The expansion of follicular lymphomas (FLs) resembles, both morphologically and functionally, normal germinal center B-cell growth. The tumor cells proliferate in networks of follicular dendritic cells and are believed to be capable of somatic hypermutation and isotype switching. To investigate the relation between somatic mutation and heavy chain isotype expression, we analyzed the variable heavy (VH) chain genes of 30 FL samples of different isotypes. The VH genes of the FLs were heavily mutated (29.3 mutations on average). In addition, isotype-switched lymphomas contained more somatic mutations than immunoglobulin M–positive lymphomas (33.8 mutations per VH gene versus 23.0, respectively). In all but one of the FLs, the ratios of replacement versus silent mutations in the framework regions were low, independent of the absolute number of somatic mutations and the level of intraclonal variation. Analysis of relapse samples of 4 FLs showed no obvious increase in somatic mutation load in most FLs and a decrease in intraclonal variation in time. In 3 of 4 cases, we obtained evidence for selection of certain subclones, rather than clonal evolution. Our findings question if intraclonal variation is always a reflection of ongoing somatic hypermutation. This may have implications for the concept of antigen-driven lymphomagenesis.
Follicular lymphoma (FL) is one of the most common B-cell non-Hodgkin lymphomas in adults in Europe and the United States.1 FLs have a relatively indolent behavior, with a reported median survival of 8 to 10 years after diagnosis.2-4 At the molecular level, FLs are characterized by the t(14;18)(q32;q21) translocation, which can be demonstrated in approximately 80% to 90% of FLs.5,6 Due to this translocation, the BCL-2 protein becomes constitutively expressed, which protects the cells against the induction of apoptosis and is thus believed to be of pathogenetic relevance.7,8 On the other hand, the t(14;18) translocation alone is not sufficient for full transformation, as has been demonstrated in bcl-2 transgenic mice.9 Moreover, the t(14;18) translocation is also found in normal B cells of healthy individuals.10-12 Additional genetic aberrations must therefore be present and have in fact been described.13 14
FLs are histologically well differentiated. Their nodular growth pattern clearly resembles the architecture of germinal centers in secondary lymphoid organs.15 The neoplastic follicles contain centroblastic and centrocytic tumor cells as well as nonneoplastic T cells, follicular dendritic cells, and few, if any, macrophages.16,17 Analyses of the variable heavy (VH) and light (VL) chain genes of the B-cell antigen receptor (BCR) of FLs have further supported their derivation from germinal center B cells: The immunoglobulin (Ig) V genes of FLs are somatically mutated,18,19 and the mutation patterns observed are reminiscent of those of normal, antigen-selected, memory B cells. In addition, intraclonal diversity in the mutated IgV genes of individual FLs is well documented 20-23 and is generally considered to be a reflection of ongoing somatic hypermutation. Finally, heavy (H) chain isotype switching has been described for 2 cases of FL,24,25 an event that is thought to take place normally in the centrocyte stage of the germinal center.26
Thus far, studies have indicated that the IgV genes of FLs contain high somatic mutation loads: Bahler et al reported an average frequency of mutation of 8.6% (amounting to approximately 25 mutations) in VH4-expressing FLs.18 This is in accordance with a study by Stamatopoulos et al in which an average mutation frequency of 9.3% (approximately 27 mutations) was observed. Also, the κ light chains used by these lymphomas were mutated, although to a lesser extent: 4.7% mutation was found on average (amounting to approximately 13 mutations per VL gene).19
In this study, we investigated the relation between the amount and patterns of somatic mutations and the expressed H chain isotypes by analyzing the BCRs of 12 IgM+(IgD+), 16 IgG+, and 1 IgA+ FL samples. One of these lymphomas contained both IgM+ and IgG+ tumor cells. Moreover, we investigated the changes over time in the VH genes of 4 patients who suffered a relapse.
Materials and methods
Patient material
Fresh tissue material of FLs was obtained from surgically removed lymph nodes at the departments of pathology of the Academic Medical Center, Amsterdam; the Westeinde Hospital, The Hague; and Leiden University Medical Center, Leiden, the Netherlands. The age of the patients ranged from 31 to 84 years (average, 63).
Immunohistochemistry
The surface immunoglobulin isotype was determined immunohistochemically. Cryostat sections were stained as described27 using monoclonal antibodies specific for human immunoglobulin heavy and light chain isotypes (Dako, Glostrup, Denmark, except for anti-IgM, -kappa, and -lambda, which were obtained from Becton Dickinson, Erembodegem-Aalst, Belgium). For FLs obtained from the Westeinde Hospital and Leiden University Medical Center, isotype expression was assessed by immunofluorescence. Briefly, acetone-fixed tissue sections were incubated at 37°C with the primary antibody (anti-IgM, -IgG, -IgA, -IgD, -kappa, or –lambda; Dako), washed 3 times in phosphate-buffered saline, and incubated for 30 minutes with swine-antirabbit labeled with fluorescein isothiocyanate (Dako).
RNA isolation and cDNA synthesis
Total cellular RNA was isolated from frozen tissue sections using the Trizol reagent (Life Technologies, Breda, the Netherlands). For complementary DNA (cDNA) synthesis, 10 μg of RNA was incubated with 5 nmol of pd(N)6 primer (Pharmacia Biotech, Roosendaal, the Netherlands) for 10 minutes at 65°C. After cooling on ice, the reaction mixture was added to a final volume of 50 μL. It contained 400 units of Moloney murine leukemia virus reverse transcriptase (RT) (Life Technologies), 8 mM of dithiothreitol, 1 mM of each dNTP, 1 × first strand buffer (50 mM of Tris-HCl, pH 8.3; 75 mM of KCl; 3 mM of MgCl2), and 60 units of RNAse inhibitor (Boehringer Mannheim, Almere, the Netherlands). The reaction was performed for 1 hour at 37°C. Subsequently, the enzyme was inactivated during 10 minutes at 95°C.
Polymerase chain reactions
The complementarity-determining region 3 (CDR3) was amplified using a forward primer with specificity for framework region 3 (FR3) in combination with reverse primers specific for JH(JHseq), Cμ, Cγ (Cγ2), Cα, or Cδ, as described.27 Either 1 μL of the cDNA reaction mixture was used, or (for a nested polymerase chain reaction [PCR]) 1 μL of PCR product from a VH family-specific PCR was used. The PCR mixture contained 1 × Taq buffer (20 mM of Tris-HCl, 50 mM of KCl, pH 8.4), 0.2 mM of each dNTP, 1.5 mM of MgCl2, 2 units of Taq polymerase (Life Technologies), and 0.5 μM of each primer. First, 10 cycles of amplification were performed in the thermal cycler (PTC-100, MJ Research Inc, Watertown, MA), ie, successively 30 seconds at 95°C, 20 seconds at 57°C, and 20 seconds at 72°C. The next 40 cycles of amplification consisted of 30 seconds at 95°C, 20 seconds at 55°C, and 20 seconds at 72°C. The reaction was completed for 6 minutes at 72°C. PCR products were analyzed on a 3% Metaphor agarose gel (FMC Bioproducts, Rockland, ME). For the VH family-specific PCR, reactions were performed with one of the VH family-specific leader primers combined with the appropriate reverse primer, either JH, Cμ, Cγ, or Cα.27 The PCR reaction mixture was the same as for the CDR3-specific PCR except that 1 unit of Taq polymerase and 0.25 μM of each primer was used. Thirty cycles of 30 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C were performed. The reaction was terminated for 6 minutes at 72°C. The PCR products were analyzed on a 1% standard agarose gel (Sigma, St. Louis, MO).
Cloning and sequencing of PCR products
PCR products were cloned into pGEM-T vectors (Promega, Leiden, the Netherlands) and transformed into DH10b bacteria (Life Technologies). Subsequently, from 4 or more colonies both strands of the inserts were sequenced to obtain the sequence of the dominant clone, the consensus sequence. Sequencing was performed with an ABI sequencer (Perkin Elmer Corp, Norwalk, CT) using the dye-terminator cycle-sequencing kit (Perkin Elmer). To determine the Taq error rate of our experimental design, 19 clones of CD79a and CD79b were sequenced. These clones were generated according to the same PCR and cloning procedures as used for the VH genes. The Taq error frequency thus established is 0.14%, which amounts to 0.4 mutation/VH clone (data not shown).
Assignment of mutations
The sequences found were compared with published germline sequences using the Vbase database28 and DNAplot29 on the Internet (http://www.mrc-cpe.cam.ac.uk/imt-doc) to identify mutations. The last nucleotide position of the V gene was excluded from the mutational analysis in view of possible nucleotide deletions at the joining sites.
Statistical analysis
To calculate whether the excess or scarcity of replacement (R) mutations in the FRs had occurred by chance, we used the binomial distribution model as proposed by Chang and Casali.30 If the B cell was selected for antibody expression, there is a counterselection for R mutations in the FRs. The ratios of R versus silent (S) mutations (R/S) found in the CDRs are often higher than expected but are not used by us as arguments for or against antigenic selection; Dörner et al showed that the R/S values of both FRs and CDRs were higher in nonproductive and, therefore, not antigen-selected rearrangements than in productive, antigen-selected rearrangements.31 Also, additional R mutations in the CDRs can be unfavorable in already selected immunoglobulin with high affinity.32 To calculate the significance of the differences in the amounts of somatic mutation in IgM+ and isotype-switched FLs, the Mann-Whitney 2-sample test was used. In this analysis, every tumor sample was included as a separate entity.
Results
Clonality assessment and isotype expression
Of the selected panel of FLs, clonality and isotype expression were established immunohistochemically. To confirm this at the molecular level, the CDR3 region was amplified with a FR3 primer in combination with primers specific for the JH gene or the 5′ regions of Cμ, Cγ (Cγ2), Cα, or Cδ.27With this PCR we can assess clonality and isotype expression with a sensitivity of about 25% clonal cells in a polyclonal background. In 30 of 59 FLs, we observed a sharp band on agarose gel with both the JH primer and 1 or 2 of the primers specific for the constant regions as downstream primers. In another 13 cases, we could establish clonality only with a Cμ, Cγ, Cα, or Cδ downstream primer and not with the JH primer. In summary, in 43 FLs (72.9%) we could confirm clonality with a CDR3-specific PCR. This percentage is similar to what has been reported by others for PCR clonality assays of FLs.33-35 Of these cases, we chose 30 FLs for our analyses of the VH gene sequences (Table1). In this selection, isotype-switched FLs were relatively overrepresented. In 3 cases, immunohistochemical determination of the expressed H chain was not clear (nos. 6, 25, and 56), whereas by RT-PCR clonal bands with the Cγ2-specific primer were obtained (not shown). In summary, 12 FLs expressed IgM, of which at least 10 coexpressed IgD at the RNA level; 13 FLs expressed IgG; and 1 FL expressed IgA (Table 1). Interestingly, in 1 of these lymphomas, no. 8-'83, both IgM- and IgG-expressing tumor cells were found by immunofluorescence (not shown). In accordance, CDR3-specific RT-PCR yielded sharp bands in the lanes corresponding to the JH, Cμ, Cδ, and Cγ downstream primers (Figure1). Sequencing of the VH-Cμ and the VH-Cγ PCR products showed that both tumor cell populations contained the same VDJ rearrangement (Table 1). A relapse sample of this patient contained only the IgG-expressing tumor cell population, assessed immunohistochemically and by CDR3-specific PCR (Figure 1).
RT-PCR analysis of the CDR3 region of FL no. 8-'83 (left panel) and its relapse, no. 8-'92 (right panel).
Upstream, the FR3 primer was used in combination with the JHseq, Cμ, Cγ2, Cα, and Cδ downstream primers, as indicated above the lanes.
RT-PCR analysis of the CDR3 region of FL no. 8-'83 (left panel) and its relapse, no. 8-'92 (right panel).
Upstream, the FR3 primer was used in combination with the JHseq, Cμ, Cγ2, Cα, and Cδ downstream primers, as indicated above the lanes.
VH, D, and JH gene usage
The complete VH genes were amplified with family-specific VH leader primers in combination with a JH primer or constant H chain isotype-specific primers. To ascertain that these VH products originated from the tumor population, we performed a nested CDR3-specific PCR on the VH products obtained. In all cases, the nested CDR3 products and the original CDR3 products had identical sizes. Next, the VH products were cloned and sequenced. The number of somatic mutations was determined by comparing the VHsequences to the germline genes with the highest homology (Table 1). Fifteen FLs used genes from the VH3 family (60%). VH4 and VH1 family genes were found in 6 FLs (24%) and 2 FLs (8%), respectively. VH5 and VH7 genes were each found once (4%). The JH4b gene was found in 9 FLs, JH6 genes were found in 6 FLs, and JH3 and JH5 genes were each used by 4 FLs. The JH1 and JH2 genes were each found once. A stretch of 7 consecutive nucleotide matches was taken as an indication that a certain D gene was used. According to this method, only 6 D genes could be assigned (Table2).
Number of somatic mutations
Most FLs, including the IgM-expressing FLs, were heavily mutated (Table 1). Still, in this selection of FLs we observed a statistically significant difference in the number of somatic mutations in IgM-expressing versus isotype-switched FL samples (P < .016; Figure 2). On average, FL samples of IgM isotype contained 23.0 somatic mutations (7.8%), ranging from 8 to 49 somatic mutations. The FL samples of IgG isotype contained an average of 34.1 somatic mutations (11.6%), with a range from 8 to 51 somatic mutations. The IgA-expressing FL harbored 29 somatic mutations (9.9%; Table 1). These differences in mutation load are also obvious from calculations on individual molecular clones of these lymphomas: When for each clone the amount of somatic mutations compared with the germline gene was calculated and when these amounts were averaged between the clones, the difference in somatic mutation loads amounted to 34.6 mutations per VH gene for isotype-switched FLs and 23.6 mutations for IgM+ FLs (data not shown). In these clonal analyses, however, Taq errors are included (≤ 0.4 mutations per clone).
The number of somatic mutations versus the H chain isotype (IgM or IgG/IgA).
The lines represent the average amount of mutations of IgM+FLs and isotype-switched FLs, respectively.
The number of somatic mutations versus the H chain isotype (IgM or IgG/IgA).
The lines represent the average amount of mutations of IgM+FLs and isotype-switched FLs, respectively.
Intraclonal variation
To assess the degree of intraclonal variation, independent clones of 21 FLs were sequenced (Table 1). In 15 of these FLs, intraclonal variation above the Taq error frequency was observed, ranging from 0.5 to 14.6 mutations per clone. Moreover, in 11 of these 15 lymphomas, mutations were found that were shared by more than 1 clone (data not shown), which argues strongly for somatic hypermutation rather than Taq error. No obvious difference in the level of intraclonal variation in IgM- and IgG-expressing FLs was observed. In 6 FLs (nos. 3-'93, 3-'95, 15, 25, 17, and 57) the intraclonal variation did not exceed the Taq error rate of 0.14%, which amounts to 0.4 mutations per clone.
Mutation patterns
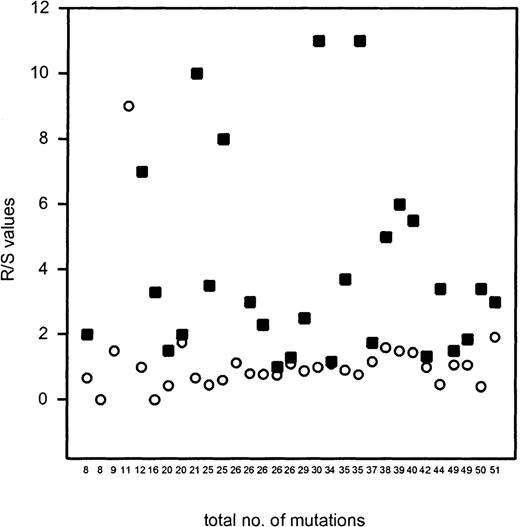
To assess whether the FLs—with variable levels of mutation and intraclonal diversity—showed signs of selection for a potentially functional BCR, the distribution of R and S mutations in FRs and CDRs was analyzed. For this purpose, the dominant or “consensus” VH sequences of individual FLs were analyzed. The ratio of R/S mutations was highly variable in the CDR1 and CDR2 (average 3.9), whereas in the FRs the R/S ratio was consistently lower than 2.0 (average 0.97), with 1 exception (FL no. 59; Table3 and Figure3). When the distribution of R and S mutations was analyzed according to the binomial distribution model by Chang and Casali,30 the number of R mutations in the FRs of most FL samples was significantly lower than would be expected if the mutations had occurred at random and in the absence of selective forces. The nature of the R mutations (conservative or nonconservative) was not different between the CDRs and FRs. These data clearly indicate that in FLs the overall structure of the BCR is maintained irrespective of the total number of somatic mutations and the degree of intraclonal variation (Table 3 and Figure 3). Only in lymphoma no. 59 was the R/S value in the FR exceptionally high: 9 R mutations versus 1 S mutation were found in the FRs, 5 of which occurred in the FR1. The absolute number of R mutations in the FRs, however, did not differ much from some other FLs. Furthermore, because expression of surface IgG was found immunohistochemically (Table 1), the high amount of R mutations in the FRs apparently did not interfere with BCR expression.
The R/S values of the CDRs and the FRs plotted against the total number of mutations of individual cases of FLs.
R/S values that amounted to infinite in the CDRs are not shown. The figure shows that R/S values in FRs are consistently low irrespective of the total number of somatic mutations, except for FL no. 59 (R/S ratio of 9; 11 mutations in total). ▪ indicates the R/S value of the CDRs; ○ indicates the R/S value of the FRs.
The R/S values of the CDRs and the FRs plotted against the total number of mutations of individual cases of FLs.
R/S values that amounted to infinite in the CDRs are not shown. The figure shows that R/S values in FRs are consistently low irrespective of the total number of somatic mutations, except for FL no. 59 (R/S ratio of 9; 11 mutations in total). ▪ indicates the R/S value of the CDRs; ○ indicates the R/S value of the FRs.
Isotype switch variants and relapse of follicular lymphoma no. 8
In FL no. 8-'83, both IgM- and IgG-expressing tumor cells were found, which harbored the same VDJ rearrangement. Compared with the germline gene V3-23, the IgM-related sequence contained 30 somatic mutations, whereas the IgG-related sequence contained 35 somatic mutations (Table 1, Figure 4). The patient was treated with chemotherapy and achieved complete remission. A local relapse 5 years later was irradiated, and 9 years after the first presentation a systemic relapse developed (no. 8-'92), which was again treated chemotherapeutically. This relapse consisted of IgG-expressing tumor cells only. Compared with the germline gene, also 35 somatic mutations were found, of which 30 somatic mutations were shared with the IgM- and IgG-related sequence of the first time point, and 5 were different (Figure 4). In all VH gene sequences of lymphoma no. 8, a deletion of codon 3 was observed (not shown), which is most likely due to somatic mutation.36 37
Schematic representation of VH sequences of patients no. 3, 6, 8, and 66.
Each line represents the consensus sequence at a certain time point. Only mutations that differ between the sequences are shown. ○ represents an S mutation compared with the germline gene; • represents an R mutation. The codon numbers in which mutations took place are indicated under these symbols. The total number of somatic mutations for each FL sample (in bold) and the intraclonal variation, in number of mutations per clone, is indicated next to each line.
Schematic representation of VH sequences of patients no. 3, 6, 8, and 66.
Each line represents the consensus sequence at a certain time point. Only mutations that differ between the sequences are shown. ○ represents an S mutation compared with the germline gene; • represents an R mutation. The codon numbers in which mutations took place are indicated under these symbols. The total number of somatic mutations for each FL sample (in bold) and the intraclonal variation, in number of mutations per clone, is indicated next to each line.
Relapses of follicular lymphomas no. 3, 6, and 66
Patient no. 3, who initially achieved complete remission after chemotherapy, developed a relapse after 2 years. In the relapse sample, the total amount of somatic mutations in the VH gene was lower than in the presentation biopsy, 21 and 26 mutations, respectively. Compared with the germline gene V3-7, 19 mutations were found that were shared between the 2 time points (Table 1 and Figure4). No significant intraclonal variation was found at either time point. Patient no. 6 presented with a stage IV, low-grade FL. No treatment was given (“watchful waiting”). After 4 years, a second lymph node was resected because of clinical progression. The total number of somatic mutations compared with the germline gene V3-23 was 44 in the presentation biopsy, compared with 50 somatic mutations in the second sample (Table 1 and Figure 4). In the samples of both time points, intraclonal variation above the Taq error was found, but in time the intraclonal variation had decreased from 2.8 to 1.8 mutations per clone. Patient no. 66 developed 2 relapses, 1 and 2 years after the presentation of the FL in 1996, respectively. Chemotherapeutic treatment started in 1997 after clinical progression, upon which only partial remission was achieved. The somatic mutation loads in the samples of these 3 time points were 38, 39, and 40 mutations, respectively, whereas the intraclonal variation gradually decreased from 3 mutations per clone to 0.5 mutations per clone. Curiously, although the absolute amount of somatic mutations increased, the location of the mutations was not the same in samples of these 3 time points (Figure 4). Compared with the germline gene, 37 somatic mutations were shared between the 3 time points. In the presentation biopsy, 1 additional R mutation was found at codon 23. This mutation was not found in the sample of the second time point. In addition, 2 other R mutations were found at codon 57 and 64. In FL no. 66-'98, except for the 37 shared mutations, a new R mutation was found at codon 5. The mutation found at codon 23 was shared with the first time point, whereas the mutation at codon 64 was shared with the second time point (Figure 4). Also, at the clonal level only a slight increase was found in the number of somatic mutations: The average number of somatic mutations of the clones compared with the germline gene V4-39 was 39.7 in the samples of the first 2 time points and 40.5 in 66-'98 (data not shown).
Discussion
In this study, the VH genes of 30 FL samples of different isotypes were analyzed. In agreement with Bahler et al,18 the VH family gene usage of the FLs studied was not obviously different from VH gene usage reported for normal B cells.38,39 Compared with normal postfollicular B cells derived from tonsil, spleen, or lymph node, the VH genes of FLs contained significantly more mutations, which is compatible with prolonged expansion in a germinal center–like environment. We observed mutation numbers ranging from 8 to 51 (2.7% and 17.2%, respectively), with an average of 29.3 (9.9%) per VH gene. Similar frequencies have been reported previously by others.18 19 In 15 of the 21 FLs studied at the clonal level, intraclonal variation above the Taq error frequency was found, varying from 0.5 to 14.8 mutations per cloned VH gene (Table 1).
The patterns of somatic mutations in most of the FLs studied are suggestive of selection for preservation of the BCR. In 26 of 31 FL samples, a significantly lower amount of R mutations was found than what would be expected in case of random mutagenesis in the absence of selection.30 This suggests that, at least at some time in the development of the lymphomas, the expression of a functional BCR has been important for cell survival. It is, however, likely that most of the “consensus” mutations were introduced before the outgrowth of the transformed clone. Therefore, the distribution of these mutations alone does not allow the conclusion that FLs need a preserved BCR structure for their survival. However, if somatic mutation occurs in the tumor stage, the finding that all FLs bear a structurally preserved BCR is significant because this must be the result of continued selection.
Several groups analyzed somatic mutation in normal B cells.37,40-44 Although variable numbers were reported—varying from 1% to 6%—all studies that analyzed both IgM- and IgG-expressing germinal center or memory B cells reported more somatic mutations in the IgV genes of isotype-switched B cells than in IgM+ B cells.40-42 It is of note that even though the overall number of somatic mutations in the IgV genes of FLs is higher than in normal postgerminal center B cells, we also find that isotype-switched FLs contain more somatic mutations than IgM+ FLs (Figure 2). Apparently, isotype-related differences in mutation frequency, which are potentially present at the moment of transformation of a single cell, are being maintained during the course of the disease.
FL no. 8-'83 contained both IgM- and IgG-expressing tumor cells. The finding that in FL no. 8-'83 the IgM-expressing cells are already heavily mutated suggests that the onset of somatic mutations occurred prior to isotype switching, like in normal B cells26 (Table1 and Figure 4). Both the immunohistochemical stainings on FL no. 8-'83, as well as the finding that the IgM- and IgG-derived VH genes contained different somatic mutations and a different amount of intraclonal variation (Table 1 and Figure 4), strongly suggest that the IgM+ and IgG+ cells represent different subpopulations. In our hands, presence of isotype switch variants in FLs is an unusual finding, because we observed it only once in this series of 43 FLs with our CDR3-specific PCR (data not shown). However, our approach may not be sufficiently sensitive to detect alternative isotypes, because we did not use clone-specific CDR3 primers but a consensus FR3 primer in combination with constant region primers.
In 4 patients, a relapse was biopsied (Table 1). Comparison of the VH gene sequences of lymphoma no. 3-'93 and its relapse, no. 3-'95, 2 years later, revealed a lower amount of somatic mutations in the VH gene of the sample of the last time point. In this respect, it is noteworthy that no significant intraclonal variation was found at either time point in this lymphoma. Apparently, in the course of disease or due to therapy, a minor subclone with a lower number of mutations, which was not detected at the time of diagnosis, had been selected. In patient no. 6 the absolute amount of somatic mutations increased significantly in 4 years, whereas the intraclonal variation decreased. In this case, the tumor population of the relapse sample could indeed have evolved by ongoing somatic mutation in combination with clonal selection. Patient no. 66 developed 2 relapses in 2 years. The number of mutations increased slightly in time, whereas the intraclonal variation decreased gradually (Figure 4, Table 1). By studying clonal relationships, there was no clear evidence of clonal evolution of this FL in time. In fact, the mutation patterns suggest that the tumor populations of the different FL samples of patient no. 66 represent different subclones that obtained a selective growth advantage at certain time points. Subclone selection rather than clonal evolution was even more apparent after the 9-year disease interval of patient no. 8. Surprisingly, despite the high intraclonal variation at the first time point (5 and 4 mutations per clone for the IgM- and IgG-derived subpopulation, respectively) and a lower, but still significant, intraclonal variation of the relapse (1.3 mutations per clone), no change in the absolute number of mutations was found compared with the IgG-derived sequence of the first time point (Figure 4). After the 9-year interval, one might have expected an accumulation of mutations, at least measurable at the clonal level. The absence of such an increase cannot be explained by clonal selection, because we would still expect an increase in S mutations, which is not the case in patient no. 8 or in patients no. 3 and 66. Thus, in 3 of 4 patients, treated or untreated, intraclonal variation had decreased in time. A decrease in intraclonal variation as well as the absence of a significant increase of somatic mutations in time seems a rule rather than an exception and has, in fact, also been documented by others22,23,45: Zhu et al described an FL that was heterogeneous at presentation, but the relapse 5 years later showed no intraclonal variation. The VH sequences at both time points were exactly the same. Four samples of an FL were described by Bahler et al with an interval spanning 2 years. Only in the first sample of the lymph node was intraclonal variation found. All lymphoma samples from different locations, except for the last relapse sample, contained 24 mutations compared with the germline gene. The tumor cells in this final relapse sample contained only 19 mutations compared with the germline gene. Most likely, a nonmutating subclone with a lower number of mutations was positively selected after chemotherapy, like in patient no. 3 of our series. This subclone must have been present at the earliest time points but not detected. Ottensmeier et al described 2 samples of an FL spanning an interval of 10 months.23 The first biopsy showed 22 somatic mutations compared with the germline gene, whereas the lymphoma population after therapy contained only 13 mutations. Intraclonal variation was present at both time points but was found to be lower in the sample of the second time point. To our knowledge, only Cleary et al observed a higher amount of intraclonal variation in the relapse of an FL.46 In addition, the absolute amount of somatic mutations increased from 35 to 38 after anti-idiotype therapy. However, in a subsequent article this group also suggested that the clones that survived the therapy might already have been present in the first biopsy as minor subclones.47
Based on the present data, it can be questioned whether intraclonal nucleotide differences in FLs are necessarily a reflection of an active mutation machinery. In general, it can be assumed that FL patients have a significant tumor load at the time of diagnosis. The fact that most patients are in Ann Arbor stage III or IV supports this notion.17,48 With the presumption that the tumor cell populations are the offspring of a single cell originally, it can be conceived that numerous cell divisions must have occurred. In view of these many cell divisions, the intraclonal differences are actually surprisingly low. The rate of somatic hypermutation, if present, must be substantially lower than in normal germinal center B cells. With respect to the decrease in clonal diversity in time in FLs, observed by us and others, it can be argued that subclones with lower or absent mutation rates are selected preferentially, possibly due to therapy. Alternatively, it can be hypothesized that, at least in cases in which the intraclonal variation decreased and no clear increase in the absolute amount of somatic mutations was found in time, the neoplastic cells do not mutate anymore. It can be envisioned that after a certain point in an ongoing transformation process, the proliferating cells are prone to acquire additional genetic alterations that give a growth advantage but interfere with the potential of somatic hypermutation or heavy chain isotype switching. Paradoxically, the somatic hypermutation process may by itself be instrumental in the process of further de-differentiation because non-Ig genes also can be targets.49 50 These genetic alterations may thus take place in several, somatically diversified, daughter cells and may even take place after isotype switch of some of the expanded clones. After cessation of active mutation, intraclonal heterogeneity may not immediately be abolished but may be maintained to a certain degree for variable periods, also depending on therapy susceptibility. This point is not trivial, because the concept of antigen-driven lymphomagenesis relies strongly on the capacity of tumor cells to somatically mutate. In other words, within the group of FLs there may be cases in which, despite intraclonal V gene diversity and mutation patterns compatible with antigen selection, BCR ligands may not play a decisive role in their propagation anymore.
Acknowledgment
The authors thank N.J. Ponne for technical assistance with sequencing, J.B.G. Mulder for assistance with immunohistochemistry, and J.W. Vaandrager and C.C.H. Vellema for kindly collecting and providing tissue material from FLs. M. Ek and Dr C.E.M. Hollak are thanked for providing clinical data. We also thank Dr R. Küppers for providing intrinsic R/S values of several VH genes, Dr A.C. Tersmette for advice on statistics, and Dr O.J. de Boer for designing a computer program to calculate the probability of the R/S distributions found.
Supported by a grant from the Dutch Cancer Society (Grant no. AMC 95-957). C.J.M.v.N. is a fellow of the Netherlands Royal Academy of Arts and Sciences.
Reprints:C. J. M. van Noesel, Department of Pathology, Academic Medical Center, Meibergdreef 9, 1105 AZ, Amsterdam, the Netherlands; e-mail: c.j.vannoesel@amc.uva.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.