Natural deletions of the region upstream of the human α-globin gene cluster, together with expression studies in cell lines and transgenic mice, identified a single element (HS −40) as necessary and perhaps sufficient for high-level expression of the α-globin genes. A similar element occupies the corresponding position upstream of the mouse (m) α-globin genes (mHS −26) and was thought to have similar functional properties. We knocked out mHS −26 by homologous recombination and observed the surprising result that instead of the expected severe α-thalassemia phenotype, the mice had a mild disease. Transcription levels of the mouse genes were reduced by about 50%, but homozygotes were healthy, with normal hemoglobin levels and only mild decreases in mean corpuscular volume and mean corpuscular hemoglobin. These results may indicate differences in the regulation of the α-globin clusters in mice and humans or that additionalcis-acting elements remain to be characterized in one or both clusters.
Introduction
The human α-globin gene cluster includes an embryonic ζ gene and 2 fetal/adult α genes lying close to the telomere of the short arm of chromosome 16 (telomere-ζ–α2-α1-centromere). This telomeric region is gene dense, and although the α genes are transcribed in an erythroid-specific manner, they are closely flanked by widely expressed genes.1,2 Extensive characterization of the chromatin structure of this region identified 4 erythroid-specific DNase I hypersensitive (HS) sites located 8 (HS −8), 10 (HS −10), 33 (HS −33), and 40 (HS −40) kilobases (kb) upstream of the ζ-globin messenger RNA (mRNA) cap site.3 Two of these sites, HS −33 and HS −40, actually lie within introns of one of the upstream, widely expressed genes (C16orf 35).
We previously described 9 patients who have inherited chromosomes with deletions that removed about 35 to 160 kb of the region between the α cluster and the telomere but left the structural α genes intact4 (D.R.H., unpublished data). All these patients have α thalassemia, and their phenotypes are consistent with severe down-regulation (< 1%-2%) of α gene expression from the affected chromosome. This suggested that the region lying between the α cluster and the telomere contains long-range, cis-acting elements that regulate α-globin gene expression. The smallest region of overlap between the upstream deletions that cause α thalassemia extends for 20.4 kb and includes HS −40 and HS −334(D.R.H., unpublished data).
Analysis of small (4-10 kb), overlapping segments of this region in stable transfectants of MEL cells or in transgenic mice showed that only HS −40 consistently acts as an enhancer of ζ- and α-globin expression.3,5-7 Even in combination, the other HS sites appear not to add substantially to this effect. Although a large (150 kb) fragment spanning all the HS sites and the α-globin cluster is expressed at relatively high levels (22%-66%) in a developmentally stable manner,4 no fragment has so far been found consistently to provide fully regulated expression of the human α cluster in the erythroid cells of transgenic mice.
Copies of a normal chromosome 16 transferred to a MEL host cell (the equivalent of early proerythroblasts) produced stable interspecific hybrids that expressed each human α gene at levels 12% to 56% that achieved by endogenous mouse α genes after induction of terminal erythroid differentiation.4 Deletion of a 1-kb fragment containing HS −40 from the human chromosome by homologous recombination severely down-regulated human α-globin gene expression in this system.8 The α genes on chromosomes from the patients with deletions of the upstream region flanking the α cluster were also expressed at very low levels (< 1%) or not at all in MEL cell hybrids.9
These results are all consistent with the HS −40 element being not only necessary but possibly sufficient to regulate human α-globin gene expression. However, the smallest of the individual upstream deletions resulting in α thalassemia is about 35 kb and the region of minimum overlap is about 20 kb, so clearly these deletions remove more than just the HS −40 element. Furthermore, expression studies in cell lines or transgenic mice are frequently subject to integration effects that may yield misleading results. Finally, analyses of intact chromosomes in hybrid cells may not re-create the normal epigenetic modifications that occur during normal development and hence could also be misleading. Only finding a discrete deletion of the HS −40 element as a natural mutation would show that this element alone is sufficient for full α-globin gene regulation.
We previously showed that the human and mouse α-globin clusters are contained within a region of conserved synteny extending for 150 kb, including the globin genes and 5 genes on the telomeric side of the cluster.10 Small segments of noncoding sequence coinciding with human HS −4 and HS −33 are also conserved between mice and humans.10 The sequence corresponding to HS −40 lies 26 kb upstream of the mouse (m) ζ gene (mHS −26) and has generally been considered to be structurally and functionally equivalent to HS −40.11,12 This mouse element also lies in an intron of the mouse orthologue of C16orf35. The core of HS −40 spans 350 base pairs (bp) and includes binding sites for GATA-1, Nfe2 (or related proteins), and 4 potential CACC boxes that bind ubiquitously expressed proteins. Several of these sites are conserved in the mouse mHS −26 element,11,13 which has also been shown to activate α-globin genes in transfected erythroid cells and transgenic mice.11 12
In this study, we deleted the mHS −26 element by homologous recombination and found that in contrast to deletion of HS −40 from the human α-globin cluster, deletion of the orthologous region in mice causes only a mild down-regulation of α gene expression and mild α thalassemia. This may indicate that there are important differences in the regulation of the human and mouse α-globin clusters and that additional cis-acting elements remain to be characterized in one or both clusters.
Materials and methods
Construction of targeting plasmid
Sequences flanking upstream (left arm, 3392 bp) and downstream (right arm, 3675 bp) of mHS −26 were produced by the Expand high-fidelity polymerase chain reaction (PCR) system (Boehringer Mannheim, Germany) from 129-strain DNA by using the following primers: left-arm forward BamHI (5′-CGCGGATCCACCTGGAGAAATGAGACCTTACCC-3′), left-arm reverseNotI (5′-AAACCACCAAAGCGGCCGCAATGTGTGTTCCCCATAGAGGACTC-3′), right-arm forward NotI (5′-AAACCACCAAAGCGGCCGCCCATCACTTGGGAGGTAGAAG-3′), and right-arm reverse MunI (5′-CCGCCAATTGCAAATCTGAAATGTGCGGC-3′), which include restriction enzyme sites (underlined). The PCR products were digested, cloned in pBluescript II SK (+/−) (Stratagene, La Jolla, CA), and sequenced. A phosphoglycerate kinase (PGK)–neomycin positive selectable marker gene flanked by loxP sites and a negative selection cassette, containing the herpes simplex virus thymidine kinase dimer with F9 enhancer and thymidine kinase promoter, were included in the construct (Figure 1).
Mapping of mouse α-globin gene locus.
Panel A shows a map of the mouse α-globin gene cluster, showing the positions of the genes and the regulatory element (HS −26) lying within an intron of the C16orf35 gene. The structures of the targeting vector, the floxed allele after homologous recombination, and the KO allele after expression of Cre recombinase are also shown. The positions of the 5′ and 3′ probes used for mapping are indicated as small black boxes. The band sizes using AflII (A) andBamHI (B) digestion are indicated. Panel B shows a Southern blot analysis of AflII- or BamHI-digested DNA from a WT mouse and heterozygotes for the floxed and KO alleles. The positions of markers (SJ5000, Amersham Pharmacia, Little Chalfont, United Kingdom) are shown along side the gels.
Mapping of mouse α-globin gene locus.
Panel A shows a map of the mouse α-globin gene cluster, showing the positions of the genes and the regulatory element (HS −26) lying within an intron of the C16orf35 gene. The structures of the targeting vector, the floxed allele after homologous recombination, and the KO allele after expression of Cre recombinase are also shown. The positions of the 5′ and 3′ probes used for mapping are indicated as small black boxes. The band sizes using AflII (A) andBamHI (B) digestion are indicated. Panel B shows a Southern blot analysis of AflII- or BamHI-digested DNA from a WT mouse and heterozygotes for the floxed and KO alleles. The positions of markers (SJ5000, Amersham Pharmacia, Little Chalfont, United Kingdom) are shown along side the gels.
Isolation of homologous recombinant clones
The plasmid (150 mg) was linearized and electroporated into 2 × 108 mouse E14Tg2a embryonic stem (ES) cells. Colonies resistant to G418 and ganciclovir were isolated. These were screened by Southern blot analysis for homologous recombinants. DNA was isolated and digested with BamHI and hybridized with appropriate probes (Figure 1) produced by PCR using the following primers: 82759-F 5′-TCTTGGTCGCTCTCCTTACAGC-3′, 83564-R 5′-GTTATCTTTCCCGCACTGGATG-3′, 95915-F 5′-ACATCTTTGACCCCAGTCCCTC-3′, and 96763-R 5′-AACTGCATGTT GACCATAGCCAG-3′. ES cells homozygous for the homologous recombination were produced by the G418 step-up technique.14
Generation of chimeras and floxed and knockout (KO) mice
Targeted ES cells with a normal karyotype were aggregated with blastocysts15 and transferred into pseudo-pregnant recipients. Germline transmission was obtained from chimeras derived from one clone. Removal of the neomycin gene insert was obtained by mating floxed heterozygous mice with mice carrying Cre recombinase driven by a GATA-1 promoter16 that expresses the recombinase very early in development. Southern blot analysis was used to assess the recombination event on DNA digested with AflII and labeled with the 3′ probe.
Cytologic and hematologic analysis
Hematologic variables were assessed by using an ABX Micros 60 counter (Block Scientific, Englewood, NJ). Slides were stained with May-Grünwald-Giemsa stain (Sigma, St Louis, MO).
DNA and RNA analysis
DNA was extracted with phenol and chloroform and analyzed by Southern blotting using standard techniques. Total RNA was prepared with TRI reagent (Sigma). RNase protection assays were performed as described previously3 by using α-phosphorus 32–guanosine triphosphate–labeled RNA probes for the mouse α-, β-, and ζ-globin genes (SP6/T7 transcription kit; Roche, Germany); 1 × 106 counts/minute was hybridized overnight with 0.16 to 1 μg total RNA.
DNase I HS sites
DNase I HS assays were performed on mouse erythroblasts obtained from the spleens of phenylhydrazine-treated adult mice17and L929 mouse fibroblasts, as described previously.3 The DNA was digested with appropriate restriction enzymes. Probes were produced by PCR with the following primers: 82758-F × 83564-R, 95915-F × 96763-R (see above), and 68959-F 5′-TCACCTTCTGAGCCTCTCCACTTC-3′; 68965-R 5′-CATCTGGGTTTTCCTTGACCG-3′; 102509-F 5′-AAGCCTATGCTGCCTC TTACTAACC-3′; 103005-R 5′-TGATGACCAGGACGGTGACTCC-3′; 73945-F 5′-AGCAGGTTCTCTGTCATCCTCTTG-3′; 74822-R 5′-GCATACCTATGTTTGCCTAATG GC-3′; 141886-F 5′-TTCTGCTGAGGTCTGAGATGGG-3′; 142293-R 5′-GCTCCATTCTTCATCACTGCATG-3′; 111354-F 5′-TCACACCAGTCGCAGAAATGC-3′; 112052-R 5′-GCCAGGGCTATCCTATGTAGAAAGC-3′; 55112-F 5′-TATT CTGTCCCCTTACCCCAA TC-3′; 56253-R 5′-CATAACCTACACTCCCAGGCTTGTC-3′; 55112-F 5′-TATTCTGTCCCCTTACCCCAATC-3′; and 56253-R 5′-CATAACCTACACTCCCAGGCTTGTC-3′ (numbers refer to coordinates in the mouse cluster [GenBank accession nos. AY016021 and AY016022]).
Analysis of C16orf35 gene expression
Total RNA was prepared with TRI reagent (Sigma). For Northern blot analysis, mRNA was selected with a PolyATtract mRNA isolation system III (Promega, Madison, WI) from 200 μg fetal liver and adult spleen RNA and from 500 mg total RNA from a pool of nonhematopoietic tissues. The polyA+ RNA was electrophoresed through a 1% agarose denaturing formaldehyde gel, blotted on a nylon membrane (Nytran N; Schleicher & Schuell, Germany), and hybridized with probes for mouse C16orf35 exon 12, obtained by PCR (Ex12-F × Exon 12-R), and human C16orf35 gene complementary DNA (cDNA) as described peviously.8 The membranes were stripped in boiling 0.5% sodium dodecyl sulfate and 0.1 × SSPE (15 mM NaCl, 1 mM NaH2PO4, 0.1 mM EDTA) and labeled with a β-actin cDNA probe (Clontech, Palo Alto, CA) as a loading control.
Construction of an HS −26 α-globin gene construct
The HS −26 α-globin gene construct was made in a single ligation reaction combining 3 fragments: (1) an 830-bpEcoRI-PvuII fragment containing HS −26 cloned into pZEr02K (Invitrogen, San Diego, CA) and released withNotI and XhoI; (2) a 577-bpNotI-NcoI fragment containing the mouse α-globin gene promoter and sequences up to the ATG translation start site; and (3) a 1.37-kb NcoI-XhoI fragment from the human α2 gene starting at the ATG translational start site and extending beyond the 3′ untranslated region. Ligation was performed by using a rapid ligation kit (Roche), with transformation into XL10 Gold cells (Stratagene). A 2.79-kb EcoRI-XhoI fragment containing the HS −26 α construct was gel purified before electroporation into MEL cells.
Results
Mice in which a floxed neomycin gene replaces the mouse α-globin regulatory element (mHS −26) have a severe form of α thalassemia
We used homologous recombination to replace a 1312-bp fragment containing mHS −26 with a PGK-neomycin gene flanked by loxP sites (Figure 1A). Colonies resistant to G418 and ganciclovir were obtained, and analysis of 96 colonies identified 5 that had been targeted correctly (Figure 1B), with removal of all elements within mHS −26 previously shown to bind transcription factors in vitro.11 Increasing the G418 concentration in the ES cell cultures resulted in production of several clones homozygous for the homologous recombinant; in vitro erythroid differentiation18 of these clones followed by RNA analysis yielded α + ζ/ε + βh1 + β mRNA ratios that were about 25% those of nonmanipulated ES cells (data not shown). Two correctly targeted ES cell clones with normal male karyotypes were aggregated with blastocysts to produce chimeric mice, one of which was used to obtain germ line transmission (floxed mHS −26+/−).
Hematologic analysis of adult floxed mHS −26+/−heterozygotes showed anisocytosis and poikilocytosis, with a significant reduction in levels of hemoglobin (Hb), mean corpuscular volume (MCV), and mean corpuscular hemoglobin (MCH) compared with those in normal littermates (Figure 2). The ratio of α-globin RNA to β-globin RNA was reduced to approximately 70.1% ± 14.2% of that observed in normal mice, consistent with the presence of α thalassemia in these mice (Figure3). Matings of heterozygous mice produced no live-born homozygotes, and analysis of timed pregnancies showed that the homozygotes died between 13.5 and 15.5 days after conception. Homozygous fetuses were recognizable from their obvious pallor resulting from severe anemia. Blood films from these mice showed marked abnormalities (Figure 2), and the fetal livers were reduced in size, with considerable dyserythropoiesis. The α/β-globin RNA ratio in these homozygotes was 35.8% ± 5.4%, whereas it was 60% ± 14.3% in heterozygous embryos and 100% ± 16.5% in normal embryonic littermates (Figure 3).
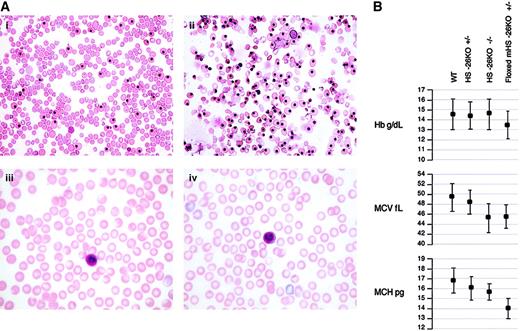
Hematologic analysis and data.
(A) Photomicrographs of 14.5-day fetal blood from a WT control (i), 14.5-day fetal blood from a floxed homozygote with marked dyserythropoiesis and an increased portion of yolk sac erythroid cells (ii), peripheral blood from a normal adult (iii), and peripheral blood from a KO adult with essentially normal red cell morphologic features (iv). Original magnifications × 400 (i and ii) and × 1000 (iii and iv). (B) Hb, MCV, and MCH data for the various HS −26 genotypes compared with age-matched control mice.
Hematologic analysis and data.
(A) Photomicrographs of 14.5-day fetal blood from a WT control (i), 14.5-day fetal blood from a floxed homozygote with marked dyserythropoiesis and an increased portion of yolk sac erythroid cells (ii), peripheral blood from a normal adult (iii), and peripheral blood from a KO adult with essentially normal red cell morphologic features (iv). Original magnifications × 400 (i and ii) and × 1000 (iii and iv). (B) Hb, MCV, and MCH data for the various HS −26 genotypes compared with age-matched control mice.
The ratio of α-globin mRNA to β-globin mRNA.
(A) Data on α/β mRNA for the various HS −26 genotypes expressed as a percentage of values in normal control mice (set at 100%). Error bars represent 1 SD. (B) Results of RNase protection assay showing expression of mouse α and β mRNAs in peripheral blood from normal, heterozygous floxed, and homozygous floxed mice.
The ratio of α-globin mRNA to β-globin mRNA.
(A) Data on α/β mRNA for the various HS −26 genotypes expressed as a percentage of values in normal control mice (set at 100%). Error bars represent 1 SD. (B) Results of RNase protection assay showing expression of mouse α and β mRNAs in peripheral blood from normal, heterozygous floxed, and homozygous floxed mice.
Removal of the floxed neomycin gene partly restores α-globin expression
Previous studies showed that insertion of an expressed, selectable marker close to the α- or β-globin genes in their natural chromosomal environments can down-regulate their expression.19-22 This effect occurs when a neomycin gene is in either orientation with respect to the globin genes.21 To analyze the phenotype of mHS −26−/− without the interference from the neomycin gene, we crossbred floxed HS −26+/− mice with mice expressing Cre recombinase very early in development. Using Southern blot analysis, we identified mice in which the neomycin gene was deleted, leaving a single loxP site in the position originally containing HS −26 (Figure 1).
Heterozygotes for the mHS −26 KO had very mild changes in hematologic values, with slightly reduced MCV and MCH levels but essentially normal levels of Hb (Figure 2). Levels of α-globin mRNA were also minimally reduced in the peripheral blood (Figure 3). Surprisingly, matings of KO heterozygotes resulted in a normal proportion of homozygous offspring—close to the expected proportion of about 25% (17 of 77). There were no obvious abnormalities in the homozygotes, and females were fully fertile and had normal litter sizes. The homozygotes had normal hemoglobin levels, but their red cells had reduced MCV and MCH values compared with those in heterozygotes and normal controls (Figure 2). Cytologic examination of the spleens from 4 or 5 homozygotes (mHS −26−/−) showed a clear increase in erythroid precursors. The α/β-globin RNA ratios in the peripheral blood were only slightly reduced in embryos and adults missing both copies of mHS −26 (Figure 3) but were significantly reduced in fetal livers (67.1% ± 9.7%; n = 4), adult spleens (52.6% ± 10%; n = 12), and bone marrow (47.8% ± 17.4%; n = 6), suggesting that selection against cells with more severe globin-chain imbalance had ameliorated the effect of α thalassemia in the peripheral blood.
Analysis of ζ-globin gene expression during development of the homozygous KO embryos showed a reduction in the ζ/ζ + α ratio from 50% at 9.5 days of gestation to 10% at 13.5 days to zero at 15.5 days (data not shown); this is similar to the pattern observed in normal animals,7 suggesting that the loss of mHS −26 does not have a differential effect on the 2 genes.
Response of mHS −26−/− mice to phenylhydrazine
Because the defect in α-globin gene expression appeared to have been partly compensated by selection during erythropoiesis, we analyzed the ability of HS −26−/− mice to adapt to erythroid stress. We therefore treated homozygous mice with acetyl-phenylhydrazine, which produces a severe hemolytic anemia. Five days after treatment, adult KO mice had a lower level of Hb (125 versus 160 g/L), MCV (47 versus 53 fL), and MCH (20.8 versus 23.4 pg) than normal controls treated in the same way. The α/β-globin RNA ratios in the blood of the treated KO mice were also lower than those in treated normal mice, and spleen α/β-globin RNA ratios were similar to those in spleens of untreated mHS −26 KO mice. Together with the data presented above, these findings show that there was a more severe defect in α-globin RNA production in mHS −26−/− mice than was indicated by the very mild phenotype in the peripheral blood.
Changes in chromatin associated with removal of HS −26 from the mouse α-globin cluster
Given the unexpectedly mild hematologic phenotype of mHS −26−/− mice, it was important to confirm that no HS had formed in the recombined locus as a result of previously unidentified transcription factor binding sites being left intact after the homologous recombination event. To study this, erythroblast nuclei from wild-type (WT) and mHS −26−/− mice treated with acetyl-phenylhydrazine were prepared. After digestion with increasing concentrations of DNase I, DNA was subsequently isolated and analyzed by blot hybridization to detect DNase I HS elements. An HS corresponding to mHS −26 was clearly present in erythroblasts from normal mice but absent in those from KO mice (Figure4A), confirming that this regulatory element was inactivated in the chromosome.
HS mapping and C16orf35 gene expression.
(A) DNase I HS site mapping of the region around the HS −26 element in normal and homozygous HS −26 KO mice. A map of the area, with the position of the BamHI sites of the limit digest, is shown at the top. The bands in the KO sample are smaller than the normal bands because of the deletion of the HS −26 element. (B) Northern blot analysis of polyA+ RNA from embryos of WT mice and mice homozygous for the floxed HS −26 KO allele. The analysis used a probe for the C16orf35 gene and showed down-regulation of this gene in the floxed mice.
HS mapping and C16orf35 gene expression.
(A) DNase I HS site mapping of the region around the HS −26 element in normal and homozygous HS −26 KO mice. A map of the area, with the position of the BamHI sites of the limit digest, is shown at the top. The bands in the KO sample are smaller than the normal bands because of the deletion of the HS −26 element. (B) Northern blot analysis of polyA+ RNA from embryos of WT mice and mice homozygous for the floxed HS −26 KO allele. The analysis used a probe for the C16orf35 gene and showed down-regulation of this gene in the floxed mice.
Upstream erythroid-specific sites were previously observed in the mouse α-globin cluster at positions mHS −45, mHS −31, mHS −29, and mHS −12 in MEL cells.23 We detected mHS −31 and mHS −12, but not mHS −45 or mHS −29, in primary erythroid cells from both normal and KO embryos. Additional erythroid-specific sites were observed at mHS −21 and mHS −8 (corresponding to human HS −33 and HS −10, respectively) in both types of embryos.
Expression of the mC16orf35 gene is down-regulated in mice in which the floxed neomycin gene replaces mHS −26
When designing the targeting construct, we took care to ensure that neither introduction of the floxed neomycin gene nor its removal by Cre interfered with the open reading frame of the highly conserved gene (mC16orf35) that normally contains mHS −26. Nevertheless, we found that expression of this gene was severely down-regulated from that in the chromosome containing the intact, functional, floxed neomycin gene (Figure 4B). However, after removal of the neomycin gene with Cre recombinase, expression of the mC16orf35 gene was normal. This finding suggests that expression of mC16orf35 is not dependent on HS −26 but that its expression is reduced by the presence of the active neomycin gene either as a result of abnormal processing of its mRNA or possibly by antisense interference.
The function of C16orf35 is unknown. However, the position and sequence of this gene has been highly conserved throughout evolution.10 Therefore, the severe down-regulation of this gene observed when an active neomycin gene was present in one of the introns could contribute substantially to the phenotype observed in the floxed mHS −26−/− homozygotes. Although these mice appeared to be developmentally normal aside from the fact that they had severe α thalassemia, further analysis of these mice is warranted.
Comparison of the expression of mHS −26α with HS −40α constructs in MEL cells
The mild phenotype of the mHS −26 KO mice indicated that the mHS −26 element is not as strong an enhancer as HS −40. To address this question directly, we compared the expression in MEL cells of an mHS −26α construct (distinguished from the endogenous genes by using the coding sequence of the human α gene) with that of an HS −40α construct studied previously.7 24 The mHS −26α construct was cotransfected into adenine phosphoribosyltransferase (APRT)–negative MEL cells with an APRT plasmid and selected in histone acetyltransferase medium. Clones positive for mHS −26α were expanded and induced to terminal differentiation. In analyses of 7 clones by RNase protection assays, the proportion of α-globin mRNA directed by mHS −26 ranged from 1% to 27% (mean, 7%), whereas levels of 5% to 62% (mean, 20%) were observed in 10 clones directed by HS −40. This result is consistent with mHS −26 being a weaker enhancer than HS −40.
Discussion
On the basis of accumulated evidence that HS −40 is the major regulator of human α-globin gene expression and the similarity in position and structure of mHS −26 in the mouse locus, we predicted that a KO of mHS −26 in situ would result in a major down-regulation of mouse α mRNA and protein production and a severe form of α thalassemia. Therefore, the very mild α-thalassemia phenotype of the mHS −26 KO homozygotes was unexpected. The Hb levels in these mice were essentially normal, and only small degrees of microcytosis and hypochromia were observed. There were no breeding problems, and females raised litters of up to 13 without difficulty. At the RNA level, output from the KO chromosome was reduced to about 50% of normal values as indicated by the α/β mRNA ratios in the bone marrow and splenic erythroblasts from untreated and anemic mice. A lesser reduction was observed in the peripheral blood, presumably because of differential cell survival with elimination of cells with the greatest chain imbalance.
Removal of the 1.3-kb fragment containing the mHS −26 element was sufficient to prevent formation of the HS, and no new HS elements or changes in the other HS elements in this region were detected. Expression of the mC16orf35 gene, in which mHS −26 resides, also appeared to be unaffected in the KO mice. Although we cannot exclude the possibility that the full HS −26 element is larger than 1.3 kb, this fragment contains all the known protein-binding motifs associated with mHS −2611, and loss of a similarly sized fragment from a human chromosome8 was found to be sufficient to down-regulate α-globin gene expression severely.
Deletion of single HS elements from the mouse β locus control region (LCR) results in mild reductions in gene expression; in this case, it is known that the 5 HS elements contribute additively to gene expression from the β-globin gene cluster.25 The implication of the finding that loss of mHS −26 results in only about a 50% decrease in mouse α gene expression is that additional elements are required for full regulation of the cluster in mice. This in turn implies that there are important differences between mice and humans in α-globin gene regulation. Alternatively, there may be redundant elements in the region upstream of the α-globin locus in both species, and a requirement for additional elements has not been recognized with the expression systems used so far to analyze human α gene expression.
Several observations support the hypothesis that the human and mouse α-globin clusters are regulated somewhat differently so that HS −40 and mHS −26 play different roles in their respective clusters. The human α-globin cluster lies very close to the telomere, whereas the mouse locus is interstitial. Although the human α-globin gene promoters are associated with CpG islands, these are almost completely eroded in mice. The mouse α-globin promoters contain GATA-1 binding sites, whereas no such sites are present in the human promoters. Furthermore, although there are broad similarities between HS −40 and mHS −26 (∼75% over 260 bp), there are many differences in the details of the binding sites of transcription factors to these regions. In particular, only 1 of the 2 potential Maf responsive element (MARE) sites in mHS −26 binds Nfe2 strongly, and in contrast to the situation with HS −40, these sites do not bind YY1 and XBP, respectively. In addition, only 3 of 4 GATA-1 binding sites and only 1 of the 4 CACC binding sites found in HS −40 are conserved in mHS −26.11 13
These differences between the mouse and human regulatory elements may be of functional importance, because the current study showed that in a MEL expression system, the levels of α gene expression under the control of mHS −26 are only about one third of those under the control of HS −40. Similarly, when transgenic founder fetuses containing a 1.6-kb mHS −26 fragment linked to the human α gene were generated, expression levels averaged 12%11 or lower,12whereas they were 40% in HS −40–human α gene transgenic fetuses.7 24 The consistently weaker enhancement with mHS −26 compared with HS −40 in these expression systems, together with the milder reduction in α gene expression observed when the mHS −26 KO was compared with loss of HS −40, suggests that these 2 elements are not equivalent.
If there are additional regulatory elements in the mouse cluster, where might they lie? A comparison of the region upstream of the α-globin locus in mice and humans is shown in Figure5. This analysis showed that these orthologous regions are very similar in terms of gene organization. A direct comparison of the region around the regulatory element using the VISTA program demonstrated high levels of sequence conservation (> 75%) in the exons of the C16orf35 gene. The HS −40/mHS −26 element also showed considerable sequence similarity (Figure 5), as did the sequences around human HS −33 which, as we showed here, has a corresponding erythroid-specific HS site at −21 in mice. We also observed an HS site at −8 in mice that corresponds to the human HS −10.
Comparison of the human and mouse α-globin gene clusters and their upstream regions.
In each cluster, genes are represented by gray boxes, with orthologous genes numbered as in the article by Flint et al,10 showing conserved gene organization. The thick black line underneath the HS −40 site represents the minimum fragment deleted in patients with α thalassemia with intact α-globin genes. The sequence of this region was compared with the mouse homologous region in the central panel by using the VISTA program.26 The percentage of sequence conservation is indicated on the right, and the exons of the C16orf35 gene are indicated as gray boxes over the sequence alignment. HS sites confirmed in this study are shown as red arrows; those reported previously in MEL cells are shown in pink.
Comparison of the human and mouse α-globin gene clusters and their upstream regions.
In each cluster, genes are represented by gray boxes, with orthologous genes numbered as in the article by Flint et al,10 showing conserved gene organization. The thick black line underneath the HS −40 site represents the minimum fragment deleted in patients with α thalassemia with intact α-globin genes. The sequence of this region was compared with the mouse homologous region in the central panel by using the VISTA program.26 The percentage of sequence conservation is indicated on the right, and the exons of the C16orf35 gene are indicated as gray boxes over the sequence alignment. HS sites confirmed in this study are shown as red arrows; those reported previously in MEL cells are shown in pink.
Along with these similarities between the 2 clusters are some differences. Erythroid-specific HS sites were found in a mouse erythroid cell line (MEL) at positions −12, −29, −31, and −4523; no corresponding sites have been observed in human cells. We confirmed that the mHS −12 and mHS −31 sites, but not the mHS −29 or HS −45 sites, occur in mouse primary erythroid cells as well as MEL cells. None of these murine-specific sites show the high levels of sequence conservation observed between HS −40/mHS −26 and HS −33/mHS −21. Differentially expressed HS sites such as these could be candidates for the additional control sequences in mice.
Although the evidence presented here is consistent with the idea that elements in addition to mHS −26 are required for complete regulation of the mouse cluster, we cannot conclude definitively that in the human cluster, HS −40 is not only necessary but also sufficient for complete regulation of the α-globin genes. No natural deletion found in patients with α thalassemia removes only HS −40. All 9 such mutations also remove HS −33. These 2 erythroid-specific HS sites are the only evolutionarily conserved noncoding sequences in this segment of DNA. Although the effect on gene expression of HS −33 alone5 and in combination with other erythroid HS sites around the α cluster7,24 has been examined, there is no evidence that HS −33 has the properties of an enhancer or LCR element. Furthermore, it should be noted that in transgenic mice, a 150-kb PAC fragment that stretches from about −56 kb through the human α cluster did not show significantly higher expression than much smaller constructs.4 27
The key observation establishing the critical role of HS −40 is that a targeted deletion of this element in a chromosome 16 hybrid MEL cell down-regulated α-globin expression to less than 3% of normal levels,8 thereby suggesting that this element alone is the major regulator of α-globin expression and that none of the remaining elements can compensate for its absence. Possible differences in epigenetic modifications of the cluster in hybrids that have not been through a normal erythroid developmental process prevent this from being a definitive observation.28-30 Developmentally regulated epigenetic phenomena have also been observed in elements of the Drosophila bithorax complex.31 Furthermore, whereas inactivation of the transcription factor NF-E2 in MEL cells down-regulates globin gene expression, a germ line KO of NF-E2 appeared to have little or no effect on globin expression.32-35 It is interesting that 2 critical MARE sites in HS −40 bind the erythroid-enriched NF-E2 protein. It remains possible, therefore, that one or more additional elements can substitute for HS −40 during the normal developmental program, but not in the context of chromosome transfer experiments.
In addition to constituting an important step toward analyzing how the mouse α-globin cluster is regulated from its natural chromosomal environment, this study highlights the caution required when comparing gene regulation in superficially similar human and mouse systems and comparing the effects of KOs in differentiated as opposed to germ line cells. Similar concerns have been raised regarding similar comparisons of the human and mouse β-globin clusters.36-38 In the future, it will be important to determine both the similarities and differences in the mechanisms by which orthologous loci that have been diverging for more than 60 million years are currently regulated.
We thank A. Smith (Centre for Genome Research, University of Edinburgh) for supplying the ES cells and thymidine kinase cassette, S. Orkin (Division of Hematology, Children's Hospital, Boston) for providing the GATA-Cre mice and the neomycin cassette, and S. Butler for assistance with the mice.
Prepublished online as Blood First Edition Paper, July 5, 2002; DOI 10.1182/blood-2002-05-1409.
Supported by a Wellcome Trust Travelling Fellowship (E.A.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
W. G. Wood, MRC Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, OX3 9DS, United Kingdom; e-mail:bwood@hammer.imm.ox.ac.uk.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal