodies specific for p300 (RW128) or MOZ (N13-2), as denoted beneath the panels. Identities of cell lines are indicated above the gel lanes. The band corresponding to predicted 430-kDa full-length MLL is indicated by a plus mark. Bands corresponding to MLL N- and C-terminal fragments, p300 and MOZ, are indicated by asterisks. Migration of molecular weight standards is shown on the left. (B) BOSC23 cells were transfected with empty pLNCX vector (vector) or pLNCX expressing full-length MLL with an HA tag at its C-terminus [MLL(H)]. Cell lysates were analyzed by immunoblot with anti-MLL N-terminal antibodies (rpN1, mmN3, mmN4, mmN6), anti-HA antibody 3F10, or anti-MLL C-terminal antibodies (mmC2, mmC3). (C) Expression of MLL and MLL-AF6 in HL-60 and ML-1 cells. Cell lysates were analyzed by immunoblot with anti-MLL N-terminal antibody (rpN1). MLL N-terminal fragment and MLL-AF6 are indicated by arrows. (D) Expression of MLL and MLL-p300 in transiently transfected BOSC23 cells. Following transfection with the constructs indicated above the gel lanes, cell lysates were analyzed by immunoblot with anti-MLL N-terminal antibody (mmN3). MLL N-terminal fragment and MLL-p300 are indicated by arrows.
To further investigate this observation, BOSC23 cells were transiently transfected with a vector encoding full-length MLL tagged with HA at its C-terminal end. Cell lysates were then immunoblotted with various anti-MLL antibodies or anti-HA antibody 3F10. All of the antibodies against the N-terminal portion of MLL (rpN1, mmN3, mmN4, and mmN6) strongly detected immunoreactive protein in the transfectant lysate [MLL(H)] that comigrated with the endogenous 300-kDa MLL protein (Figure 1B, lanes 1-8). In the same manner, anti-MLL antibodies raised against the C-terminal portion of MLL (mmC2 and mmC3) also detected endogenous and exogenous MLL proteins of the same mobility (180 kDa) as detected with the anti-HA antibody 3F10 (Figure 1B, lanes 9-14). Therefore, we conclude that MLL is proteolytically processed into at least 2 fragments, a 300-kDa N-terminal fragment and a 180-kDa C-terminal fragment (hereafter referred to as MLLN and MLLC, respectively).
To determine whether leukemia-associated MLL-fusion proteins are processed, expression of MLL-AF6 was examined in ML-1 cells, which lack a germline MLL locus. In these cells, one MLLallele is involved in a t(6;11)(q27;q23) chromosomal translocation that results in the expression of a chimeric MLL-AF6 protein, whereas the other MLL allele is deleted.39 Intact MLL-AF6, but not normal MLL, was expressed in ML-1 cells (Figure 1C, lane 2). Similarly, no processing of MLL-p300 was observed in BOSC23 cells that were transfected with an MLL-p300 expression construct (Figure 1D, lane 3), although endogenous MLLN was detected.
MLL is specifically cleaved at amino acids D2666 and D2718
To determine the specific site where MLL is cleaved, we transiently transfected BOSC23 cells with an expression vector that encodes MLL containing an HA epitope at its C-terminal end. Processed MLLC was affinity purified with an anti-HA antibody and sequenced from its N-terminus by Edman degradation. The 8–amino acid sequence obtained (GVDDXTES, X represents an unidentified amino acid) matched residues 2719 to 2726 of MLL, suggesting that MLL is cleaved at D2718. To confirm this, we constructed an MLL substitution mutant in which aspartic acid 2718 was converted into alanine (D2718A). Western blot analysis with anti-MLL N-terminal and anti-HA antibodies showed that, relative to cells expressing wild-type MLL, most of the mutant MLL protein remained unprocessed (Figure2C, lanes 4 and 9), indicating that D2718 is indeed a cleavage site. However, a considerable amount of the mutant protein was still cleaved. Furthermore, the MLLC fragment released from the D2718A mutant MLL protein and recognized by the anti-HA antibody appeared to be slightly larger than that processed from wild-type MLL (Figure 2C, lanes 3 and 4). This suggested that there might be another processing site upstream of D2718. We thus searched the amino acid sequence of MLL for sequences resembling residues 2715 to 2726 and found a similar sequence at residues 2663 to 2674 (Figure 2B). To determine if cleavage occurs at this sequence, we constructed a substitution mutant whose aspartic acid at D2666 was converted to alanine (D2666A). A double-substitution mutant whose D2666 and D2718 residues were both converted into alanines (DD2666/2718AA) also was constructed. While the D2666A mutant appeared to be cleaved as well as wild-type MLL, cleavage of the DD2666/2718AA double mutant was completely abolished (Figure 2C, lanes 5 and 10). Identical results were obtained when COS-7 or HeLa cells were transfected with the same vectors (data not shown). Thus, MLL appears to be proteolytically processed at 2 sites, D2666 and D2718, with D2718 being the major cleavage site.
Identification of MLL processing sites.
(A) Purification of MLL proteins. BOSC23 cells were transfected with an MLL-expression vector [pLNCX MLL(H)], lysed 48 hours later, and subjected to immunoprecipitation with an anti-HA antibody 3F10. An aliquot of the immunoprecipitate was separated in a denaturing gel and stained by the silver stain method. Full-length MLL, the purified C-terminal 180-kDa fragment (MLLC), and the copurified 300-kDa band are indicated. Migration of molecular weight markers is indicated on the left. (B) Amino acid sequences of MLL processing sites. The sequences around the 2 processing sites of human MLL (hMLL) and putative processing sites of mouse MLL (mMLL), pufferfish MLL (fMLL), Drosophila TRX (dTRX), and human MLL2 (hML2) are aligned.1,2,25,40 41 Conserved amino acids are shaded and marked with an asterisk. The processing site is indicated by an arrow in the consensus sequence below. The numbers refer to positions relative to the first amino acid of each protein. (C) Inhibition of processing by mutations in the cleavage sites. BOSC23 cells were transfected with expression constructs encoding proteins indicated above the gel lanes and harvested 48 hours after transfection. All MLL proteins were tagged with HA at their C-termini. The cell lysates were analyzed by immunoblot with anti-HA antibody (left panel) or anti-MLL N-terminal antibody (right panel).
Identification of MLL processing sites.
(A) Purification of MLL proteins. BOSC23 cells were transfected with an MLL-expression vector [pLNCX MLL(H)], lysed 48 hours later, and subjected to immunoprecipitation with an anti-HA antibody 3F10. An aliquot of the immunoprecipitate was separated in a denaturing gel and stained by the silver stain method. Full-length MLL, the purified C-terminal 180-kDa fragment (MLLC), and the copurified 300-kDa band are indicated. Migration of molecular weight markers is indicated on the left. (B) Amino acid sequences of MLL processing sites. The sequences around the 2 processing sites of human MLL (hMLL) and putative processing sites of mouse MLL (mMLL), pufferfish MLL (fMLL), Drosophila TRX (dTRX), and human MLL2 (hML2) are aligned.1,2,25,40 41 Conserved amino acids are shaded and marked with an asterisk. The processing site is indicated by an arrow in the consensus sequence below. The numbers refer to positions relative to the first amino acid of each protein. (C) Inhibition of processing by mutations in the cleavage sites. BOSC23 cells were transfected with expression constructs encoding proteins indicated above the gel lanes and harvested 48 hours after transfection. All MLL proteins were tagged with HA at their C-termini. The cell lysates were analyzed by immunoblot with anti-HA antibody (left panel) or anti-MLL N-terminal antibody (right panel).
MLLN interacts with MLLC
Immunoprecipitation of MLLC from transfected BOSC23 cells revealed the presence of a 300-kDa coprecipitating band (Figure2A, lane 2), suggesting that MLLN may copurify with MLLC. Furthermore, affinity purification of MLL tagged at its N-terminus with the FLAG epitope from transfected BOSC23 cells using an anti-FLAG antibody yielded both the expected 300-kDa protein as well as a broad 180-kDa band (Figure3A, lane 1). Taken together, these purification data strongly suggested that MLLN and MLLC interact with each other.
N-terminal MLL interacts with C-terminal MLL.
Copurification of MLLC with MLLN. (A) BOSC23 cells were transfected with pLNCX vectors encoding MLL or the DDAA mutant tagged with FLAG at their N-termini. Proteins were immunoprecipitated with anti-FLAG antibody, eluted by incubation with FLAG peptide, separated in a denaturing gel, and stained with Coomassie brilliant blue (CBB). The purified fragments are indicated by asterisks (MLLN and MLLC in lane 1, unprocessed MLL in lane 2). Molecular weight markers are on the left. (B) BOSC23 cells were transfected with pLNCX vector alone or pLNCX encoding MLL protein tagged with His and FLAG at its N-terminus and HA at its C-terminus [denoted (Hi-FL)MLL(HA)]. Cell lysates were immunoprecipitated with anti-FLAG antibody or anti-HA antibody. The whole cell lysates (WCE) and the immunoprecipitates (denoted as IP:anti-HA and IP:anti-FL) were analyzed by immunoblot with anti-His antibody (left panel) or anti-HA antibody (right panel). The purified fragments are indicated by asterisks (MLLN in left panel, MLLC in right panel). (C) Interaction of endogenous MLLN and MLLC. Cell lysates were immunoprecipitated with nonspecific mouse IgG, mouse anti-MLL N-terminal (mmN4), or anti-MLL C-terminal (mmC2) antibodies. The precipitates were analyzed by immunoblot with a mouse anti-MLL C-terminal (mmC2) antibody. MLLC is indicated by an asterisk.
N-terminal MLL interacts with C-terminal MLL.
Copurification of MLLC with MLLN. (A) BOSC23 cells were transfected with pLNCX vectors encoding MLL or the DDAA mutant tagged with FLAG at their N-termini. Proteins were immunoprecipitated with anti-FLAG antibody, eluted by incubation with FLAG peptide, separated in a denaturing gel, and stained with Coomassie brilliant blue (CBB). The purified fragments are indicated by asterisks (MLLN and MLLC in lane 1, unprocessed MLL in lane 2). Molecular weight markers are on the left. (B) BOSC23 cells were transfected with pLNCX vector alone or pLNCX encoding MLL protein tagged with His and FLAG at its N-terminus and HA at its C-terminus [denoted (Hi-FL)MLL(HA)]. Cell lysates were immunoprecipitated with anti-FLAG antibody or anti-HA antibody. The whole cell lysates (WCE) and the immunoprecipitates (denoted as IP:anti-HA and IP:anti-FL) were analyzed by immunoblot with anti-His antibody (left panel) or anti-HA antibody (right panel). The purified fragments are indicated by asterisks (MLLN in left panel, MLLC in right panel). (C) Interaction of endogenous MLLN and MLLC. Cell lysates were immunoprecipitated with nonspecific mouse IgG, mouse anti-MLL N-terminal (mmN4), or anti-MLL C-terminal (mmC2) antibodies. The precipitates were analyzed by immunoblot with a mouse anti-MLL C-terminal (mmC2) antibody. MLLC is indicated by an asterisk.
Further support for their interaction was obtained by reciprocal immunoprecipitations. BOSC23 cells were transiently transfected with a vector encoding full-length MLL tagged with His and FLAG epitopes at its N-terminal end and HA at its C-terminal end. Immunoprecipitation was performed with either anti-HA or anti-FLAG antibodies, followed by immunoblotting with anti-His or anti-HA antibodies. Under these conditions, MLLN coprecipitated with MLLC and vice versa (Figure 3B, lanes 4 and 12), confirming their in vivo interaction. We also examined if endogenous MLLN and MLLC were complexed with each other. As shown in Figure 3C, MLLC coprecipitated with MLLNin BOSC23, Raji, and Jurkat cells.
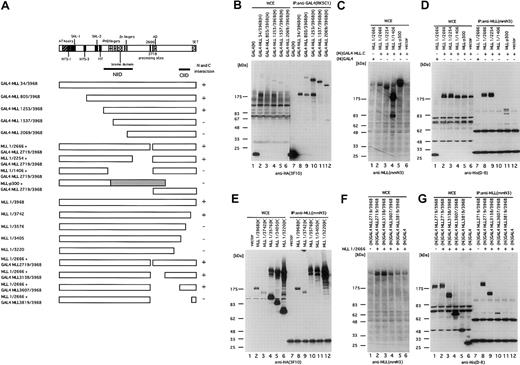
Mapping of MLL intramolecular interaction domains
To define potential domains responsible for interaction of MLLN and MLLC, a series of deletion mutants (Figure 4A) was constructed and tested in coprecipitation assays. N-terminal mutants were fused to the GAL4 DNA binding domain to ensure their nuclear localization. All mutants were tagged with HA at their C-terminal ends. To define the interaction domain from the internal direction, the N-terminal fragments were cotransfected together with His-tagged GAL4-MLLC or its deletion mutants. The N-terminal deletion mutants were immunoprecipitated with an anti-GAL4 antibody, while C-terminal deletion mutants or coexpressed N- and C-terminal fragments were immunoprecipitated with an anti-MLL N-terminal antibody. The precipitates were separated in denaturing gels and immunoblotted with anti-HA or anti-His antibodies. Analysis of progressive N- and C-terminal MLLN deletions defined the region required for interaction with MLLC to reside in residues 1253 to 2254 containing the MLL PHD fingers, bromo domain, and zinc finger (Figure4B,D). Consistent with this determination, MLL-p300,34 a leukemic fusion protein that contains residues 1 to 1493 of MLL, did not interact with MLLC (Figure 4D, lane 11). Similarly, progressive deletions of MLLC defined amino acids 3607 to 3742 to be necessary for interaction with MLLN (Figure 4E-G). We refer to these 2 domains as the N-terminal intramolecular interaction domain (NIID, residues 1253-2254) and the C-terminal intramolecular interaction domain (CIID, residues 3607-3742).
Mapping of the intramolecular interaction domains.
(A) Schematic representation of the various MLL deletion mutants tagged with HA at their C-termini and various GAL4-MLL C-terminal fragments tagged with His at their N-termini. The known motifs and domains of the AT hook (171-309), NTS-1 (244-314), SNL-1 (388-432), NTS-3 (727-762), SNL-2 (1050-1089), the MT domain (1136-1203), NIID (1253-2254), the PHD fingers (1418-1628), the bromo domain (1633-1769), the Zn fingers (Zn: 1870-1977), the processing sites (D2666 and D 2718), the transactivation domain (denoted as AD, 2829-2883), CIID (3607-3742), and the SET domain (3818-3969) are shown.40 46 Interaction between N- and C-terminal fragments is also shown on the right as present (+) or absent (–). (B) Determination of N-terminal edge of the N-terminal intramolecular interaction domain (NIID). BOSC23 cells were transfected with the pM vector or one of the pM MLL(H) vectors expressing N-terminal deletion mutants of MLL fused with GAL4 at their N-termini and tagged with HA at their C-termini. The cells were harvested and lysed in lysis buffer 48 hours after transfection. The GAL4-MLL proteins were immunoprecipitated with the anti-GAL4 antibody. The whole cell extracts (WCE, lanes 1-6) and the immunoprecipitates (IP:GAL4, lanes 7-12) were analyzed by immunoblot with anti-HA antibody 3F10. Molecular weight markers are on the left. (C, D) Determination of the C-terminal edge of the N-terminal intramolecular domain. BOSC23 cells were transfected with pLNCX vectors expressing C-terminal deletion mutants of MLL and pLNCX vector expressing GAL4-MLL 2719/3968 tagged with His at the N-termini. The MLL proteins were immunoprecipitated with anti-MLL N-terminal antibody (mmN3). WCE and the precipitates were separated in denaturing gels and immunoblotted with anti-MLL N-terminal antibody (mmN3) (WCE only; C) or anti-His antibody (D-8; D). (E) Determination of the C-terminal edge of the C-terminal intramolecular interaction domain. BOSC23 cells were transfected with the pLNCX vectors expressing C-terminal deletion mutants of MLL tagged with HA at their C-termini. The MLL proteins were immunoprecipitated with the anti-MLL N-terminal antibody (mmN3) antibody. WCE (lanes 1-6) and the immunoprecipitates (lanes 7-12) were immunoblotted with the anti-HA antibody 3F10. Panels F and G show the determination of the N-terminal edge of the C-terminal intramolecular interaction domain. BOSC23 cells were transfected with pLNCX vectors expressing MLL1/2666 and pLNCX vectors expressing N-terminal deletion mutants of GAL4-MLL 2719/3968 tagged with His at the N-termini. The MLL proteins were immunoprecipitated with anti-MLL N-terminal antibody (mmN3). The whole cell extracts and the precipitates were separated in denaturing gels and immunoblotted with anti-MLL N-terminal antibody (mmN3) (WCE only; F) or anti-His antibody (D-8; G).
Mapping of the intramolecular interaction domains.
(A) Schematic representation of the various MLL deletion mutants tagged with HA at their C-termini and various GAL4-MLL C-terminal fragments tagged with His at their N-termini. The known motifs and domains of the AT hook (171-309), NTS-1 (244-314), SNL-1 (388-432), NTS-3 (727-762), SNL-2 (1050-1089), the MT domain (1136-1203), NIID (1253-2254), the PHD fingers (1418-1628), the bromo domain (1633-1769), the Zn fingers (Zn: 1870-1977), the processing sites (D2666 and D 2718), the transactivation domain (denoted as AD, 2829-2883), CIID (3607-3742), and the SET domain (3818-3969) are shown.40 46 Interaction between N- and C-terminal fragments is also shown on the right as present (+) or absent (–). (B) Determination of N-terminal edge of the N-terminal intramolecular interaction domain (NIID). BOSC23 cells were transfected with the pM vector or one of the pM MLL(H) vectors expressing N-terminal deletion mutants of MLL fused with GAL4 at their N-termini and tagged with HA at their C-termini. The cells were harvested and lysed in lysis buffer 48 hours after transfection. The GAL4-MLL proteins were immunoprecipitated with the anti-GAL4 antibody. The whole cell extracts (WCE, lanes 1-6) and the immunoprecipitates (IP:GAL4, lanes 7-12) were analyzed by immunoblot with anti-HA antibody 3F10. Molecular weight markers are on the left. (C, D) Determination of the C-terminal edge of the N-terminal intramolecular domain. BOSC23 cells were transfected with pLNCX vectors expressing C-terminal deletion mutants of MLL and pLNCX vector expressing GAL4-MLL 2719/3968 tagged with His at the N-termini. The MLL proteins were immunoprecipitated with anti-MLL N-terminal antibody (mmN3). WCE and the precipitates were separated in denaturing gels and immunoblotted with anti-MLL N-terminal antibody (mmN3) (WCE only; C) or anti-His antibody (D-8; D). (E) Determination of the C-terminal edge of the C-terminal intramolecular interaction domain. BOSC23 cells were transfected with the pLNCX vectors expressing C-terminal deletion mutants of MLL tagged with HA at their C-termini. The MLL proteins were immunoprecipitated with the anti-MLL N-terminal antibody (mmN3) antibody. WCE (lanes 1-6) and the immunoprecipitates (lanes 7-12) were immunoblotted with the anti-HA antibody 3F10. Panels F and G show the determination of the N-terminal edge of the C-terminal intramolecular interaction domain. BOSC23 cells were transfected with pLNCX vectors expressing MLL1/2666 and pLNCX vectors expressing N-terminal deletion mutants of GAL4-MLL 2719/3968 tagged with His at the N-termini. The MLL proteins were immunoprecipitated with anti-MLL N-terminal antibody (mmN3). The whole cell extracts and the precipitates were separated in denaturing gels and immunoblotted with anti-MLL N-terminal antibody (mmN3) (WCE only; F) or anti-His antibody (D-8; G).
MLL processing occurs rapidly after translation
To examine the kinetics of MLL processing, we performed pulse-chase experiments. MLL tagged with HA at its C-terminal end was transiently expressed in BOSC23 cells, which were then metabolically labeled with 35S-methionine for 3 hours and further cultured in medium without 35S-methionine for up to 2 days. Cell lysate proteins were immunoprecipitated with anti-MLL N-terminal antibody. The precipitates were separated in denaturing gels and visualized by autoradiography or immunoblotted with anti-HA or anti-MLL N-terminal antibodies. At the start of the chase period (0-hour time point), fully half of the labeled MLL that had been synthesized during the 3-hour labeling period had already been processed (Figure5A, lane 4). The levels of MLLN and MLLC increased after 3 and 9 hours of chase, while the levels of unprocessed MLL concomitantly decreased (Figure 5A, lanes 5 and 6). The difference in the levels of processed and unprocessed MLL strongly suggests that processing occurs as a consequence of maturation and not in the lysis procedure. A small amount of unprocessed MLL was still detected 24 hours after cessation of the labeling period, but this completely disappeared by 48 hours (Figure 5B, lanes 5 and 6). Thus, it appears that processing of MLL initiates within a few hours of its synthesis and is virtually complete within 24 hours.
MLL processing occurs within a few hours of its translation.
BOSC23 cells were transfected with empty vector (vector) or a construct encoding full-length MLL tagged with HA at its C-terminus [MLL(H)]. The cells were metabolically labeled with [35S]-methionine for 3 hours. After labeling, the cells were cultured in nonradioactive medium for the indicated time periods and then lysed in lysis buffer. MLL proteins were immunoprecipitated with anti-MLL antibody, separated in denaturing gels, and visualized by autoradiography (A and B) or immunoblotted with anti-HA (C and D) or anti-MLL N-terminal (E and F) antibodies. In the left panels (A, C, and E), the cells were harvested at 0, 3, and 9 hours after labeling. In the right panels (B, D, and F), the cells were harvested at 0, 1, and 2 days after labeling. Unprocessed MLL, the N-terminal fragment, and the C-terminal fragment are indicated by arrows.
MLL processing occurs within a few hours of its translation.
BOSC23 cells were transfected with empty vector (vector) or a construct encoding full-length MLL tagged with HA at its C-terminus [MLL(H)]. The cells were metabolically labeled with [35S]-methionine for 3 hours. After labeling, the cells were cultured in nonradioactive medium for the indicated time periods and then lysed in lysis buffer. MLL proteins were immunoprecipitated with anti-MLL antibody, separated in denaturing gels, and visualized by autoradiography (A and B) or immunoblotted with anti-HA (C and D) or anti-MLL N-terminal (E and F) antibodies. In the left panels (A, C, and E), the cells were harvested at 0, 3, and 9 hours after labeling. In the right panels (B, D, and F), the cells were harvested at 0, 1, and 2 days after labeling. Unprocessed MLL, the N-terminal fragment, and the C-terminal fragment are indicated by arrows.
MLLC is phosphorylated after proteolytic processing
We noted that after 3 and 9 hours of chase, the bands corresponding to MLLC became broader than at 0 hour (Figure5A, lanes 4-6), apparently due to the presence of slower migrating proteins. In addition, MLLC complexed with MLLNmigrated more slowly than uncomplexed MLLC (Figure 4D, lanes 2-6). This suggested that MLLC may undergo posttranslational modification after processing and complex formation. We assessed whether the observed migration differences might result from phosphorylation by performing metabolic labeling with [32P]-orthophosphate and in vitro dephosphorylation assays. BOSC23 cells expressing MLL tagged with HA at its C-terminal end [MLL(H)] were labeled with [32P]- orthophosphate, and MLL proteins were then immunoprecipitated with the anti-HA antibody. The precipitates were then immunoblotted with either anti-MLL N-terminal or anti-HA antibodies or visualized by autoradiography (Figure 6A). Both MLLN and MLLC were phosphorylated in vivo. Treatment of the anti-MLL N-terminal immunoprecipitates with calf intestinal alkaline phosphatase resulted in a faster migrating and sharper MLLC band (Figure 6B, lane 3 versus 4). Thus, we conclude that the mobility shift of MLLC observed 3 hours after chasing is mainly due to phosphorylation. In addition, since the newly synthesized MLLC band broadens over time after chasing, it appears that phosphorylation occurs after MLL is processed.
MLLC is highly phosphorylated.
(A) MLL is phosphorylated in vivo. BOSC23 cells were transfected with the empty pLNCX vector (vector) or pLNCX MLL(H) [denoted as MLL(H)]. 48 hours after transfection, the cells were metabolically labeled with [32P]-orthophosphate for 3 hours and lysed in lysis buffer. MLLC was immunoprecipitated with the anti-HA antibody 3F10. The precipitates were analyzed by immunoblot with anti-MLL N-terminal antibody (mmN3; left panel) or anti-HA antibody 3F10 (middle panel) or visualized by autoradiography (right panel). Molecular weight markers are on the left. (B) Effects of phosphatase treatment on the mobility of MLLC. BOSC23 cells were transfected with the empty pLNCX vector or pLNCX MLL(H). The cells were lysed in lysis buffer 48 hours after transfection. MLL proteins were immunoprecipitated with the anti-MLL N-terminal antibody (rpN1). Immunoprecipitates were treated with or without calf intestine alkaline phosphatase for 30 minutes and subjected to immunoblotting with the anti-HA antibody 3F10.
MLLC is highly phosphorylated.
(A) MLL is phosphorylated in vivo. BOSC23 cells were transfected with the empty pLNCX vector (vector) or pLNCX MLL(H) [denoted as MLL(H)]. 48 hours after transfection, the cells were metabolically labeled with [32P]-orthophosphate for 3 hours and lysed in lysis buffer. MLLC was immunoprecipitated with the anti-HA antibody 3F10. The precipitates were analyzed by immunoblot with anti-MLL N-terminal antibody (mmN3; left panel) or anti-HA antibody 3F10 (middle panel) or visualized by autoradiography (right panel). Molecular weight markers are on the left. (B) Effects of phosphatase treatment on the mobility of MLLC. BOSC23 cells were transfected with the empty pLNCX vector or pLNCX MLL(H). The cells were lysed in lysis buffer 48 hours after transfection. MLL proteins were immunoprecipitated with the anti-MLL N-terminal antibody (rpN1). Immunoprecipitates were treated with or without calf intestine alkaline phosphatase for 30 minutes and subjected to immunoblotting with the anti-HA antibody 3F10.
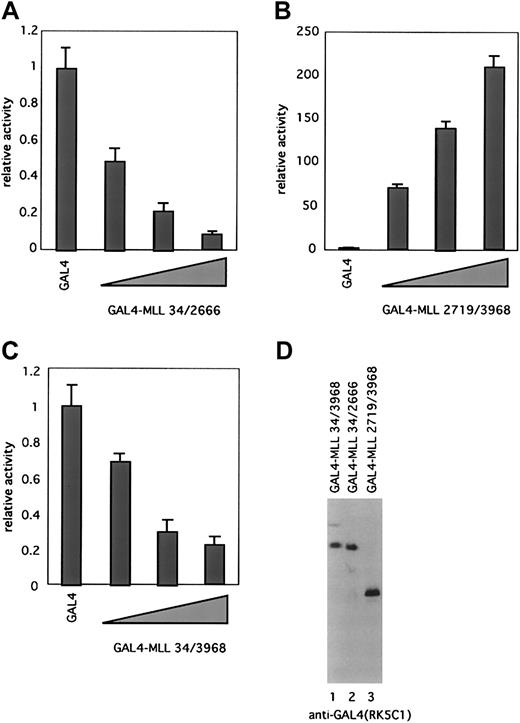
MLLN and MLLC display divergent transcriptional properties
To investigate the role of MLLN and MLLCin transcription, we performed transactivation assays with GAL4 fusion proteins containing either full-length MLL (residues 34-3968), MLLN (residues 34-2666), or MLLC (residues 2719-3968). These were expressed transiently in BOSC23 cells together with a reporter construct containing 5 tandem GAL4-binding elements upstream of a minimal E1B promoter. Equivalent expression of fusion proteins was confirmed by immunoblotting with the anti-GAL4 antibody (Figure 7D). Transactivation activity was measured by the luciferase assay. MLLN (GAL4-MLL 34/2666) displayed a remarkable ability to repress transcription in a dose-dependent fashion (Figure 7A). In contrast, MLLC(GAL4-MLL 2719/3968) displayed strong transactivation activity (Figure7B). These results are consistent with previous data showing that the MT homology domain of MLL (within MLLN) possesses repression activity, whereas a transcriptional activation domain maps to a region between amino acids 2829 to 2883 in the portion of MLL that we have isolated as MLLC.28 29 Full-length MLL showed an intermediate transcriptional property with weak repression activity (Figure 7C).
Processing divides MLL into fragments with opposite transcriptional properties.
(A-C) Transcriptional properties of MLL and its cleavage products were evaluated as GAL4 fusion proteins. BOSC23 cells were transiently cotransfected with pFR-luc, pRL-tk, and expression vectors encoding GAL4-MLL 34/2666 (A), GAL4-MLL2719/3968 (B), or GAL4-MLL34/3968 (C). The cells were lysed 48 hours later and assayed for luciferase activity. The activity of each expression vector was divided by the luciferase activity of pLNCX-GAL4 to give the fold-activation. Results are presented as the averages ± SD of 2 independent experiments, each of which was performed in duplicate. (D) Expression of GAL4-MLL fusion proteins. BOSC23 cells were transfected with expression vectors indicated above the gel lanes, harvested 48 hours later, and analyzed by immunoblot with the anti-GAL4 antibody.
Processing divides MLL into fragments with opposite transcriptional properties.
(A-C) Transcriptional properties of MLL and its cleavage products were evaluated as GAL4 fusion proteins. BOSC23 cells were transiently cotransfected with pFR-luc, pRL-tk, and expression vectors encoding GAL4-MLL 34/2666 (A), GAL4-MLL2719/3968 (B), or GAL4-MLL34/3968 (C). The cells were lysed 48 hours later and assayed for luciferase activity. The activity of each expression vector was divided by the luciferase activity of pLNCX-GAL4 to give the fold-activation. Results are presented as the averages ± SD of 2 independent experiments, each of which was performed in duplicate. (D) Expression of GAL4-MLL fusion proteins. BOSC23 cells were transfected with expression vectors indicated above the gel lanes, harvested 48 hours later, and analyzed by immunoblot with the anti-GAL4 antibody.
Discussion
MLL is proteolytically processed
MLL is a large protein with an estimated molecular mass of 431 kDa. We show here that MLL is proteolytically processed in all cell lines examined and that this processing results in 2 large fragments whose amino acid sequences predict molecular weights of 297 kDa and 134 kDa, although the latter migrates anomalously in SDS-PAGE at 180 kDa. Cleavage sites are located at D2666 and D2718, with D2718 being the major site used in BOSC cells. A comparison of sequences at the 2 sites predicts a possible consensus sequence for processing as QXD/GZDD (X is a hydrophobic amino acid, Z is alanine or valine). This sequence is conserved in MLL homologs of mouse, pufferfish (Fugu rubripes), Drosophila, and another human MLL family member, MLL2 (Figure 2B).25,40 41 The consensus sequence is not found in other proteins currently present in data bases, suggesting that a hypothetical processing enzyme could be specific for MLL family proteins.
Previous studies raised the possibility that TRX, the Drosophilahomolog of MLL, may be cleaved under some conditions.20 Although the cleavage site in TRX has not been biochemically determined, an amino acid sequence that matches the predicted consensus sequence for MLL processing was found in TRX at residues 2264 to 2275 (see Figure 2B). This prediction is supported by the observation that the TRX protein of trxE3mutant embryos, in which residues 2164 to 2443 have been deleted, is not processed.20 Thus, TRX seems to be processed proteolytically in the same fashion and at similar sites as MLL, suggesting that this processing event is evolutionally conserved. ThetrxE3 mutation is also associated with abnormal expression of homeotic genes and lethality in homozygotes,21,42-44 which suggests that the region deleted in the trxE3 mutant is required for proper regulation of homeotic gene expression by TRX. Since the deleted region encompasses 280 amino acids that include not only the putative processing sites but also a presumptive zinc (Zn)–finger domain and several other potential functional domains,44 it is not clear if the phenotype conferred by the trxE3mutation can be attributed simply to the lack of processing. However, it remains possible that processing of TRX and MLL is critical for their function and that abrogation of processing could cause aberrant development and embryonic lethality.
The role of TRX processing in development is suggested by the phenotypic differences between the trxE3 mutant and the trxB11 mutant, which lacks almost all of its TRX protein due to a frame shift mutation.21 In the embryos of both mutants, the expression of ANT-C genes such asScr and Antp is reduced in similar ways.21,42 In contrast, the trxE3mutation has no effect on the expression of BX-C genes in the embryonic nervous system, whereas this expression is reduced in thetrxB11 mutant. Supporting this is that in late homozygous trxB11 embryos, the first and third midgut constrictions are shifted posteriorly, whereas no substantial changes can be seen in trxE3embryos.21,42 Thus, it can be concluded that the region deleted in trxE3 is not required to maintain the level of BX-C transcripts but is necessary to maintain the expression of ANT-C genes.21 This suggests that TRX regulates ANT-C gene expression via a mechanism that is different from that used for BX-C gene expression.21 It is possible that the former mechanism requires TRX processing, whereas the latter does not.
We found by immunoprecipitation purification experiments that the N- and C-terminal fragments resulting from MLL processing interact with each other. Reciprocal immunoprecipitation analysis verified that this interaction occurs. These observations suggest that after MLLN and MLLC are generated by proteolytic cleavage, they remain in a complex. The interaction domains that we refer to as NIID and CIID contain evolutionary conserved sequences such as PHD fingers and a portion of ATA2.40 41 Therefore, the interaction between N- and C-terminal fragments of MLL/TRX family proteins is likely to be evolutionary conserved. In addition, loss of either NIID or CIID results in a decrease in the processing efficiency of MLL (Figure 4B,E).
MLL processing is followed by phosphorylation
Our pulse/chase experiments showed that MLL processing occurs, at the earliest, within a few hours of translation. Following processing, MLLC is phosphorylated by unknown mechanisms. However, formation of an MLLN/MLLC complex seems to be one of the requisites for phosphorylation of MLLC. As shown in Figure 4D, free uncomplexed MLLC displayed a faster migration and sharper resolution in denaturing gels, which suggests the relative absence of phosphorylation. The functional consequences of MLLCphosphorylation are not known. However, a candidate MLL-interacting protein, Sbf1, which also forms a biochemical complex with TRX, has been implicated in the regulation of phosphorylation. Sbf1 interacts with the SET domain13 at the C-terminus of MLLC. Sbf1 is related to myotubularin, a lipid and dual-specificity protein phosphatase, but Sbf1 has a leucine substitution of a critical cysteine residue in the phosphatase signature motif, resulting in the absence of phosphatase activity. As a consequence, Sbf1 is proposed to antagonize the activity of phosphatases (possibly myotubularin). Thus, myotubularin and Sbf1 may be involved in regulation of MLL phosphorylation. Since Sbf1 mutants inhibit myoblast differentiation and stimulate the growth of B-cell precursors,13 45 MLLC phosphorylation may play an important role in regulating differentiation and growth control.
MLL processing generates functionally distinct transcriptional modules
Why is it necessary for MLL to be proteolytically processed into 2 fragments that otherwise interact with each other? One possibility is that the observed processing is simply an initial step in a destruction pathway, perhaps as a consequence of hyperexpression. We believe that this is unlikely since the processed MLLN and MLLC polypeptides are the major endogenous forms of MLL in all cell lines examined and are much more stable (half-lives of at least several days) compared to full-length MLL (half-life of a few hours). We propose that MLL processing may regulate its transcriptional properties, which appear to be bifunctional. MLLN contains the AT hook region, which is a putative DNA binding domain, and also the MT domain, which has strong transcriptional repression activity.28 29 We found that MLLN indeed has repressive activity. On the other hand, MLLC has strong transactivation properties. Therefore, processing divides MLL into 2 noncovalently interacting fragments that possess opposite transcriptional properties. The intact MLL protein, as a complex of MLLN and MLLC, displays intermediate activity that continues to read out as mild repression on the promoter and conditions used for our experiments. These data suggest that the relative stoichiometry of MLLC and MLLN may modulate the expression state of MLL target genes, a possibility that warrants further investigation.
We thank Dr A. D. Miller for the LNCX vector, Dr D. Baltimore for the BOSC23 cells, and Dr H. Hirai for the partial cDNA of MLL. We also are grateful to S. Mitani, M. Mori, C. Hatanaka, and Y. Aikawa for technical assistance.
Prepublished online as Blood First Edition Paper, June 28, 2002; DOI 10.1182/blood-2002-04-1015.
Supported in part by Grant-in-Aids for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology; and in part by grants from the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research of Japan; and a grant from the National Institutes of Health (CA55029).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Issay Kitabayashi, Chromatin Function in Leukemogenesis Project, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo 104-0045, Japan; e-mail:ikitabay@gan2.ncc.go.jp.

![Fig. 1. MLL is proteolytically processed in vivo. / (A) Expression of MLL proteins in various cell lines. Cell lysates were analyzed by immunoblot with anti-MLL N-terminal (rpN1, mmN3) or C-terminal antibodies (mmC2, mmC3) or antibodies specific for p300 (RW128) or MOZ (N13-2), as denoted beneath the panels. Identities of cell lines are indicated above the gel lanes. The band corresponding to predicted 430-kDa full-length MLL is indicated by a plus mark. Bands corresponding to MLL N- and C-terminal fragments, p300 and MOZ, are indicated by asterisks. Migration of molecular weight standards is shown on the left. (B) BOSC23 cells were transfected with empty pLNCX vector (vector) or pLNCX expressing full-length MLL with an HA tag at its C-terminus [MLL(H)]. Cell lysates were analyzed by immunoblot with anti-MLL N-terminal antibodies (rpN1, mmN3, mmN4, mmN6), anti-HA antibody 3F10, or anti-MLL C-terminal antibodies (mmC2, mmC3). (C) Expression of MLL and MLL-AF6 in HL-60 and ML-1 cells. Cell lysates were analyzed by immunoblot with anti-MLL N-terminal antibody (rpN1). MLL N-terminal fragment and MLL-AF6 are indicated by arrows. (D) Expression of MLL and MLL-p300 in transiently transfected BOSC23 cells. Following transfection with the constructs indicated above the gel lanes, cell lysates were analyzed by immunoblot with anti-MLL N-terminal antibody (mmN3). MLL N-terminal fragment and MLL-p300 are indicated by arrows.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/10/10.1182_blood-2002-04-1015/4/m_h82223465001.jpeg?Expires=1769091364&Signature=vsCyNCAEJ1o3kxDKSSKSQqe8Fk5OIuth6melDSygdIvurEOQ2-CNxfzcb24lQdUSQxQCF2PdI5jcqi2yTF39L196fPdwS2ZFGTAOVi~vHh6wghf0L-iIFwXTf48XnLBXfSS3pnLDjcWo-U673iM-h9x4V0NabD4xpgwNRS9MSSGePPsFlSdlm4lHbeIa9gOkzcPFEZ42tSBuG3QKByt9MzgWAYfCs5aYkfobNEqTK0jEWx~K8Q4d9PW2wZl3JAq~hC4wS-I-1Ji-Q3B6pNDwnVzYeZdOKfuiOT93jThWXlQVBDNW~lgT7XmWQE3Nsu-NpJr4vydSvNJIowajMyKZtw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Identification of MLL processing sites. / (A) Purification of MLL proteins. BOSC23 cells were transfected with an MLL-expression vector [pLNCX MLL(H)], lysed 48 hours later, and subjected to immunoprecipitation with an anti-HA antibody 3F10. An aliquot of the immunoprecipitate was separated in a denaturing gel and stained by the silver stain method. Full-length MLL, the purified C-terminal 180-kDa fragment (MLLC), and the copurified 300-kDa band are indicated. Migration of molecular weight markers is indicated on the left. (B) Amino acid sequences of MLL processing sites. The sequences around the 2 processing sites of human MLL (hMLL) and putative processing sites of mouse MLL (mMLL), pufferfish MLL (fMLL), Drosophila TRX (dTRX), and human MLL2 (hML2) are aligned.12254041 Conserved amino acids are shaded and marked with an asterisk. The processing site is indicated by an arrow in the consensus sequence below. The numbers refer to positions relative to the first amino acid of each protein. (C) Inhibition of processing by mutations in the cleavage sites. BOSC23 cells were transfected with expression constructs encoding proteins indicated above the gel lanes and harvested 48 hours after transfection. All MLL proteins were tagged with HA at their C-termini. The cell lysates were analyzed by immunoblot with anti-HA antibody (left panel) or anti-MLL N-terminal antibody (right panel).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/10/10.1182_blood-2002-04-1015/4/m_h82223465002.jpeg?Expires=1769091364&Signature=jCYWX-feGgr1b3pk7k1WLUb1xHEjojoB-8rzHHEsOlY3QDjPeHgjpMV3mvhQLo~lz7Gv9yba72HltGqjHtoaCHIbpjaJSieH86gExa2sqPZeRxq0wZ-AMvP4cF9iuiKZlHzKLSCRbJRO4G5raSVYE50GJKvXotkUUNuJRzfQCsqIlBdq7JlwzmclAItnSiVPpFtc5Wil8hJvE7nQkGYATHycUxUXB9o-XZTFlfK0Nqag1ESFBRQlNTK8Dlm0SewsKrzruShQI7LT~Fp-7h3Wnfb5a9iLCtRM5iiz2KfhP85zvy8ilW9xJ3SZOeVjWScUoM247zs-sqFAwnV6k0~-YQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. N-terminal MLL interacts with C-terminal MLL. / Copurification of MLLC with MLLN. (A) BOSC23 cells were transfected with pLNCX vectors encoding MLL or the DDAA mutant tagged with FLAG at their N-termini. Proteins were immunoprecipitated with anti-FLAG antibody, eluted by incubation with FLAG peptide, separated in a denaturing gel, and stained with Coomassie brilliant blue (CBB). The purified fragments are indicated by asterisks (MLLN and MLLC in lane 1, unprocessed MLL in lane 2). Molecular weight markers are on the left. (B) BOSC23 cells were transfected with pLNCX vector alone or pLNCX encoding MLL protein tagged with His and FLAG at its N-terminus and HA at its C-terminus [denoted (Hi-FL)MLL(HA)]. Cell lysates were immunoprecipitated with anti-FLAG antibody or anti-HA antibody. The whole cell lysates (WCE) and the immunoprecipitates (denoted as IP:anti-HA and IP:anti-FL) were analyzed by immunoblot with anti-His antibody (left panel) or anti-HA antibody (right panel). The purified fragments are indicated by asterisks (MLLN in left panel, MLLC in right panel). (C) Interaction of endogenous MLLN and MLLC. Cell lysates were immunoprecipitated with nonspecific mouse IgG, mouse anti-MLL N-terminal (mmN4), or anti-MLL C-terminal (mmC2) antibodies. The precipitates were analyzed by immunoblot with a mouse anti-MLL C-terminal (mmC2) antibody. MLLC is indicated by an asterisk.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/10/10.1182_blood-2002-04-1015/4/m_h82223465003.jpeg?Expires=1769091364&Signature=YwsxxoLd8VS8TrS~CR-C7fMJJRr4wpp1XwwHbPcdddN-bSUMO2QuZupG7M5jUsEL6xorsJyY5L0q5aDhc9KghkQFVzO9LleKgRHu~jcL4ncygi~S7zsfbt68Zvyp1d~e2hhh8SYDLRf-f~z7znyukxNuE3WZ~OH2OgFVRn9Lr34dv-m-kyH-S-lUwaeZ~FqHpgl3tkFxApYDYqBzST-hR2rr46FQPRutPhe2-1rc4NWgu8s5c6gPZBMBtN~45erSi0DfawzOuj0Xi4IrvLYrnaOYk5XH3Gt7m3MsFIuPc9vNuclLG3iOXfkRNkESz6z7rmsuSciUzO5tyfRTr1g~3w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. MLL processing occurs within a few hours of its translation. / BOSC23 cells were transfected with empty vector (vector) or a construct encoding full-length MLL tagged with HA at its C-terminus [MLL(H)]. The cells were metabolically labeled with [35S]-methionine for 3 hours. After labeling, the cells were cultured in nonradioactive medium for the indicated time periods and then lysed in lysis buffer. MLL proteins were immunoprecipitated with anti-MLL antibody, separated in denaturing gels, and visualized by autoradiography (A and B) or immunoblotted with anti-HA (C and D) or anti-MLL N-terminal (E and F) antibodies. In the left panels (A, C, and E), the cells were harvested at 0, 3, and 9 hours after labeling. In the right panels (B, D, and F), the cells were harvested at 0, 1, and 2 days after labeling. Unprocessed MLL, the N-terminal fragment, and the C-terminal fragment are indicated by arrows.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/10/10.1182_blood-2002-04-1015/4/m_h82223465005.jpeg?Expires=1769091364&Signature=k0Af-lF15mgrhCKukjAmXsPCiBecGHuS1DgmYIelK67xA~wY21bD0PGIdIUdqmjtPkbrPgL9hsglnUDLJFLu8oLO7Sdb1~fdtBCpzg439Yy-jmECTIgMOlsQSVCE0eqeM30xbdIKXMBbBB82m9LzpGSn8W6JO9iN1xYDI8MhVP405-BS0ivhmQm4S07LudrQd3UMzd1UluXesUqZXafDTTt7JKivM6yG4pjazFeCT9Fb4GFaKgWi7NpbRjZ9IqQaYa7axzfnkbgSByY-eW~zokWbjgxhrdoRyZmavkZUxV37HCOvbocSrbK60KqI0~YxQ4OXkjtvfkNxv1hSQ00R3g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. MLLC is highly phosphorylated. / (A) MLL is phosphorylated in vivo. BOSC23 cells were transfected with the empty pLNCX vector (vector) or pLNCX MLL(H) [denoted as MLL(H)]. 48 hours after transfection, the cells were metabolically labeled with [32P]-orthophosphate for 3 hours and lysed in lysis buffer. MLLC was immunoprecipitated with the anti-HA antibody 3F10. The precipitates were analyzed by immunoblot with anti-MLL N-terminal antibody (mmN3; left panel) or anti-HA antibody 3F10 (middle panel) or visualized by autoradiography (right panel). Molecular weight markers are on the left. (B) Effects of phosphatase treatment on the mobility of MLLC. BOSC23 cells were transfected with the empty pLNCX vector or pLNCX MLL(H). The cells were lysed in lysis buffer 48 hours after transfection. MLL proteins were immunoprecipitated with the anti-MLL N-terminal antibody (rpN1). Immunoprecipitates were treated with or without calf intestine alkaline phosphatase for 30 minutes and subjected to immunoblotting with the anti-HA antibody 3F10.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/10/10.1182_blood-2002-04-1015/4/m_h82223465006.jpeg?Expires=1769091364&Signature=IHovYlAISjIt2BBEnC-ymTpBuSeWorehxqNdgZh9OgYmfqRGVYfIXJqJy3kmxuif5y-3EIiaGHfQB6ay3psFOvlCCp~v7o43hpRG5AZcG001nDf4~EhyNaaHoS8aaRohFHI9b~rTf-UUPAczGi5BRutlNBblYbd50AkmAZCn~O~OF~6pkgLF4Idz8WOsVLQxAzDzvig32u79-jgc-NLuyXnzqdfGOFoiLD6wWTFUWPbL3ZE4JWNEIrw3KzOGKnIjLMjDHa~PdsGZKPZG2mtVqnzSpV8~4tTsWeCXS26ki5~86kHRDMceWCi3c5XMAo7kG8iFQcornppyIwSBdWcokQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal