Patients with paroxysmal nocturnal hemoglobinuria (PNH) have blood cells deficient in glycosyl phosphatidylinositol (GPI)–linked proteins owing to a somatic mutation in the X-linked PIGA gene. To target Piga recombination to the erythroid/megakaryocytic lineage in mice, the Cre/loxP system was used, and Cre was expressed under the transcriptional regulatory sequences ofGATA-1. Breeding ofGATA1-cre (G) transgenic mice with mice carrying a floxed Piga (L) allele was associated with high embryonic lethality. However, double-transgenic (GL) mice that escaped early recombination looked healthy and were observed for 16 months. Flow cytometric analysis of peripheral blood cells showed that GL mice had up to 100% of red cells deficient in GPI-linked proteins. The loss of GPI-linked proteins on the cell surface occurred late in erythroid differentiation, causing a proportion of red cells to express low residual levels of GPI-linked proteins. Red cells with residual expression of GPI-linked proteins showed an intermediate sensitivity toward complement and thus resemble PNH type II cells in patients with PNH. Recombination of the floxed Piga allele was also detected in cultured megakaryocytes, mast cells, and eosinophils, but not in neutrophils, lymphocytes, or nonhematopoietic tissues. In summary, GATA1-Cre causes high-efficiency Pigagene inactivation in a GATA-1–specific pattern. For the first time, mice were generated that have almost 100% of red cells deficient in GPI-linked proteins. These animals will be valuable to further investigate the consequences of GPI-anchor deficiency on erythroid/megakaryocytic cells.
Introduction
In patients with paroxysmal nocturnal hemoglobinuria (PNH), a somatic mutation in the X-linkedPIGA gene causes a proportion of blood cells to be deficient in all proteins that are linked to the membrane via a glycosyl phosphatidylinositol (GPI) molecule.1,2PIGAencodes a protein subunit of the N-acetyl glucosaminyltransferase, an enzyme essential in GPI-anchor biosynthesis.3 The lack of GPI-linked proteins on blood cells is responsible for some of the clinical symptoms seen in patients with PNH. For example, intravascular hemolysis and hemoglobinuria are caused by the lack of CD55 decay accelerating factor (DAF) and CD59 membrane inhibitor of reactive lysis (MIRL) on red cells. CD55 and CD59 are GPI-linked complement-regulatory molecules. Their absence increases the sensitivity of PNH red cells toward complement-mediated lysis, which, when the complement becomes activated, leads to intravascular hemolysis and hemoglobinuria.
PNH is believed to be a disorder of the hematopoietic stem cell. Cells deficient in GPI-linked proteins are found not only within the red cells but also within the granulocytes, monocytes, platelets, and lymphocytes (reviewed by Rosse4). An inherited form of PNH has never been described. Indeed, Piga gene inactivation in murine embryonic stem cells is lethal in very early embryonic development.5,6 To study the functional role of GPI-linked proteins in hematopoiesis, we therefore generated a mouse line in which Piga gene inactivation can be targeted specifically to hematopoietic cells by means of the Cre/loxPsystem.7 Targeting Piga gene inactivation to the early embryo, we previously reported on mice that were mosaic for PIGA− cells (EIIa-cre ×loxPiga [EL] mice).8 Mosaic animals showed only a small proportion of PIGA− circulating blood cells, limiting our studies on the functional consequences of PIGA− red blood cells in these mice. Here we targetedPiga gene inactivation to the erythroid/megakaryocytic lineage by expressing the Cre recombinase under GATA-1 transcription regulatory sequences.
GATA-1 was the first transcription factor shown to be necessary for erythroid-specific gene expression.9-11 It is expressed at low levels in early CD34+ hematopoietic stem cells12 and at high levels in erythroblasts, megakaryocytes, mast cells, and eosinophils.13-16Experiments using targeted gene mutation showed that GATA-1 is required for terminal differentiation in both primitive and definitive erythropoiesis17 and plays an important role in megakaryocyte proliferation and differentiation.18Transcriptional regulatory sequences necessary for expression in primitive and definitive erythroid cells have been identified.19,20 Recent expression studies usinglacZ, Scl, and Gfp complementary DNAs in a transgene under these GATA-1 regulatory sequences suggested that GATA-1 regulatory sequences are able to mediate tissue-specific gene expression in transgenic mice.19,21 22 Here we used the transcription regulatory elements of GATA-1 to drive the expression of the Cre recombinase. GATA1-Cre transgenic animals were crossbred with mice carrying a floxed Piga allele. For the first time we generated offspring with up to 100% of red cells deficient in GPI-linked proteins. Owing to the time point of loxPiga gene recombination, a proportion of red cells resembled PNH type II cells with a residual expression of GPI-linked proteins. Recombination of the floxed Piga allele was also found in megakaryocytes, mast cells, and eosinophils, but not in any other blood cell lineage or in nonhematopoietic tissue.
Materials and methods
Mice
The production of the loxPiga (L) mice has been described previously (Figure1A).8 TwoGATA1-cre transgenic founder lines (GATA1-cre1, GATA1-cre2) were crossbred with L mice. The GATA1-cre1transgene was obtained by replacing the lacZ coding region in the UHR-5,3,-lacZ construct19 with the coding region of cre. GATA1-cre1 encompasses all 3 DNase hypersensitive regions and all 6 GATA-1 exons, with the exception of testis-specific exon 1. For the GATA1-cre2 construct (Figure1B), a 2-kb DNA fragment containing the chicken β-globin insulator sequence23 (5′HS4) (generously provided by Hurry Mead and Yann Echelard, Genzyme Transgenics, Boston, MA) was inserted at the 3′Sal I site. Transgenic mice were generated by pronuclear injection of zygotes from the CD-1 mouse strain.24 InGATA1-cre1 × L offspring, the proportion of red cells deficient in GPI-linked proteins was low and these cells disappeared within the first 4 months of life, suggesting silencing of the transgene (data not shown). Studies described here were all performed with offspring from the GATA1-cre2 transgenic founder line (G mice). L mice were all C57BL/6.L (N ≥ 10); G mice were C57BL/6.G (N ≥ 3). Single transgenic G and L mice were born at the expected frequency. Gender-matched G or L single-positive littermates were used as control mice.
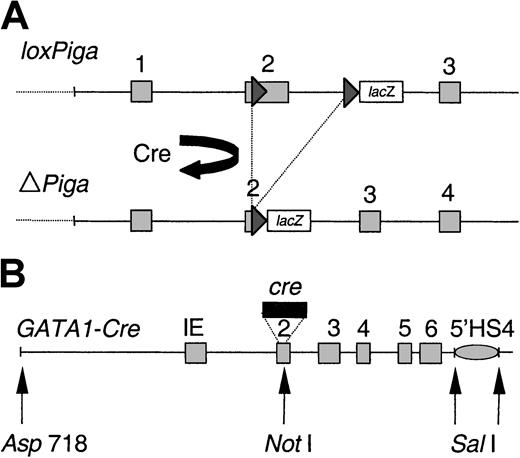
Genomic structure of the transgenes.
(A) Genomic structure of the floxed Piga gene (L) before(loxPiga) and after (ΔPiga) Cre-mediated loxP recombination. Cre-mediatedloxPiga gene recombination deletes 662 base pairs ofPiga exon 2 and 1.6 kilobases (kb) of the adjoining intron 2, which completely abrogates the function of PIGA.8 (B) Map of the GATA1-cre transgene in G mice. IE refers to the erythroid GATA-1 promoter, 5′HS4, a 5′ element of the chicken β-globin locus that functions as an insulator (gray oval).23 Exons are boxed and numbered; loxP sites are shown as triangles; restriction endonuclease sites are indicated.
Genomic structure of the transgenes.
(A) Genomic structure of the floxed Piga gene (L) before(loxPiga) and after (ΔPiga) Cre-mediated loxP recombination. Cre-mediatedloxPiga gene recombination deletes 662 base pairs ofPiga exon 2 and 1.6 kilobases (kb) of the adjoining intron 2, which completely abrogates the function of PIGA.8 (B) Map of the GATA1-cre transgene in G mice. IE refers to the erythroid GATA-1 promoter, 5′HS4, a 5′ element of the chicken β-globin locus that functions as an insulator (gray oval).23 Exons are boxed and numbered; loxP sites are shown as triangles; restriction endonuclease sites are indicated.
DNA analysis
Blood cell analysis
Red blood cell counts, hemoglobin levels, and platelet counts were determined with a Hemavet 850 blood cell counter (CDC Technology, Oxford, CT). Reticulocyte counts were assessed by flow cytometry.8
Flow cytometric analysis
Flow cytometric analysis of fetal and adult blood cells was performed as described.6 Bone marrow cells from mouse femora were washed in phosphate-buffered saline plus 2% bovine calf serum and analyzed by flow cytometry. The following chromophore-labeled monoclonal antibodies against GPI-linked surface antigens were used: CD24 (30-F1), Ly6G (RB6-8C5), and CD48 (HM48-1). Alexa Fluor 488–labeled mutant aerolysin (FLAER) that binds to the GPI-anchor protein complex26 was a generous gift from Thomas Buckley (University of Victoria, BC, Canada). Lineage specificity was determined by means of chromophore-labeled monoclonal antibodies against CD11b (M1/70), B220 (RA3-6B2), TcR β chain (H57-597), CD4 (GK1.5), CD8 (53-6.7), Ter119 (TER-119), CD71 (C2), c-kit (CD117, 2B8), and CD34 (RAM34). Lineage commitment was determined with a mixture of phycoerythin-labeled antibodies against CD3e (145-2C11), CD4, CD5 (53-7.3), CD8, B220, CD11b, and Ter119. All antibodies were purchased from Pharmingen (San Diego, CA).
Analysis of primitive red cells and fetal liver hematopoiesis
Fetuses at embryonic day (E) 10.5 to 13.5 were isolated. Red blood cells from the umbilical cord were collected and analyzed for the expression of the GPI-linked protein CD24 by flow cytometry.6 Fetal livers were disintegrated, the cells resuspended in 2% fetal calf serum–Iscoves modified Dulbecco medium and analyzed for the expression of CD24.
Cell cultures
For methylcellulose cultures, 104/mL fetal liver cells or 2 × 104/mL bone marrow cells were plated in 1% methylcellulose Methocult M3434 (Stemcell Technologies, Vancouver, BC, Canada) and cultured for 7 days. For erythroid burst-forming unit (BFU-E) colonies, 2 × 104 fetal liver cells and 5 × 104 bone marrow cells were plated in Methocult M3230 (Stemcell Technologies) supplemented with 50 ng/mL rat stem cell factor (rat SCF) and 3 U/mL erythropoietin (both a gift from Amgen, Thousand Oaks, CA). After 8 days of culture, colonies were counted and individually analyzed by flow cytometry.
Cultures enriched in megakaryocytes were obtained by plating 0.8 × 106/mL bone marrow cells in Myelocult M5300 medium (Stemcell Technologies) supplemented with 5 μg/mL recombinant human megakaryocyte growth and development factor (PEG-rHuMGDF) (a generous gift from Amgen) for 10 days. Cytospin preparations were stained with May-Grünwald-Giemsa27 and for acetylcholinesterase activity.28 DNA was isolated from the bulk culture and analyzed by PCR.
Eosinophil cultures were obtained by plating 0.4 × 106/mL bone marrow cells in Myelocult M5300 medium supplemented with 10 ng/mL recombinant murine interleukin 3 (rmIL-3) (R&D Systems, Minneapolis, MN) and 10 ng/mL recombinant murine interleukin 5 (rmIL-5) (R&D Systems) for 7 to 10 days. Cytospin preparations from 10-day cultures were stained with May-Grünwald-Giemsa and Luxol Fast Blue.29 DNA was isolated from the bulk culture and analyzed by PCR.
Liquid cultures for granulocytes were performed in Myelocult M5300 medium supplemented with 50 ng/mL rat SCF and glycosylated recombinant human granulocyte colony-stimulating factor (r-metHuG-CSF) (generously provided by Amgen). DNA was isolated from the bulk culture and analyzed by PCR.
Mast cell cultures were obtained as described.30 In brief, bone marrow cells were cultured in RPMI 1640 supplemented with 1 mM sodium pyruvate, 1 mM nonessential amino acids, 5.5 × 105 2-mercaptoethanol, 0.075% sodium bicarbonate, 2 mM L-glutamine, 10% fetal bovine serum, 15% Wehi-3 conditioned medium, and 5% pokeweed mitogen–stimulated spleen-conditioned medium (Stemcell Technologies). A homogeneous population of mast cells was obtained after 4 weeks as determined by Toluidine Blue staining and the expression of CD117. DNA was isolated from the bulk culture and analyzed by PCR.
Complement-sensitivity test
Complement sensitivity was assessed as previously described.8 Acidified human serum was used as a source of complement. The hemoglobin released was determined by measuring absorbance at 412 nm.
Results
Generation of GATA1-cre ×loxPiga offspring
To restrict Piga gene inactivation in vivo to hematopoietic cells, we crossbred mice that had a floxedPiga allele (L mice, Figure 1A) with transgenic mice expressing Cre under the regulatory sequences required for erythroid- and megakaryocyte-specific expression of the mouse GATA-1gene (G mice, Figure 1B).19 Of 168 newborn mice, 16 carried a GATA1-cre and a loxPiga transgene (GL mice). Table 1 summarizes the numbers of GL offspring obtained and expected from the first generation of GL breeding. Interestingly, double-transgenic mice were born only from crosses in which the GATA1-cre transgene was provided by the mother and the loxPiga allele came from the father (47% of expected). No offspring carrying both transgenes were obtained from the opposite breeding. Analysis of embryos collected at different stages of development showed embryos with complex developmental abnormalities and extensive recombination of the floxed Piga allele in nonhematopoietic tissues as determined by Southern blot analysis (data not shown). Highly efficient excision of floxed DNA sequences in early embryogenesis has been observed previously in offspring of G mice bred with Rosa26 carrying a floxed β-galactosidase–neomycin phosphotransferase fusion gene and was believed to be caused by leaky Cre expression in early embryogenesis.31 In these experiments, Cre-mediated DNA excision was equally efficient whether the target gene was of paternal or maternal origin. Pigamaps to the X chromosome. We find the loxPiga allele escapes early excision only when it is on the paternal X chromosome, suggesting some difference in Cre accessibility between paternal and maternal X chromosomes during the crucial period in embryogenesis.
Double-transgenic mice that escaped early recombination were born alive, looked healthy, and were indistinguishable from littermates that carried only one of the transgenes. Owing to the breeding strategy, GL mice of the first generation were all females. Male mice hemizygous (G+/−L+) and female mice homozygous for theloxPiga gene (G+/−L+/+) were generated in the second generation by breeding G+/−L+/− females with L+ males.
GL mice have circulating red blood cells that are deficient in GPI-linked proteins
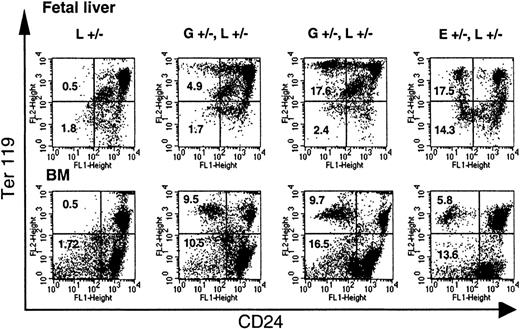
Circulating red blood cells from GL mice were analyzed for the expression of GPI-linked proteins. Figure2A shows representative examples from 2 newborn and two 4-month-old GL animals. Animals heterozygous forloxPiga (G+/−L+/−) have 2 populations of red cells, 1 with normal expression of CD24 and 1 deficient in CD24. In contrast, males hemizygous (G+/−L+) and females homozygous for theloxPiga allele (G+/−L+/+) showed close to 100% of red blood cells deficient in CD24. This is consistent with Piga's being on the X chromosome. In male offspring, the inactivation of the one Piga allele is sufficient to cause the loss of GPI-linked proteins. In females, the inactivation of the Piga allele when it is on the active X chromosome will lead to the loss of GPI-linked proteins on the cell surface. In females heterozygous for L, owing to random X-inactivation, this is expected to occur in about half of the cells. Long-term follow-up showed a decrease in the proportion of red cells deficient in GPI-linked proteins in G+/−L+/− animals within the first 4 months of life (40% ± 13% at birth [no. = 15] to 31% ± 7% at the age of 4 months [no. = 13]; P < .05). Thereafter, the proportion of GPI-anchor–deficient red blood cells was stable over the observation period of 16 months (Figure 2B). In G+/−L+ and G+/−L+/+mice, the proportion of red blood cells deficient in GPI-linked proteins increased in the first months of life and remained stable at 99% ± 0.5% (no. = 8) throughout the entire observation period of up to 8 months (Figure 2B).
GATA1-Cre–mediated loxPiga recombination in erythroid cells.
GATA1-Cre–mediated loxPiga recombination leads to primitive and definitive red blood cells that are completely or partially deficient in GPI-linked proteins. (A) Flow cytometric analysis for the expression of CD24 on red blood cells from L+/− control mice, G+/−L+/−, G+/−L+/+, and E+/−L+/− at birth and at the age of 4 months (4M). E+/−L+/− mice are offspring from theEIIa-cre × L breeding and are mosaic for a nonfunctional PIGA protein.8 Note that while a proportion of red cells in E+/−L+/− mice is completely deficient in CD24, in GL mice the affected proportion of red cells have residual expression of CD24. Numbers indicate the percentage of cells with normal decreased expression of CD24. (B) Time course of the relative proportion of blood cells deficient in GPI-linked proteins in G+/−L+/− and G+/−L+/+ mice during an observation period of 16 and 8 months, respectively. Means and SDs are shown. Note that in G+/−L+/− mice, the proportion of reticulocytes deficient in CD24 is always higher than the proportion of GPI-anchor–deficient red cells (P ≤ .05). (C) Decreasing levels of residual CD24 expression on the affected red cell population in GL mice during the observation period. The y-axis represents the mean fluorescence intensity of red cells deficient in GPI-linked proteins normalized to the mean fluorescence intensity determined at birth. (D) Expression of CD24 on red cells from 4 representative fetuses at E13.5. At this stage in development, 2 circulating red cell populations can be identified in healthy fetuses by flow cytometry. One population has high side scatter characteristics (SSChigh) and a low level of CD24 expression (CD24low), which corresponds to circulating primitive red cells. Red cells derived from definitive hematopoiesis are SSClow and CD24high (left panel). Note that both E+/−L+/− and GL mice have a proportion of primitive and definitive red cells deficient in CD24. However, in contrast to E+/−L+/− mice, in whom PIGA− red cells completely lack the expression of CD24,8 the affected primitive and definitive red blood cell populations in GL mice show a wide distribution of residual expression of CD24. SSC indicates side scatter; L+/−, healthy control mouse. E+/−L+/− mice are offspring from the EIIa-cre × L breeding.8 Numbers indicate the percentage of cells with normal decreased expression of CD24.
GATA1-Cre–mediated loxPiga recombination in erythroid cells.
GATA1-Cre–mediated loxPiga recombination leads to primitive and definitive red blood cells that are completely or partially deficient in GPI-linked proteins. (A) Flow cytometric analysis for the expression of CD24 on red blood cells from L+/− control mice, G+/−L+/−, G+/−L+/+, and E+/−L+/− at birth and at the age of 4 months (4M). E+/−L+/− mice are offspring from theEIIa-cre × L breeding and are mosaic for a nonfunctional PIGA protein.8 Note that while a proportion of red cells in E+/−L+/− mice is completely deficient in CD24, in GL mice the affected proportion of red cells have residual expression of CD24. Numbers indicate the percentage of cells with normal decreased expression of CD24. (B) Time course of the relative proportion of blood cells deficient in GPI-linked proteins in G+/−L+/− and G+/−L+/+ mice during an observation period of 16 and 8 months, respectively. Means and SDs are shown. Note that in G+/−L+/− mice, the proportion of reticulocytes deficient in CD24 is always higher than the proportion of GPI-anchor–deficient red cells (P ≤ .05). (C) Decreasing levels of residual CD24 expression on the affected red cell population in GL mice during the observation period. The y-axis represents the mean fluorescence intensity of red cells deficient in GPI-linked proteins normalized to the mean fluorescence intensity determined at birth. (D) Expression of CD24 on red cells from 4 representative fetuses at E13.5. At this stage in development, 2 circulating red cell populations can be identified in healthy fetuses by flow cytometry. One population has high side scatter characteristics (SSChigh) and a low level of CD24 expression (CD24low), which corresponds to circulating primitive red cells. Red cells derived from definitive hematopoiesis are SSClow and CD24high (left panel). Note that both E+/−L+/− and GL mice have a proportion of primitive and definitive red cells deficient in CD24. However, in contrast to E+/−L+/− mice, in whom PIGA− red cells completely lack the expression of CD24,8 the affected primitive and definitive red blood cell populations in GL mice show a wide distribution of residual expression of CD24. SSC indicates side scatter; L+/−, healthy control mouse. E+/−L+/− mice are offspring from the EIIa-cre × L breeding.8 Numbers indicate the percentage of cells with normal decreased expression of CD24.
The deficiency of CD24 on the affected red blood cells was not complete in all cells, but showed a wide distribution ranging from almost normal to complete deficiency (Figure 2A). The proportion of red cells with a residual expression of CD24 and the level of residual expression were highest after birth but decreased in the first 4 months of life. Thereafter, only a small proportion of red blood cells had a residual expression of GPI-linked proteins, the remainder being completely deficient in CD24. Figure 2C shows the decrease in mean fluorescence intensity of the affected red cell population in GL mice over time.
GATA1-Cre mediates loxPiga gene inactivation in primitive erythropoiesis
To test whether GATA1-Cre mediates loxPiga gene recombination during primitive erythropoiesis, blood samples from genotyped fetuses between E10.5 and E13.5 were analyzed for the expression of CD24. At E10.5, all circulating red blood cells are nucleated primitive red blood cells derived from erythropoiesis of the yolk sac, whereas at E13.5 about 50% of circulating red blood cells are primitive red blood cells and 50% are definitive red cells. In all GL fetuses analyzed (no. = 12), GPI-linked proteins were deficient on red cells derived from primitive and definitive hematopoiesis. Figure2D shows 2 representative G+/−L+/− fetuses, each with primitive and definitive red cells deficient in GPI-linked proteins.
Recombination of the loxPiga allele occurs late in erythroid differentiation
To investigate the stage in erythroid maturation when CD24 expression is lost, liver cells of E12.5 to E13.5 fetuses and bone marrow cells of adult mice were analyzed for the expression of CD24 and Ter119.32 33 On fetal liver cells, which at the age of E12.5 to E13.5 consist of 80% erythroid cells, a decreased expression of CD24 was detected only on Ter119high but not on Ter119low or Ter119neg cells (Figure3, top panel). Similarly, on bone marrow cells, CD24 deficiency was detected only on cells that simultaneously expressed Ter119 at high levels (Figure 3, bottom panel). No CD24-deficient cells were detected in progenitor cells expressing c-kit+ or on CD34+Lin− cells (data not shown). This suggests that the loss of CD24 occurs at the time of Ter119 expression. In mice hemizygous or homozygous forloxPiga, which had almost 100% of circulating red blood cells deficient in CD24, a small proportion (6%) of Ter119+CD24+ cells was detected in the bone marrow (data not shown), suggesting that the final loss of CD24 occurs as these cells mature into circulating red cells.
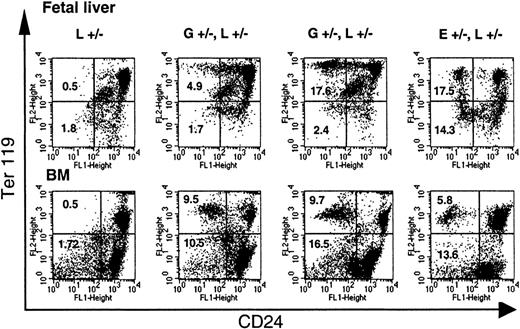
Loss of CD24 expression during erythroid differentiation.
The loss of GPI-linked proteins on red cell precursors from GL mice occurs with the expression of Ter119. Single-cell suspensions of fetal liver cells from E13.5 fetuses and bone marrow cells (BM) from adult mice were analyzed for the expression of Ter119 and CD24. Note that while in E+/−L+/− mice a distinct proportion of Ter119high and Ter119low cells are deficient in CD24, in GL mice a significant proportion of CD24-deficient cells are found only in the Ter119high cell population. L+/− indicates control mouse. E+/−L+/− mice are offspring from theEIIa-cre × L breeding8 and are mosaic for a nonfunctional PIGA protein. Numbers indicate the percentage of cells in the left upper and left lower quadrant (cells deficient in CD24). Vertical marker shown delineates CD24− from CD24+ Ter119low cells.
Loss of CD24 expression during erythroid differentiation.
The loss of GPI-linked proteins on red cell precursors from GL mice occurs with the expression of Ter119. Single-cell suspensions of fetal liver cells from E13.5 fetuses and bone marrow cells (BM) from adult mice were analyzed for the expression of Ter119 and CD24. Note that while in E+/−L+/− mice a distinct proportion of Ter119high and Ter119low cells are deficient in CD24, in GL mice a significant proportion of CD24-deficient cells are found only in the Ter119high cell population. L+/− indicates control mouse. E+/−L+/− mice are offspring from theEIIa-cre × L breeding8 and are mosaic for a nonfunctional PIGA protein. Numbers indicate the percentage of cells in the left upper and left lower quadrant (cells deficient in CD24). Vertical marker shown delineates CD24− from CD24+ Ter119low cells.
To further investigate the time point of loxPiga gene recombination, single BFU-Es grown in methylcellulose from bone marrow or fetal liver cells were analyzed for the expression of CD24. Flow cytometric analysis of 86 BFU-Es from 7 GL mice showed that 26.7% had reduced expression of CD24. The deficiency of CD24 on BFU-Es ranged from a partial (20.9% of BFU-Es) to an almost complete deficiency of CD24 (5.8% of BFU-Es), suggesting that the loss of CD24 on red cells occurs mainly after the BFU-E maturation stage (Figure4).
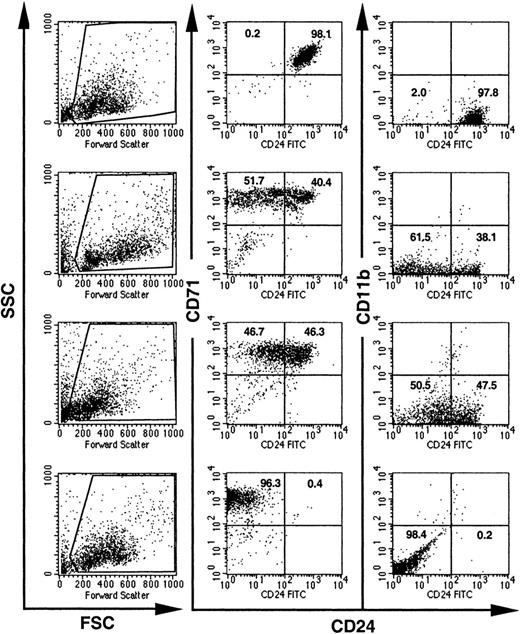
Expression of CD24 on individual BFU-Es from GL mice.
BFU-E colonies grown in methylcellulose from bone marrow cells of GL mice were individually picked and analyzed for the expression of CD24. Visual inspection, high CD71 expression, and the lack of CD11b were used to distinguish BFU-Es from mast cell and myeloid colonies. Dead cells and debris were excluded by their light scatter characteristics (the gates used are shown on the left). Numbers indicate the percentage of cells with normal decreased expression of CD24.
Expression of CD24 on individual BFU-Es from GL mice.
BFU-E colonies grown in methylcellulose from bone marrow cells of GL mice were individually picked and analyzed for the expression of CD24. Visual inspection, high CD71 expression, and the lack of CD11b were used to distinguish BFU-Es from mast cell and myeloid colonies. Dead cells and debris were excluded by their light scatter characteristics (the gates used are shown on the left). Numbers indicate the percentage of cells with normal decreased expression of CD24.
GATA1-Cre mediates recombination of the loxPiga allele in megakaryocytes, mast cells, and eosinophils but not in granulocytes, monocytes, or nonhematopoietic tissues
To test whether GATA1-Cre mediates recombination of theloxPiga allele in all blood cell lineages that express GATA-1 at high levels, liquid cultures were obtained from fetal liver cells and bone marrow cells from adult mice and cultured under conditions that support the growth of megakaryocytes, mast cells, eosinophils, or granulocytes.
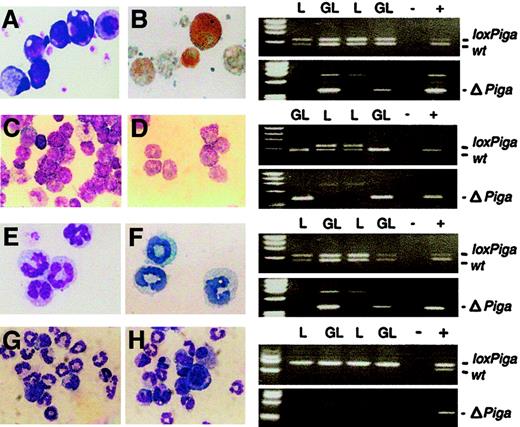
Megakaryocytes were obtained from fetal liver or bone marrow cell cultures in the presence of PEG-rHuMGDF (Figure5A-B). A PCR amplification product specific for ΔPiga was obtained from all cultures from GL mice (no. = 4) but not from control mice (G or L mice), indicating that loxPiga recombination occurred at the molecular level. None of the available antibodies against GPI-linked antigens stained platelets or megakaryocytes sufficiently to discriminate stained from unstained cells. Thus, the degree of GPI-anchor deficiency on platelets and megakaryocytes could not be determined.
GATA1-Cre–mediated recombination of the
loxPiga allele in megakaryocytes, mast cells, and eosinophils. Liquid cultures enriched in megakaryocytes (panels A and B), mast cells (panels C and D), eosinophils (panels E and F), and granulocytes (panels G and H) were performed as described. Bulk cultures were analyzed for the wild-type (wt), loxPiga, and ΔPiga allele by PCR. Panels on the right show the PCR analysis for each culture system. Panels A, C, E, G, and H are May-Grünwald-Giemsa stains from cultures obtained from GL animals. Panel B shows acetylcholinesterase activity in GL megakaryocytes; panel C shows Toluidine Blue–positive GL mast cells; and panel E shows Luxol Fast Blue–positive GL eosinophils. L stands for DNA isolated from cultured cells from L+/− control mice; GL for DNA isolated from cultured cells from G+/−L+/− mice; + and − represent positive and negative PCR controls. For granulocyte cultures G+/−L+/+ mice and L+/+ control mice were used instead. Amplification products representing the wt,loxPiga, and ΔPiga allele are indicated.
GATA1-Cre–mediated recombination of the
loxPiga allele in megakaryocytes, mast cells, and eosinophils. Liquid cultures enriched in megakaryocytes (panels A and B), mast cells (panels C and D), eosinophils (panels E and F), and granulocytes (panels G and H) were performed as described. Bulk cultures were analyzed for the wild-type (wt), loxPiga, and ΔPiga allele by PCR. Panels on the right show the PCR analysis for each culture system. Panels A, C, E, G, and H are May-Grünwald-Giemsa stains from cultures obtained from GL animals. Panel B shows acetylcholinesterase activity in GL megakaryocytes; panel C shows Toluidine Blue–positive GL mast cells; and panel E shows Luxol Fast Blue–positive GL eosinophils. L stands for DNA isolated from cultured cells from L+/− control mice; GL for DNA isolated from cultured cells from G+/−L+/− mice; + and − represent positive and negative PCR controls. For granulocyte cultures G+/−L+/+ mice and L+/+ control mice were used instead. Amplification products representing the wt,loxPiga, and ΔPiga allele are indicated.
Homogeneous mast cell cultures were obtained by culturing bone marrow cells for 4 to 6 weeks in conditioned medium enriched in IL-3 and SCF30 (Figure 5C-D). The loss of the loxPigaallele and amplification of the product specific for ΔPiga indicated that loxPigarecombination in cultured mast cells occurred to 100%. Cultured mast cells from G+/−L+ mice did not bind FLAER and lacked CD48, a GPI-linked protein expressed on the majority of normal cultured mature mast cells (data not shown).
Cultures enriched in eosinophils were obtained from bone marrow cells in the presence of IL-3 and IL-5 (Figure 5E-F). PCR analysis specific for ΔPiga demonstrated thatloxPiga recombination occurs in eosinophils as well. However, cultured eosinophils from G+/−L+ mice bound FLAER equally when compared with eosinophils cultured from control mice (data not shown), indicating that recombination of theloxPiga allele occurs late in eosinophil maturation at a time when most GPI-linked proteins are already expressed on the cell surface.
No recombination of the loxPiga allele was detected in granulocyte cultures or in monocyte/macrophage cultures either by PCR or by flow cytometric analysis (Figure 5F-G and data not shown).
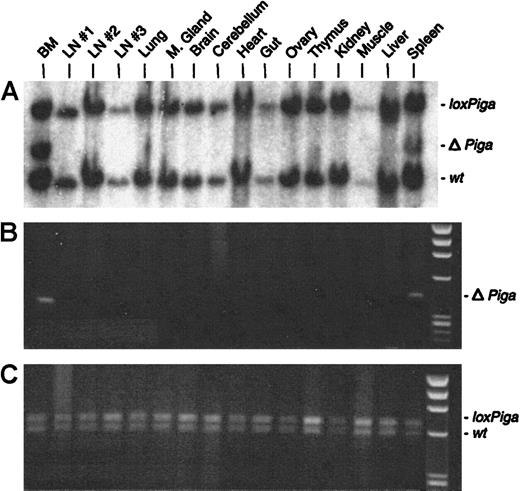
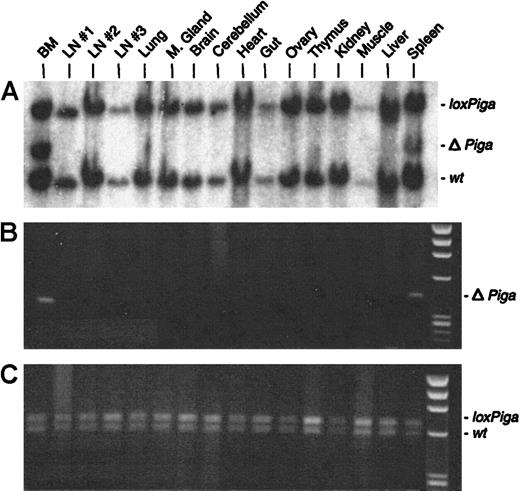
No or very low numbers (in 2 mice) of granulocytes, B cells, or T cells deficient in GPI-linked proteins were detected (Figure 2B). Southern blot and PCR analysis with DNA isolated from a variety of tissues from 3 representative GL animals showed recombination of theloxPiga allele only in bone marrow and spleen cells but not in any other tissue (representative data are displayed in Figure6). The sole exception was DNA isolated from the testis of a G+/−L+ mouse, which only by PCR revealed a faint amplification product specific for ΔPiga (data not shown).
Cell type–specific recombination of the
loxPiga allele in GL mice. Southern blot analysis (A) and PCR analysis (B and C) were used to examine recombination of the loxPiga allele in various tissues from G+/−L+/− mice.25 Restriction fragments and amplification products representing the wt,loxPiga, and ΔPiga allele are indicated.
Cell type–specific recombination of the
loxPiga allele in GL mice. Southern blot analysis (A) and PCR analysis (B and C) were used to examine recombination of the loxPiga allele in various tissues from G+/−L+/− mice.25 Restriction fragments and amplification products representing the wt,loxPiga, and ΔPiga allele are indicated.
Red blood cells with residual expression of GPI-linked proteins have an intermediate sensitivity toward complement-mediated lysis
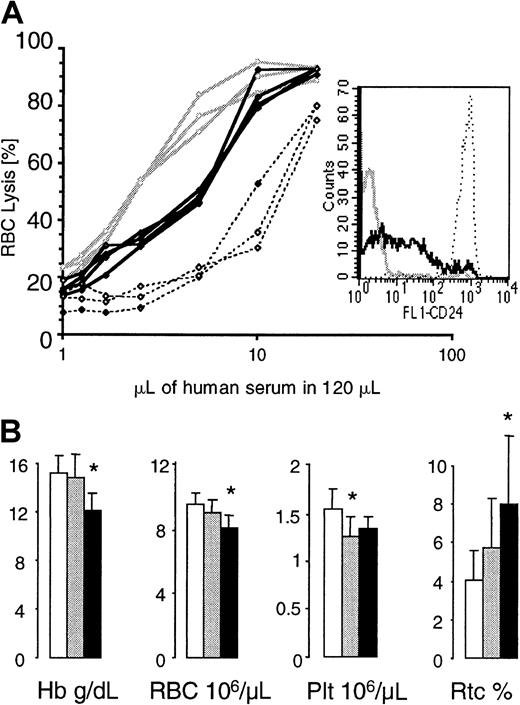
To assess the complement sensitivity of red blood cells with a residual expression of GPI-linked proteins, we incubated red cells from G+/−L+/+ animals with serial dilutions of acidified human serum.8 Red cells from healthy mice and from mice whose blood cells completely lack GPI-linked proteins34 were used as controls. At all complement concentrations, more GL red cells were lysed when compared with normal red cells, but fewer GL red cells were lysed when compared with red cells that completely lack GPI-linked proteins (Figure7A). This indicates that red blood cells with a residual expression of GPI-linked proteins have an intermediate sensitivity to complement.
Red blood cell values in GL mice and complement sensitivity of GL red blood cells.
(A) Complement sensitivity of GL red blood cells. Red blood cells from control mice (no. = 3, stippled lines), G+/−L+ mice (no. = 3, bold lines), and mice with blood cells that completely lack GPI-linked proteins (no. = 3, gray lines) were exposed to serial dilutions of acidified human serum. The insert shows one representative example of CD24 expression on red cells from each group of mice analyzed. (B) Hemoglobin levels (Hb) and red blood cell (RBC), platelet (Plt), and reticulocyte (Rtc) counts were obtained from control (empty bars), G+/−L+/− (gray bars), and G+/−L+(/+) (black bars) mice at the age of 4 months. * indicates a statistical significant difference compared with age-matched control mice (P < .05).
Red blood cell values in GL mice and complement sensitivity of GL red blood cells.
(A) Complement sensitivity of GL red blood cells. Red blood cells from control mice (no. = 3, stippled lines), G+/−L+ mice (no. = 3, bold lines), and mice with blood cells that completely lack GPI-linked proteins (no. = 3, gray lines) were exposed to serial dilutions of acidified human serum. The insert shows one representative example of CD24 expression on red cells from each group of mice analyzed. (B) Hemoglobin levels (Hb) and red blood cell (RBC), platelet (Plt), and reticulocyte (Rtc) counts were obtained from control (empty bars), G+/−L+/− (gray bars), and G+/−L+(/+) (black bars) mice at the age of 4 months. * indicates a statistical significant difference compared with age-matched control mice (P < .05).
G+/−L+/− mice and G+/−L+(/+) mice looked healthy and were well during the entire observation period of 16 and 8 months, respectively. Although red blood cell values were within normal limits, the hemoglobin and red cell counts in G+/−L+/− mice were always lower then in control mice. In contrast, the reticulocyte counts were highest in G+/−L+(/+) animals (counts in G+/−L+(/+) mice were greater than in G+/−L+/− mice, which were greater than in control mice). Figure 7B shows red blood cell values, platelet counts, and reticulocyte counts in G+/−L+(/+), G+/−L+/−, and control mice at the age of 4 months.
Discussion
The deficiency of GPI-linked proteins on the cell surface is the hallmark of blood cells in patients with PNH. The lack, or decreased expression, of these proteins is believed to be responsible for the clinical symptoms of patients with this disease. A mouse that has blood cells deficient in GPI-linked proteins and closely mimics the human disease would provide a powerful tool for studying the pathophysiology of PNH and the role of GPI-linked proteins on blood cells. Here we report on the generation of mice that have almost 100% of red blood cells deficient in GPI-linked proteins. In contrast to our previously reported mice, which were mosaic for a nonfunctional Pigagene and had only a small proportion of red cells deficient in GPI-linked proteins,8 the mice described here show highly efficient Piga gene recombination limited to hematopoietic cells of the erythroid/megakaryocytic lineage. Owing to the time point of loxPiga gene recombination, a proportion of red blood cells in our GL mice copied the phenotype that in humans has previously been described as PNH type II cells.
GATA1-Cre–mediated Piga gene inactivation was associated with a high embryonic lethality owing to leaky Cre expression in early embryogenesis leading to loxPiga gene recombination in multiple nonhematopoietic tissues. However, a proportion of GL mice escaped early embryonic gene recombination, were born alive, looked healthy, and were fertile. All double-transgenic mice born alive had circulating red blood cells deficient in GPI-linked proteins. In males hemizygous and females homozygous for the loxPiga allele, the proportion of red cells deficient in GPI-linked proteins was almost 100% in all animals tested, suggesting that GATA1-Cre–mediated recombination of the loxPiga allele in erythroid cells occurs at very high efficiency throughout adult life. A decreased expression of GPI-linked proteins was also detected on circulating primitive red cells of GL fetuses, indicating that recombination of theloxPiga allele also occurs in primitive erythropoiesis originating from the blood cell islands of the yolk sack.
Red cells in patients with PNH have been classified as PNH type I, II, and III cells depending on their sensitivity toward complement.35 PNH type I cells have a normal sensitivity toward complement, whereas PNH type III cells are highly sensitive. PNH type II cells are complement sensitive as well, but to a lesser extent.35 In humans, complement sensitivity of red cells depends directly on the expression level of CD59 and CD55. Thus, type I cells have a normal, and type II cells a decreased, expression of CD55 and CD59, while type III cells completely lack these 2 proteins. Point mutations in the PIGA gene that only impair but do not abrogate the function of the glycosyltransferase are mainly responsible for the PNH type II phenotype of red cells from patients with PNH.36 In GL mice, Cre-mediated DNA excision ofPiga exon 2 completely abolishes PIGA function.8 The residual expression of GPI-linked proteins on red blood cells from GL mice was therefore a surprise. The expression of GPI-linked proteins ranged from close to normal levels to the complete lack of these proteins (Figure 2A). As in humans, in GL mice, red cells with a residual expression of GPI-linked proteins have an intermediate sensitivity to complement when compared with healthy red cells and with red cells that completely lack GPI-linked proteins (Figure 7A). Thus, GL red blood cells with residual expression of GPI-linked proteins phenocopy PNH type II cells from patients with PNH. The loss of GPI-anchored proteins on GL blood cells afterPiga gene inactivation depends on the half-life of the PIGA protein, the half-life of the anchor precursors, and the half-life of the GPI-linked proteins on the cell surface. Investigation of the time point of Piga recombination in erythroid maturation revealed that the loss of GPI-linked proteins in GL mice occurs rather late in erythropoiesis, after the BFU-E stage, in cells that already express Ter119, which is a late erythroid differentiation marker.33 Thus, although the red cells in GL mice copy the PNH type II phenotype from patients with PNH, the molecular mechanism that gives rise to these cells is very different. In our animals, the PNH type II phenotype is caused by continuous Piga gene inactivation in late erythroid cells that do not undergo sufficient cell divisions to completely lose the expression of GPI-linked proteins. However, clinical symptoms in PNH are caused by the deficiency of GPI-linked proteins on blood cells. Thus, despite the difference in the molecular mechanisms of Piga gene inactivation, GL mice will be a valuable tool for further investigating the consequence of GPI-anchor deficiency specifically on red blood cells and possibly also on platelets.
PCR analysis of DNA isolated from cultured megakaryocytes, mast cells, and eosinophils demonstrated that GATA1-Cre mediates loxPigarecombination in these blood cell lineages. Previous experiments using the GATA-1 transcription regulatory sequences showed transgene expression in megakaryocytes but not in mast cells or eosinophils.19 A possible explanation might be a short time period of transgene expression during eosinophil or mast cell maturation, which might be missed in assaying for the transgene product. In contrast, the irreversibility of loxP-dependent DNA excision and the sensitivity of the assay to screen forPiga gene inactivation in our animals provide us with a very sensitive tool to trace the earliest time point of transgene expression during maturation in an individual blood cell lineage.
GATA-1 expression has been found in CD34+ hematopoietic progenitor cells enriched for hematopoietic stem cells.12However, no evidence was found that would suggest that in GL mice recombination of the loxPiga allele occurs in hematopoietic stem cells. The small proportion of GPI-anchor–deficient white blood cells in 2 GL animals is most likely due to low-level mosaicism caused by incomplete recombination in early embryogenesis. However, low-levelPiga gene inactivation in a committed pluripotent hematopoietic progenitor cell in these animals cannot be excluded. The lack of GPI-anchor–deficient cells within myeloid cells, T cells, and B cells in the majority of GL mice suggests either that GATA1-Cre expression is not sufficient or that additional expression regulatory sequences are required to cause recombination of the loxPigaallele in multipotent progenitor cells.
GL mice looked healthy with no obvious signs of anemia, hemoglobinuria, or thrombosis. However, in all GL mice, red blood cell values, although within normal limits, were lower than red blood cell values from age- and sex-matched control mice (Figure 7B). Furthermore, in GL mice heterozygous for loxPiga, the proportion of mature red blood cells deficient in GPI-linked proteins was below the expected 50%. In contrast, the proportion of reticulocytes deficient in GPI-linked proteins was around 50% (Figure 2B). These findings suggest that red cells deficient in GPI-linked proteins have a decreased half-life in circulation. The most likely explanation is that this is due to the increased sensitivity of GL red cells to complement. PNH patients with only PNH type II cells have mild hemolysis and are rarely transfusion dependent. Similarly, the anemia in our mice is very mild and the reticulocytosis is moderate.
In summary, findings in our GL mice demonstrate that recombination of the loxPiga allele occurs at high efficiency in a pattern specific for GATA-1 expression. Circulating red cells from GL mice share many characteristics of red cells from patients with PNH. GL mice thus promise to be an interesting tool for modeling the human disease in order to study the functional role of GPI-anchor deficiency in hemolysis and thrombosis.
We thank Patty Sipes for the excellent care of our mouse colony; Martin Rogers for technical support; Peter Besmer, Memorial Sloan Kettering Cancer Institute (New York, NY), for advice in mast cell cultures; Dan Link and Philip J. Mason for stimulating discussions; Hurry Mead and Yann Echelard, Genzyme Transgenics (Boston, MA), for their kind provision of chicken β-globin insulator; Thomas Buckley, University of Victoria (BC, Canada), for his generous gift of FLAER; and Amgen (Thousand Oaks, CA), for its generous provision of erythropoietin, h-metHuG-CSF, rat SCF, and PEG-rHuMGDF.
Supported by The Barnes Jewish Hospital Foundation, Howard Hughes Medical Institute, the Mallinckrodt Foundation, the McDonnell Foundation, and grant RO1-HL63208 from the National Institutes of Health (M.B.). S.H.O. is an investigator of the Howard Hughes Medical Institute. P.K. is the recipient of a fellowship from the Akademische Nachwuchsförderung ZH, Switzerland.
M.J. and P.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Monica Bessler, Division of Hematology, Department of Internal Medicine, Washington University School of Medicine, 660 S Euclid Ave, Box 8125, St Louis, MO 63110-10093; email:mbessler@im.wustl.edu.