Activation and dysfunction of endothelial cells play a prominent role in patho-physiological processes such as atherosclerosis. We describe the identification by differential display of 106 cytokine-responsive gene fragments from endothelial cells, activated by monocyte conditioned medium or tumor necrosis factor-. A minority of the fragments (22/106) represent known genes involved in various processes, including leukocyte trafficking, vesicular transport, cell cycle control, apoptosis, and cellular protection against oxidative stress. Full-length cDNA clones were obtained for five novel transcripts that were induced or repressed more than 10-fold in vitro. These novel human cDNAs CA2_1, CG12_1, GG10_2, AG8_1, and GG2_1 encode inhibitor of apoptosis protein-1 (hIAP-1), homologues of apolipoprotein-L, mouse rabkinesin-6, rat stannin, and a novel 188 amino acid protein, respectively. Expression of 4 novel transcripts is shown by in situ hybridization on healthy and atherosclerotic vascular tissue, using monocyte chemotactic protein-1 as a marker for inflammation. CA2_1 (hIAP-1) and AG8_1 are expressed by endothelial cells and macrophage foam cells of the inflamed vascular wall. CG12_1 (apolipoprotein-L like) was specifically expressed in endothelial cells lining the normal and atherosclerotic iliac artery and aorta. These results substantiate the complex change in the gene expression pattern of vascular endothelial cells, which accompanies the inflammatory reaction of atherosclerotic lesions.
ENDOTHELIAL CELLS play a central communicative role between the blood in the lumen of the vessel and the surrounding vascular tissue.1,2 These cells form the nonthrombogenic lining of the vessel and play an active role in maintaining the hemostatic balance, eg, via the expression of the anticoagulant cofactor thrombomodulin, the secretion of the profibrinolytic tissue-type plasminogen activator, and the production of prostanoids that attenuate platelet activation. Invasion of the vessel wall by circulating monocytes and other blood-borne cells is actively promoted by the endothelial cells via the regulated expression of adhesion molecules. On the abluminal side there is constant communication between the endothelial cells and the underlying smooth muscle cells, causing them to contract or relax in response to altering blood pressure and flow via the production of signaling molecules such as prostanoids, endothelins, and nitric oxide. As a result of these functions, activation and dysfunction of endothelial cells play a crucial role during the initiation and progression of atherosclerosis.1-3 Furthermore, activation of endothelial cells plays a definitive role in many other processes, such as tumor-induced angiogenesis, allergic inflammation, adult respiratory distress syndrome, and a variety of inflammatory and allergic disease states.4-6 The altered properties of the endothelial cells results from altered patterns of gene expression, eg, invasion of the vessel wall by leukocytes can only occur after the induced expression by the endothelial cells of adhesion molecules such as E-Selectin and vascular cell adhesion molecule-1 (VCAM-1).1,7 Only a small percentage of the total human gene repertoire will be subject to change in a given pathological situation, because the majority of genes plays a more basal housekeeping role within any given cell type. Many genes and proteins have been implicated in atherosclerosis, based on studies of single specific genes and proteins.2 However, the mere fact that specific functions have been assigned to only 5% to 10% of the estimated total of 50,000 to 100,000 human genes indicates that a full understanding of the atherosclerotic process is far from established, because it is conceivable that many of the presently unknown genes might also play a prominent role in this disease.8 9 The identification and characterization of these novel atherogenesis-related genes is the goal of our studies to define the difference between a resting (nonatherogenic) and an activated (atherogenic) endothelial cell at the level of gene expression. Next, the specific functional roles of the corresponding gene-products in atherosclerosis will increase our knowledge about the patho-physiology of the atherosclerotic vascular wall and may identify targets for noninvasive early diagnosis and/or therapeutic agents.
The infiltration of the vessel wall by monocytes is a hallmark of the initiation and progression of atherosclerosis. Activated monocytes and their resulting macrophage foam cells will secrete a complex mixture of cytokines,3,10,11 including tumor necrosis factor-α (TNF-α), which greatly affects endothelial cell function.12 Indeed, one of the cellular mediators of the TNF-α response, NF-κB, has recently been identified in atherosclerotic lesions.13 The secretion products of monocytes, including TNF-α, are known to have a profound influence on the expression levels of certain endothelial cell genes, of which the adhesion molecules E-selectin, VCAM-1, and intercellular adhesion molecule-1 (ICAM-1) have been studied in great detail.1,2 Little is known about other cytokine responsive genes, although some important ones have been studied with respect to the influence of endothelial cells on the hemostatic balance, such as tissue factor, plasminogen activator inhibitor-1, tissue-type plasminogen activator, and thrombomodulin.1 We have set out to expand our knowledge of changes in endothelial cell gene expression patterns in response to monocyte stimulation to describe the role of endothelium in initiation and progression of atherosclerosis at the molecular level. Unfortunately, the composition and architecture of atherosclerotic lesions are extremely diverse: these lesions have greatly varying cell-type composition and occur in many different vessels, which are embedded in different tissues with cells derived from different embryonic origin. Therefore, we have taken an unbiased approach to identify candidate atherogenesis-involved known and novel genes in pure cell cultures. Next, in vivo significance is tested by studying expression of these genes in a variety of atherosclerotic lesions. The differential display of gene expression technique by random-primed reverse transcriptase-polymerase chain reaction (DD/RT-PCR)14 15 was used to detect variations of mRNA levels upon activation of cultured endothelial cells. This method allowed us to identify genes that are either induced or repressed by a given stimulus. Furthermore, the isolation of total RNA from resting and cytokine-stimulated endothelial cells at various time points enabled us to identify genes of different temporal kinetics, ie, immediate early, delayed early, and late genes. In this report, we describe the identification, in an unbiased way, of a large panel of candidate genes that are potentially involved in atherosclerosis by mimicking inflammatory conditions in vitro. The full-length cDNA for 5 novel genes is reported, and we started the localization of gene expression for these novel genes in vascular endothelial cells by in situ hybridization on human healthy and diseased vascular material.
MATERIALS AND METHODS
Cell culture and fluorescence-activated cell sorting (FACS) analysis.
Endothelial cells were isolated from nontraumatized human umbilical veins as described.16 These human umbilical vein endothelial cells (HUVEC) were cultured in gelatin-coated tissue-culture flasks (Nunc, Roskilde, Denmark) in medium composed of equal parts of Medium-199 and RPMI-1640 (GIBCO-BRL, Paisley, Scotland), supplemented with 20% (vol/vol) heat-inactivated, pooled human serum, 2 mmol/L glutamine, and a 1/100 dilution of antibiotic/antimyotic mix (GIBCO-BRL; final concentrations: 100 U/mL penicillin, 100 U/mL streptomycin, and 2.5 mg/mL fungizone) at 37°C in a 5% CO2 humidified air incubator. HUVEC were passaged once with trypsin/EDTA (GIBCO-BRL) and only secondary cultures, which had been confluent for 2 or 3 days, were used for all experiments described in this report. Identity of the HUVEC was confirmed by their cobblestone morphology and positive staining for von Willebrand factor. Primary human monocytes were isolated from fresh buffy coats (Central Laboratory of the Netherlands Blood Transfusion Service, Amsterdam, The Netherlands) by Ficoll gradient centrifugation and attachment to tissue-culture plastics. The monocytes (±106 cells per flask) were washed twice with serum-free Iscove’s modified Dulbecco’s modified Eagle’s medium (DMEM; GIBCO-BRL) and were cultured overnight at 37°C in a humidified, 5% CO2/air incubator in 8 mL of Iscove’s supplemented with 2% human albumin, insulin/transferin/selenite growth supplement (Sigma, St Louis, MO), and antibiotic/antimyotic. The conditioned medium was collected after 16 hours and stored in aliquots at −70°C. HUVEC received fresh full-growth medium 16 hours before the beginning of the activation experiment. Human recombinant TNF-α was lot no. DOE 247/91 (Bayer AG, Wuppertal, Germany). Stimulation was performed either with TNF-α added to the full growth medium (20% human serum) or with serial dilutions of the monocyte-conditioned medium in serum-free medium. At different time points the cells were washed with phosphate-buffered saline (PBS) and trypsinized for flow cytometric analysis. Suspended cells were washed twice in cold PBS and subsequently fixed in ice-cold PBS containing 1% bovine serum albumin (BSA), 0.3 mmol/L EDTA, 0.01% (wt/vol) sodium azide, and 0.1% (wt/vol) p-formaldehyde at a final concentration of 5 × 106 cells/mL. The primary antibodies used were directed against tissue factor and ICAM-1, respectively. After the addition of the primary monoclonal antibody to the suspension, the cells were incubated for 30 minutes at 4°C and washed twice in cold PBS containing 1% (wt/vol) BSA, 0.3 mmol/L EDTA, and 0.01% (wt/vol) sodium azide. Subsequently, R-phycoerythrin (RPE)-conjugated F(ab′)2 fragments of rabbit-antimouse Igs (R 0439; Dako A/S, Glostrup, Denmark) were added, and cells were incubated for another 30 minutes at 4°C. After two washes, HUVEC were gated by forward scatter and side scatter using a FACScan (Becton Dickinson, Cowley, Oxford, UK) and 5,000 cells were counted. This established that maximal stimulation, as evidenced by maximal fluorescence shift, was obtained for both tissue factor and ICAM-1 at concentrations of 12 nmol/L TΝF-α and the 1/10 diluted monocyte-conditioned medium.
RNA isolation and DD/RT-PCR.
Total RNA was isolated from resting and activated cells with TRIZOL (GIBCO-BRL) with a typical yield of 80 μg/8 × 106cells and stored at −70°C. The DD/RT-PCR reactions were performed with 12 different anchored primers (T11XY; X = A, C, or G; Y = A, C, G, or T) and arbitrary decamers no. 1 through 12 as described.15 In brief, 0.2 μg total RNA was reverse transcribed with 20 U Superscript-RTII (GIBCO-BRL) in the presence of 2.5 μmol/L anchored primer and a dNTP concentration of 20 μmol/L in a volume of 20 μL first-strand buffer. After the addition of 10 μL water, 2 μL of first-strand cDNA was subjected to DD/RT-PCR in a Perkin-Elmer 9600 thermocycler (Perkin Elmer, Norwalk, CT) using GeneAmp tubes in a total volume of 20 μL under the following conditions: 1× PCR-II buffer, 1.5 mmol/L MgCl2, 2 μmol/L dNTPs, 1 μmol/L anchored primer, 0.5 μmol/L decamer, 2 μCi (60 nmol/L) α-33P-dATP (Redivue, Amersham, UK), and 1 U AmpliTaq (Perkin Elmer) for 40 cycles of 30 seconds at 94°C, 2 minutes at 40°C, and 30 seconds at 72°C, followed by 10 minutes at 72°C. After completion of the reaction, 14 μL loading mix (90% [vol/vol] formamide, 1 mmol/L EDTA, Xylene cyanol, and BromoPhenol Blue) was added and a 2- to 5-μL sample was analyzed by standard 6% sequencing polyacrylamide gel electrophoresis (PAGE; 8 mol/L urea) at 70 W. Subsequently, the gel was transferred to Whatman paper (Whatman, Maidstone, UK) and vacuum-dried without prior fixation. The gel was marked at the 4 corners with radioactive ink and analyzed by autoradiography after exposure for 16 to 72 hours at room temperature to BioMax film (Eastman Kodak, Rochester, NY). Maximal comparability of the reactions that were to be analyzed in concert was ensured by using core mixes for both RT-reactions and DD/RT-PCR reactions and performing all reactions in concert in 96-well format for a single anchored primer, all 12 decamers, and the 10 individual RNA preparations. These RNAs were isolated from unstimulated HUVEC (0 and 20 hours) and HUVEC exposed to TNF-α or monocyte-conditioned medium for 3, 6, or 20 hours each.
Cloning and sequencing of DD/RT-PCR fragments.
DD/RT-PCR fragments of interest were recovered from sequencing gels and reamplified by the DD/RT-PCR protocol at a dNTP concentration of 40 μmol/L. Reactions were analyzed on 1.5% agarose gel using the 100-bp marker (GIBCO-BRL), and the bands of appropriate length were excised and purified using QIAEX (Qiagen GmbH, Hilden, Germany). These inserts were TA-cloned by ligation to either the pCR-II vector (Invitrogen, Carlsbad, CA) or pGEM-T (Promega, Madison, WI) according to the manufacturer’s instructions. Sequencing of DD/RT-PCR fragment clones was performed on purified plasmid DNA using the AutoRead Sequencing-kit and Cy5-labeled T7 or SP6 oligonucleotides and analyzed on the ALF-express automatic sequencer (materials and protocol: Pharmacia, Uppsala, Sweden). Sequence files from the ALF-express were exported in GCG-format and analyzed and stored using the GCG-program (Wisconsin Package Version 9.1, Genetics Computer Group [GCG], Madison, WI) run on a mainframe UNIX computer. A GCG database was constructed using DATASET and all sequences were cross-analyzed by the FASTA algorithm. Sequence identity was confirmed by BLAST searches on the combined Genbank/EMBL nonredundant (nr) and expressed sequence tag libraries (dbEST), accessed through the National Center for Biotechnology Information homepage (http://www.ncbi.nlm.nih.gov./). Protein homologies were traced using the Blitz algorithm run at the EMBL/European Bioinformatics Institute (http://www.ebi.ac.uk), and functional protein domains were traced using the BLOCKS WWW-server (http://www.blocks.fhcrc.org/). Multiple protein sequence alignments were performed using the ClustalW service at EBI.
Northern blotting analysis.
Formamide and heat-denatured 10 μg total RNA was electroforesed on a standard formaldehyde 1% agarose gel and blotted to Hybond-N nylon membranes (Amersham, Amersham, UK) according to the manufacturer’s protocol. Filters were prehybridized in 5× SSPE (20× SSPE is 3.6 mol/L NaCl, 0.2 mol/L sodium phosphate, pH 7.7, 0.02 mol/L EDTA), 0.5% sodium dodecyl sulfate (SDS; wt/vol), 5× Dennhardts’ (100× Denhardt’s solution is 2% [wt/vol] BSA, 2% [wt/vol] Ficoll, and 2% [wt/vol] polyvinylpyrrolidone), and 50% (vol/vol) deionized formamide at 42°C in a hybridization oven for at least 4 hours. Hybridizations were performed at greater than 1 × 106 cpm/mL for 16 to 24 hours at 42°C in a solution of 5× SSPE, 0.5% SDS (wt/vol), 10% dextrane-sulfate (wt/vol), and 50% (vol/vol) deionized formamide. As probes, we used agarose-purified restriction fragments corresponding to the original DD/RT-PCR fragment or IMAGE-clones labeled to high specific radioactivity using the random oligo labeling kit (GIBCO-BRL) and α-32P-dATP (Redivue; Amersham); unincorporated nucleotides were removed by the Qiaquick nucleotide removal kit (Qiagen GmbH). Filters were washed for 15 minutes in 2× SSPE at room temperature, followed by 30 minutes in 1× SSPE at 65°C and twice for 10 minutes in 0.2× SSPE at 65°C. Filters were analyzed by autoradiography after exposure to Xomat-R films (Eastman Kodak) for various periods at −70°C using intensifying screens. Radioactivity was quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Construction and screening of an activated EC cDNA library.
Total RNA was isolated with Trizol from HUVEC that had been stimulated for 6 hours with the conditioned medium from elutriated monocytes that had been activated overnight with limitedly oxidized human low density lipoprotein (LDL) (Sigma). First-strand cDNA synthesis was performed with 10 μg of total RNA, using Superscript RT-II reverse transcriptase and oligo-dT. The second-strand synthesis was performed by the modified procedure of Gubler and Hoffmann.17Subsequently, the cDNA was treated with T4 DNA polymerase and ligated to an excess of nonpalindromic BstXI-linkers. Finally, the cDNA was size fractionated on a low melting type agarose gel and cDNA exceeding 600 bp was divided in two pools of sizes less than 1,500 bp and greater than 1,500 bp. Both pools were separately ligated into theBstXI sites of the vector prcCMV (Invitrogen). Ligation mixtures were electroporated into Escherichia coli strain MC 1061/P3 and the cDNA library was plated as 1 × 105and 6 × 104 independent colonies for sizes less than 1,500 bp and greater than 1,500 bp, respectively. The cDNA library was screened by colony hybridization using radioactive probes, identical to those used for Northern blotting analysis (see above). Hybridization of the probe was performed at 65°C for 16 hours in the presence of 0.1% (wt/vol) SDS, 5× Denhardts’ solution, 0.1 mg/mL herring sperm DNA, and 6× SSC (20× SSC is 3 mol/L NaCl, 0.3 mol/L sodium citrate). The Hybond-N membranes were washed with increasing stringency down to 0.2× SSC. IMAGE Consortium (LLNL) cDNA-clones48 (http://www-bio.llnl.gov/image/) were obtained from the American Type Culture Collection (ATCC; Rockville, MD; http://www.atcc.org/) and their identity was confirmed by resequencing of purified plasmid from individual colonies.
Vascular tissue collection, morphological analysis, and immuno-histochemistry.
Human vascular tissue specimens displaying various stages of atherosclerosis were collected after obtaining informed consent during organ transplantations from multiorgan donors who did not have a prior history of vascular disease (approved by the AMC Medical Ethical Committee #95/146). Tissues were fixed in saline, 3.8% (wt/vol) p-formaldehyde, or formalin within 5 minutes after resection and subsequently paraffin-embedded. Paraffin sections (5 μm) of vascular tissue were mounted on 3-aminopropyl-triethoxysilane–coated slides. Morphological analysis was performed by using the Masson Trichrome staining. Cell-type specific staining was performed using Ulex europaeus lectin for endothelial cells and antibodies 1A4 or HAM-56 (DAKO) directed against smooth muscle cell specific α-actin or monocytes/macrophages, respectively. Counterstaining was performed with haematoxylin/eosin according to standard procedures.
In situ hybridization.
Riboprobes for various gene transcripts, synthesized as described below, contained the following sequences: human von Willebrand factor: 8239-8442 (192 bp; GB: X04385); monocyte chemotactic protein-1 (MCP-1): 86-741 (656 bp; GB: M24545); ferritin: 302-451 (149 bp; GB: M97164); CA2_1: 2331-3317 (996 bp; this study; corresponding to 303-1289 of MIHC/hIAP-1 GB: U37546.); GG2_1: 151-868 (718 bp; this study); GG10_2: 2375-2945 (571 bp; this study); AG8_1: 194-845 (652 bp; this study); and CG12_1: 1687-2298 (611 bp; this study). Riboprobes were synthesized by in vitro transcription of cDNA fragments cloned in various pGEM vectors (Promega), containing T7 and SP6 RNA polymerase transcription initiation sites. In brief, the constructs were linearized and riboprobes were synthesized for 1 hour at 37°C in RNA polymerase buffer according to the manufacturer’s instructions for SP6 RNA polymerase (Promega) or T7 RNA polymerase (Stratagene, La Jolla, CA) and labeled with [35S]-UTP (Amersham). Paraffin sections (5 μm) of vascular tissue were mounted on 3-aminopropyl-triethoxysilane–coated slides. In situ hybridization was performed as described,18 with minor modifications. The sections were pretreated with proteinase K (20 μg/mL) for 5 minutes, refixed in 4% (wt/vol) p-formaldehyde, and treated for 10 minutes with 0.25% (wt/vol) acetic anhydride in 0.1 mol/L triethanolamine (pH 8.0). The riboprobes were added to and stored in hybridization mixture, which consisted of 40% (vol/vol) formamide, 8% (wt/vol) dextrane sulfate, 0.8× Dennhardts’, 0.5 mg/mL yeast tRNA, 4 mmol/L EDTA, 16 mmol/L Tris-HCl (pH 8.0), and 0.24 mol/L NaCl. Hybridizations were performed overnight at 50°C in 8 μL (0.5 μCi probe) per section under a coverslip in a moist chamber. After hybridization, coverslips were removed in 5× SSC, 10 mmol/L dithiothreitol (DTT) at 50°C (30 to 60 minutes), followed by a high stringency wash for 30 minutes at 65°C in 50% (vol/vol) formamide, 2× SSC, 10 mmol/L DTT. RNAse A digestion (20 μg/mL) was performed for 30 minutes at 37°C in 10 mmol/L Tris-HCl (pH 8.0), 5 mmol/L EDTA, 500 mmol/L NaCl. The high stringency wash was repeated, followed by washing for 15 minutes with 2× SSC. After dehydration, autoradiography emulsion was applied as a 1:1 dilution of Ilford G5 emulsion (Ilford, Paramus, NJ) with 2% (vol/vol) glycerol. After an exposure of 1 to 6 weeks, slides were developed in Kodak D19 (Eastman Kodak), fixed in Kodak UNIFIX (Eastman Kodak), and counterstained with haematoxylin and eosin.
RESULTS
Endothelial cell model system.
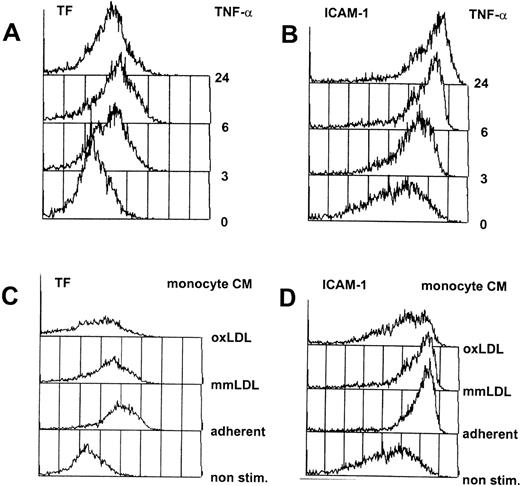
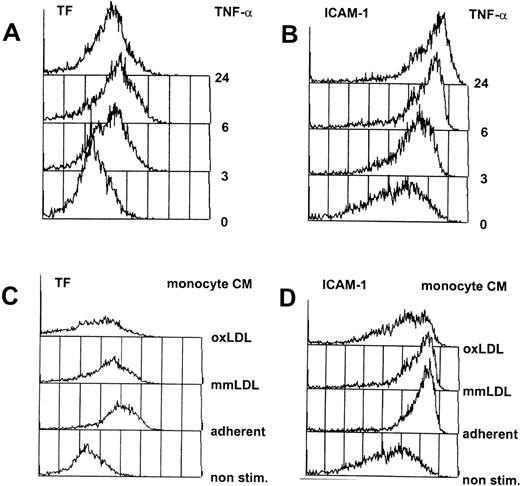
Cultured primary endothelial cells from the human umbilical vein (HUVEC) were chosen as model cells to identify cytokine-responsive vascular endothelial cell genes. To mimic the situation in the fatty streak, the earliest detectable onset of atheromas where monocytes have entered the vessel wall and start to accumulate lipid, we have incubated HUVEC with the conditioned medium from activated human primary monocytes, containing a complex mixture of cytokines and chemokines.3,10,11 As a second, independent activator, we used purified, recombinant human TNF-α, a cytokine released from activated monocytes and macrophages, which is believed to play an important role during inflammatory processes including atherogenesis (reviewed in Sherry and Cerami19). Titration of these stimuli was performed by FACScan analysis of the expression of an immediate early gene (tissue factor) and a late gene (ICAM-1) at different dilutions of either TNF-α or the conditioned medium from activated monocytes. A uniform activation of the total population of cells, necessary for unambiguous DD/RT-PCR interpretation, was obtained at 12 nmol/L TNF-α or the 1/10 dilution of the conditioned medium of adherence-activated monocytes,10 as shown in Fig 1.
Flow cytometric analysis of cytokine-activated HUVEC. HUVEC were continuously exposed to 12 nmol/L TNF- (A and B) or the 1/10 diluted conditioned medium from activated human monocytes (C and D), and at various timepoints aliquots of the cells were analyzed by flow cytometry as described in Materials and Methods. In each case, 5,000 cells were counted (y-axis) and fluorescence intensity is exponentially plotted starting from 100 (x-axis). Indicated are the FACscan readouts for the immediate response gene tissue factor (TF; A and C), and ICAM-1 (B and D), a late induced gene that is also present at low amounts on nonstimulated cells. (C) and (D) show nonactivated HUVEC (non stim.) or HUVEC activated by conditioned medium from monocytes activated by adherence to plastic tissue culture flasks with the addition of growth medium without additions (adherent) or supplemented with 50 μg/mL of minimally modified LDL (mmLDL) or oxidized LDL (oxLDL).
Flow cytometric analysis of cytokine-activated HUVEC. HUVEC were continuously exposed to 12 nmol/L TNF- (A and B) or the 1/10 diluted conditioned medium from activated human monocytes (C and D), and at various timepoints aliquots of the cells were analyzed by flow cytometry as described in Materials and Methods. In each case, 5,000 cells were counted (y-axis) and fluorescence intensity is exponentially plotted starting from 100 (x-axis). Indicated are the FACscan readouts for the immediate response gene tissue factor (TF; A and C), and ICAM-1 (B and D), a late induced gene that is also present at low amounts on nonstimulated cells. (C) and (D) show nonactivated HUVEC (non stim.) or HUVEC activated by conditioned medium from monocytes activated by adherence to plastic tissue culture flasks with the addition of growth medium without additions (adherent) or supplemented with 50 μg/mL of minimally modified LDL (mmLDL) or oxidized LDL (oxLDL).
Differential display of gene expression.
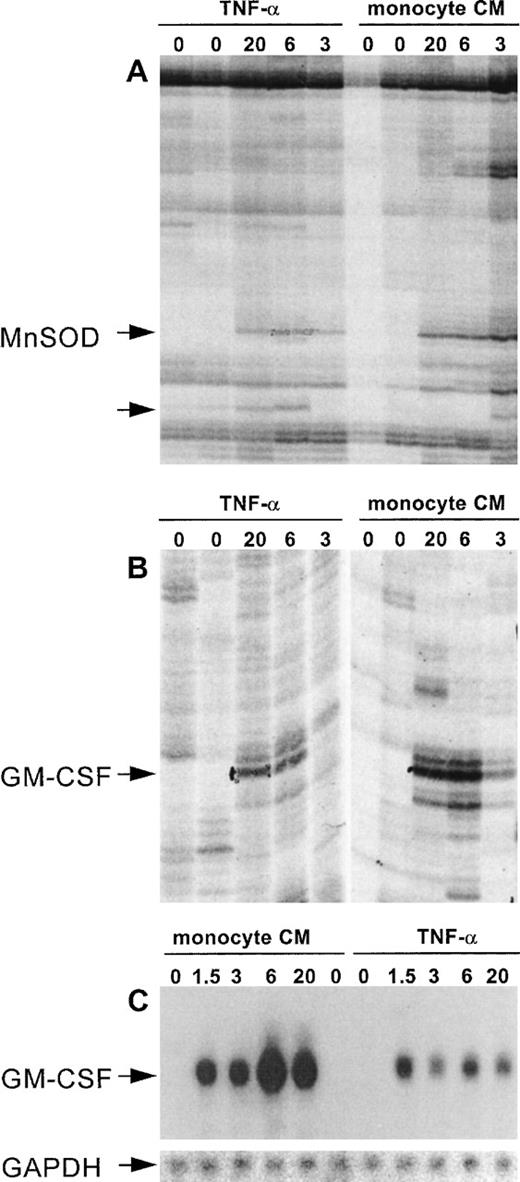
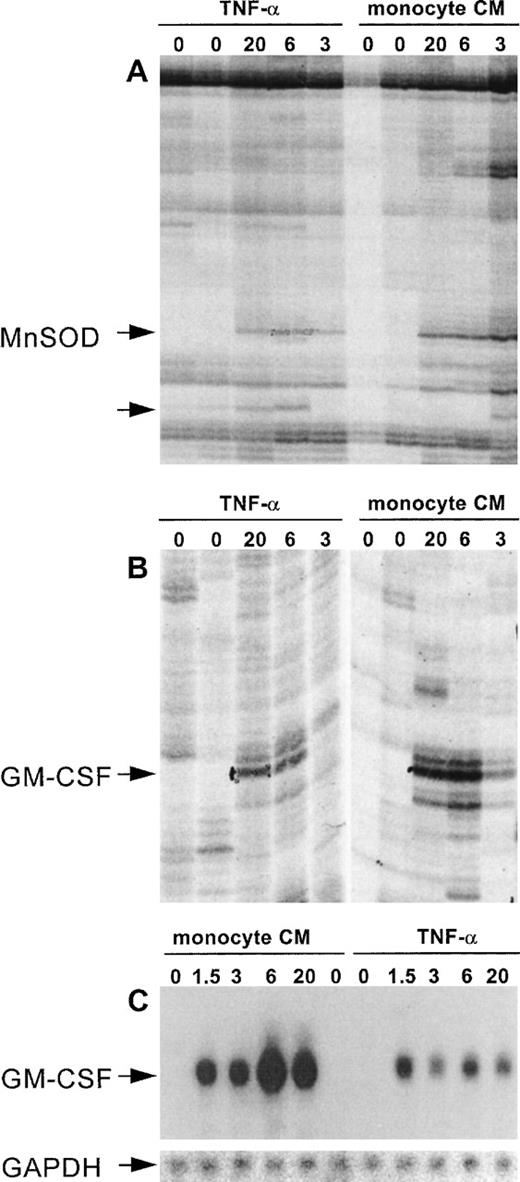
An efficient and reproducible protocol for DD/RT-PCR was established, based on published methods.14,15 This protocol enabled the simultaneous identification of genes that are either induced or repressed. Furthermore, by using RNA samples isolated at appropriate time points, immediate early, delayed early, and late genes were identified in a single experiment. Therefore, total RNA was isolated from resting cells (0 and 20 hours) and from cells activated by either monocyte supernatant or purified human recombinant TNF-α for various time periods (1.5, 3, 6, and 20 hours). The quality and suitability of each RNA preparation was assessed by Northern blotting using specific probes for tissue factor, an early induced protein, and for thrombomodulin, expression of which is repressed by TNF-α (not shown). Isolated total RNA was subjected to DD/RT-PCR and analyzed. The specific set of 144 oligonucleotides we used was designed by Bauer et al15 and is expected to display approximately 80% of the different mRNA species that are expressed in a single cell based on statistical grounds. Each primer combination yielded 50 to greater than 150 visible bands on gel for each RNA sample, and differentially expressed bands were distributed randomly over the different primer combinations, with an average of 16 (range, 8 to 23) for each anchored primer with 12 arbitrary decamers.15 Differentially displayed bands were identified by rigorous selection to prevent inclusion of false positives, a problem frequently encountered during DD/RT-PCR analyses (Fig 2).
DD/RT-PCR reproducibly identifies cytokine-responsive genes in HUVEC. (A) and (B) show differential display patterns for the primer sets T11GA with decamer 11 (A) and decamer 7 (B). These represent the selection criteria we used to exclude false-positives by only pursuing bands that show reproducible differential expression on DD/RT-PCR gels. (A) and (B) show enlargements for bands that are reproducibly induced at 3, 6, and 20 hours by both TNF- and monocyte-conditioned medium, whereas they are not detectable in unstimulated cells. After cloning and sequencing, these bands were shown to represent Mn-SOD and GM-CSF, respectively. (A) also shows an unknown gene fragment that is specifically induced by TNF- but not by monocyte-conditioned medium (unlabeled arrow) and was excluded from our analysis. (C) shows confirmation of differential expression of GM-CSF using the DD-fragment as probe and a Northern blotting analysis of the same RNA sample as used for the DD/RT-PCR. The insert shows the specific hybridization signal for GAPDH as internal control for equal RNA loading.
DD/RT-PCR reproducibly identifies cytokine-responsive genes in HUVEC. (A) and (B) show differential display patterns for the primer sets T11GA with decamer 11 (A) and decamer 7 (B). These represent the selection criteria we used to exclude false-positives by only pursuing bands that show reproducible differential expression on DD/RT-PCR gels. (A) and (B) show enlargements for bands that are reproducibly induced at 3, 6, and 20 hours by both TNF- and monocyte-conditioned medium, whereas they are not detectable in unstimulated cells. After cloning and sequencing, these bands were shown to represent Mn-SOD and GM-CSF, respectively. (A) also shows an unknown gene fragment that is specifically induced by TNF- but not by monocyte-conditioned medium (unlabeled arrow) and was excluded from our analysis. (C) shows confirmation of differential expression of GM-CSF using the DD-fragment as probe and a Northern blotting analysis of the same RNA sample as used for the DD/RT-PCR. The insert shows the specific hybridization signal for GAPDH as internal control for equal RNA loading.
Sequence analysis of DD/RT-PCR fragments.
The gene fragments that showed differential expression in a consistent and reproducible way after stimulation of HUVEC by both TNF-α (193 bands) and monocyte-conditioned medium (122 bands) were cloned and sequenced in triplicate, totalling 119 fragments, 9 of which represented repressed genes. The sequences obtained from DD/RT-PCR fragments were cross-analyzed to trace redundancy: indeed, on 3 occasions a sequence appeared 3 times, whereas duplication was traced 6 times. In all cases, fragments of identical length and sequence had been amplified by different anchored primers but identical arbitrary decamer, with the exception of tissue factor, which was amplified with the anchored primer only (Table 1). Only in 5 of 119 cases did we observe colonies of different sequence, indicating contamination of the differentially displayed band by other display products of the same length, whereas in only 3 of 119 cases the sequence of the DD/RT-PCR fragment proved identical to mitochondrial DNA. The resulting 106 unique sequences were compared with the nonredundant Genbank/EMBL database at NCBI and EBI, using BLAST and FASTA algorithms, showing that 22 of 106 gene fragments represented mRNAs that encode known proteins (listed in Table 1). The remaining 84 sequences, representing novel genes, were compared with the Expressed Sequence Tag-libraries at the National Center for Biotechnology Information (dbEST) and The Institute for Genomic Research (TIGR-HCI).16 We found that 33 of our novel sequences were represented in these databases. The remainder of 51 DD-fragments represent sequences not previously described.
Verification of differential expression by Northern blotting analysis.
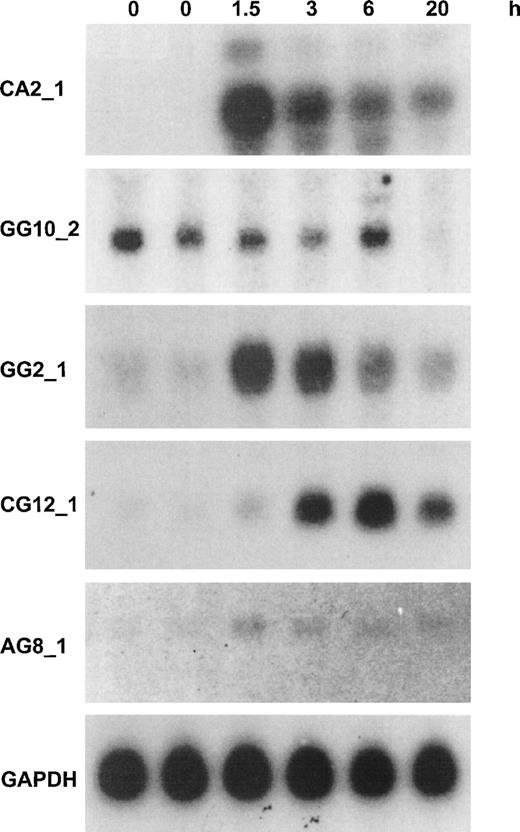
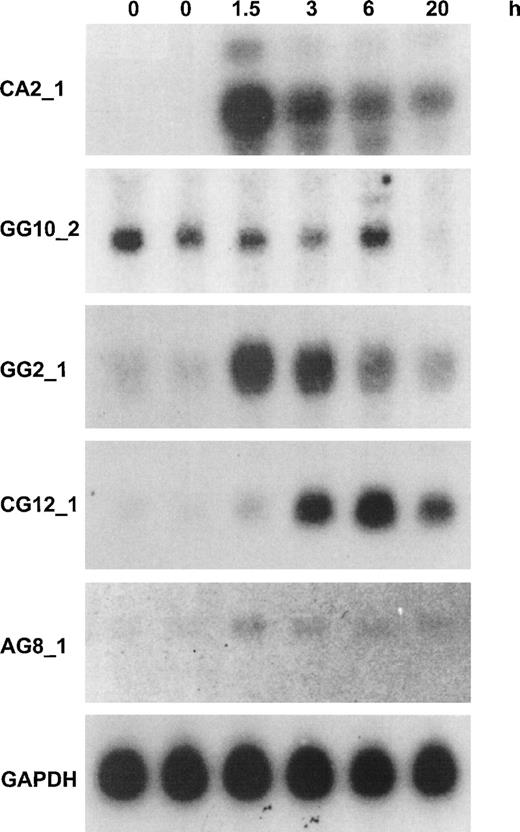
The fragments that were identified by differential display were analyzed by Northern blotting analysis. We produced a series of 20 identical filters loaded with 10 μg of total RNA per lane of HUVEC stimulated for 0, 0, 1.5, 3, 6, or 20 hours with 12 nmol/L human recombinant TNF-α. Filters were probed with highly radiolabelled purified DD/RT-PCR fragments, showing that all of the DD-fragments corresponding to known mRNAs and most of the novel DD-fragments corresponding to ESTs gave a signal on Northern blot. Only in 2 cases did fragments turn out to be nondifferential, whereas others showed induction or repression of threefold to greater than 10-fold. None of the novel, non-EST transcripts could be detected in this way; these await further analysis with more sensitive methods. This is not unexpected, because most average abundant transcripts should at present be represented in dbEST (NCBI). Furthermore, it should be emphasized that DD/RT-PCR fragments do not constitute optimal probes for Northern blotting analysis, because they are derived from the 3′-UTR of mRNAs, which are rich in repetitive sequences and secondary structure. Figure 3 shows the results of Northern blotting analysis for 5 novel transcripts that are represented in dbEST (NCBI) and for which the full-length mRNA was isolated as described below. Four probes were chosen that showed greater than 10-fold induction by Northern blotting analysis, whereas the fifth probe represented a gene that was totally repressed by 20 hours (Fig 3).
Northern blotting analysis for the expression of 5 novel cytokine-responsive genes in resting and TNF-–activated HUVEC. Northern blotting analysis of 10 μg HUVEC total RNA with probes from DD-fragments or corresponding EST-clones was performed as described in Materials and Methods. Time periods of continuous stimulation by TNF- are indicated, and GAPDH analysis is given as control for equal loading. The approximate length of the transcripts was determined from the position of 28S, 18S, and 5S ribosomal RNA at the following: CA2_1: an abundant band of 5.2 kb and two minor bands of 6 and 4.4 kb, respectively; GG10_2: 2.8 kb; GG2_1: 1.8 kb; CG12_1: 2.3 kb; and AG8_1: 3.2 kb. A detailed analysis of these 5 transcripts is given in the text and in Fig 4. In the case of CA2_1, identical patterns and intensities were found with radiolabelled probes from bases 639-1032 (DD-fragment) and 2331-3317 (hIAP-1) from the full-length sequence. The other probes that were used represent the following parts of the full-length sequences: GG10_2: 2697-2883; GG2_1: 670-1882; CG12_1, 1690-2298; AG8_1: 2880-3289; and GAPDH: 360-1070 (GB: M33197).
Northern blotting analysis for the expression of 5 novel cytokine-responsive genes in resting and TNF-–activated HUVEC. Northern blotting analysis of 10 μg HUVEC total RNA with probes from DD-fragments or corresponding EST-clones was performed as described in Materials and Methods. Time periods of continuous stimulation by TNF- are indicated, and GAPDH analysis is given as control for equal loading. The approximate length of the transcripts was determined from the position of 28S, 18S, and 5S ribosomal RNA at the following: CA2_1: an abundant band of 5.2 kb and two minor bands of 6 and 4.4 kb, respectively; GG10_2: 2.8 kb; GG2_1: 1.8 kb; CG12_1: 2.3 kb; and AG8_1: 3.2 kb. A detailed analysis of these 5 transcripts is given in the text and in Fig 4. In the case of CA2_1, identical patterns and intensities were found with radiolabelled probes from bases 639-1032 (DD-fragment) and 2331-3317 (hIAP-1) from the full-length sequence. The other probes that were used represent the following parts of the full-length sequences: GG10_2: 2697-2883; GG2_1: 670-1882; CG12_1, 1690-2298; AG8_1: 2880-3289; and GAPDH: 360-1070 (GB: M33197).
Cloning and analysis of 5 novel cytokine-responsive transcripts.
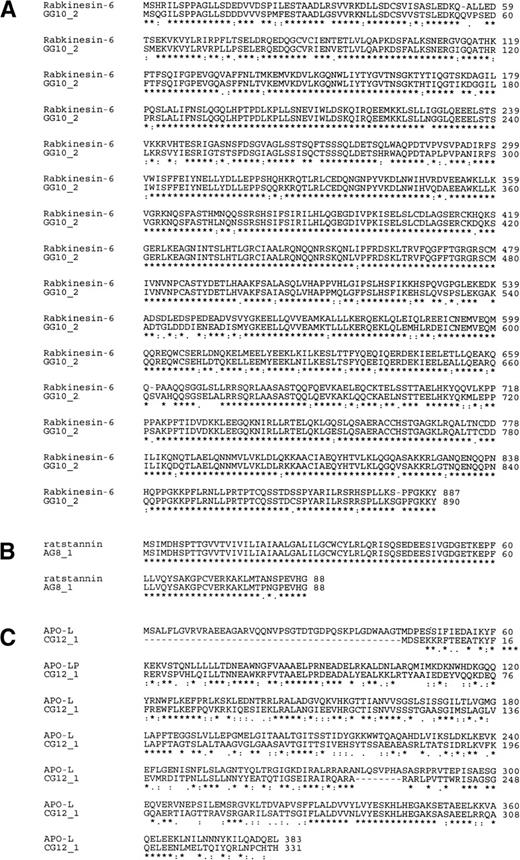
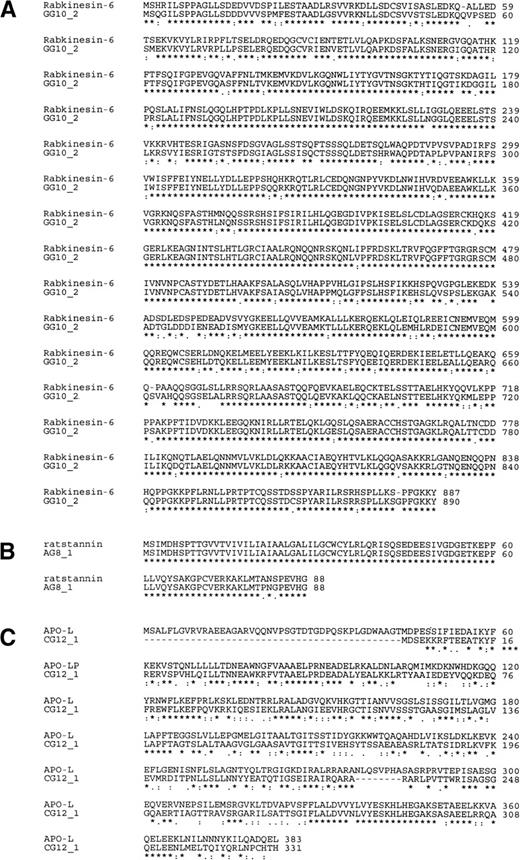
Specific details of each gene transcript can only be shown by full-length cDNAs, despite the rapidly increasing information on overlapping EST sequences at Unigene (NCBI). The IMAGE Consortium cDNA Clones48 from which EST-sequences were derived can be obtained through various distributors, but usually represent partial cDNAs as a result of their average sizes ranging from 0.5 to 2 kb. Therefore, a cDNA library was constructed from the mRNA isolated from HUVEC that had been activated for 6 hours by monocyte-conditioned medium. Next, differential display probes or corresponding IMAGE-clones were used to screen this cDNA library for the 5 mRNAs represented in Fig 3, resulting in the isolation of several full-length and partial clones. Full-length sequences for these five cDNAs can be obtained through their Genbank accession numbers.2 Novel transcript CA2_1 (5,212 bp, GB:AF070674) contained the identical DD/RT-PCR fragments CA2_1_3, AC2_1_4, and GC2_1_5 (397 bp) at position 639-1032, which resulted from oligo-dT priming at a 27-bp polyA stretch in the 5′-UTR of the full-length message, rather than at the genuine polyA-site at the 3′-end. A mRNA length of 5.4 kb has been extensively documented for hIAP-1, but only partial cDNA sequences had been deposited in Genbank. The sole open reading frame (ORF) encodes the known protein human inhibitor of apoptosis protein-1 (hIAP-1; GB:U45878), also known by the names MIHC (GB: U37546) and cIAP-2. This protein blocks TNF-α receptor-mediated cellular apoptosis by direct inhibition of various caspases.22 The novel cDNA GG10_2 (2,872 bp, GB: AF070672) contains DD/RT-PCR fragment GG10_2 (188 bp) at position 2697-2872 and represents a TNF-α–repressed mRNA. The full-length sequence was constructed from the largest partial cDNA obtained from our cDNA library (1.5 kb) and the overlapping IMAGE Consortium (LLNL) cDNA Clone (clone ID: 613195).48 The predicted 890 amino acid sequence is 86% identical (91% homologous; Fig 4A) to the recently described murine protein Rabkinesin-6 (GB: Y09632); therefore, this cDNA represents the human homolog.23 The novel cDNA AG8_1 (3,295 bp, GB: AF070673) contains DD/RT-PCR fragment AG8_1 (403 bp) at position 2880-3289. We have isolated a partial cDNA of 2.0 kb from our activated HUVEC library and completed the sequence with the overlapping IMAGE Consortium cDNA Clone (clone ID: 0969636). The predicted ORF (Fig 4B) encodes the human homolog of rat stannin25,26 (GB: M81639). The novel cDNA CG12_1 (2,298 bp, GB: AF070675) contains DD/RT-PCR fragment CG12_1 (180 bp) at the extreme 3′-end at position 2127-2298. The full-length cDNA sequence (2,298 bp) showed it to encode a 331 amino acid protein that is 70% homologous (50% identical) to the recently described human apolipoprotein-L, an HDL-associated lipoprotein produced by the pancreas24 (Fig 4C). Finally, the novel cDNA GG2_1 (1,889 bp, GB: AF070671) contained DD/RT-PCR fragment GG2_1_2 (303 bp) at the very 3′-end at position 1595-1889. The cDNA encodes an 188 amino acid protein and shows no homology at either the nucleotide, amino acid, or structural level with any protein or gene present in the combined NCBI and EMBL databases. Therefore, we are at present unable to assign a function to this gene.
Amino-acid sequence homology for 3 novel full-length human cDNAs obtained from an activated HUVEC cDNA library. Amino acid sequences from the coding sequence of 3 novel human cDNAs were compared to known proteins using the ClustalW algorithm for (A) the novel human cDNA GG10_2 and murine Rabkinesin-640; (B) the novel human cDNA AG8_1 and rat stannin39; and (C) the novel human cDNA CG12_1 and the human apolipoprotein-L.41
Amino-acid sequence homology for 3 novel full-length human cDNAs obtained from an activated HUVEC cDNA library. Amino acid sequences from the coding sequence of 3 novel human cDNAs were compared to known proteins using the ClustalW algorithm for (A) the novel human cDNA GG10_2 and murine Rabkinesin-640; (B) the novel human cDNA AG8_1 and rat stannin39; and (C) the novel human cDNA CG12_1 and the human apolipoprotein-L.41
In situ hybridization on human vascular tissue.
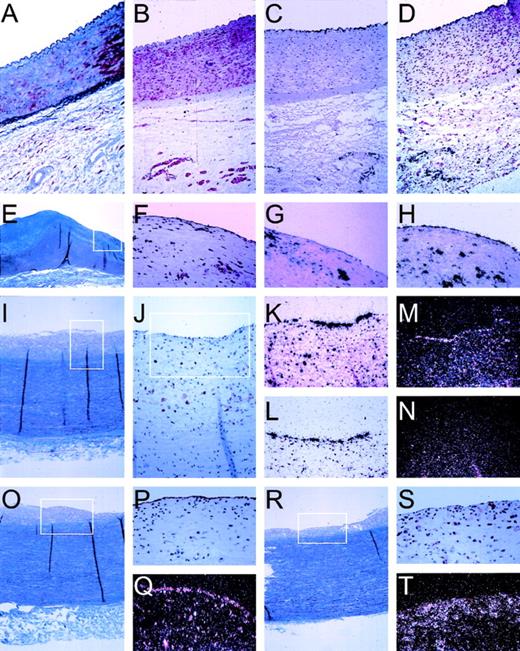
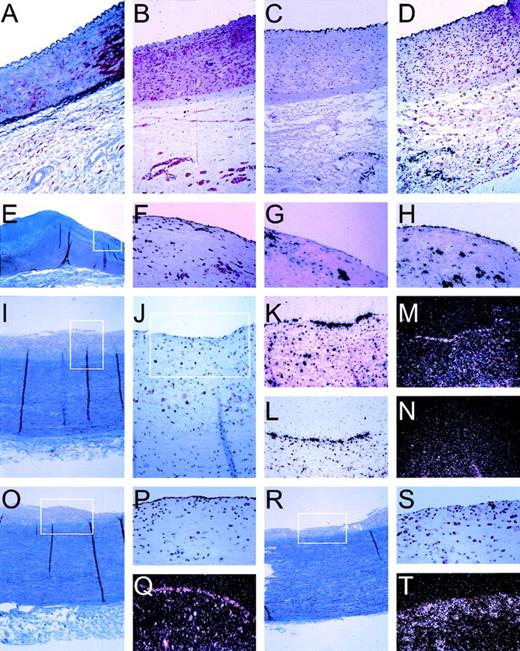
The isolation of candidate genes as described so far gives detailed information about the response of cultured HUVEC to cytokine stimulation. To determine whether this approach resulted in the isolation of novel genes that are actually expressed in arterial vascular endothelial cells, we performed in situ hybridization studies on human vascular tissue. Endothelial cells are thought to play an essential role, especially during the onset of lesion formation. Therefore, vascular specimens displaying early stages of inflammatory lesion formation were obtained from organ donors who did not have a prior history of vascular disease. A series of vascular specimens was analyzed by (immuno)histochemical staining and selected for displaying either normal vascular wall or early and advanced lesions. Integrity of the endothelial lining and its mRNA was substantiated by using probes for human von Willebrand factor, being solely expressed in the endothelial cells (Fig 5). Cytokine activation of the endothelial lining was checked by a probe for MCP-1, a cytokine-responsive gene that was identified in our differential display approach and extensively described as a marker for endothelial cell activation.3,11,27 28 MCP-1 is shown to be an excellent marker both for endothelial cell activation and more generally for inflammation of the vessel wall by virtue of its high expression in macrophages (Fig 5). Expression by arterial endothelial cells is exclusively observed when macrophages are present close to the endothelium (Fig 5), whereas macrophages that have deeply infiltrated into the vessel wall do not elicit such expression in endothelial cells (not shown). Next, we tested expression of our 5 novel genes. Expression of CA2_1 (hIAP-1) could be readily detected only in the endothelial lining of atherosclerotic aorta and iliac arteries, coinciding with MCP-1 expression, whereas we never detected hIAP-1 in normal arteries or in arteries that did not display substantial monocyte/macrophage infiltrates. Subsequently, we determined in vivo expression of CG12_1, which shows low expression in unstimulated cultured cells but is induced approximately 20-fold upon cytokine activation in vitro (Fig 3). The mRNA for this apolipoprotein-L–like protein can be readily detected in the normal endothelial lining of nonatherosclerotic iliac artery (Fig 5). In addition, expression is seen in capillaries in the adventitia but not in smooth muscle cells. Also, in atherosclerotic vascular material, expression is only seen in endothelial cells and not in macrophages. This indicates that the expression of this novel gene seems limited to endothelial cells in the tissues examined so far. The novel human cDNA AG8_1 (stannin), expressed at low levels in cultured cells, is detected in the endothelial cells and in intimal macrophages of atherosclerotic lesions, but was undetectable in the normal vessel wall (Fig 5). Interestingly, GG2_1, which is expressed at high levels both in unstimulated and cytokine-activated cultured cells (Fig 3), can be detected only with great difficulty in the arterial vascular material screened so far (Fig 5). The novel cDNA gene GG10_2 (Rabkinesin-6), being constitutively expressed in HUVEC and repressed by TNF-α in vitro, could not be detected in the vessel wall so far, indicating low steady-state mRNA levels in vivo.
Pattern of in situ hybridization of mRNA expression in human vascular tissue. In situ hybridization and immuno-histochemical staining was performed on serial sections (5 μm) of formaline-fixed and paraffin-embedded vascular tissue as described in Materials and Methods. Tissues were derived from the normal iliac artery of a 12-year-old male (A through D), 0.5 cm past the aortic bifurcation and the abdominal aorta of a 49-year-old woman (E through H), or a 39-year-old woman (I through T), both 1 cm before the bifurcation. (A) Masson Trichrome staining showing an overview through the iliac artery with the lumen on top, endothelial cells lining the vessel wall, the internal elastic lamina, the smooth muscle cell-containing media, and the spongy-appearing adventitia. (B) Specific staining of smooth muscle cells, using an antibody directed against SMC-specific -actin (1A4). (C) Autoradiographic image of the specific hybridization of an antisense probe for the endothelial cell specific protein von Willebrand factor, showing the integrity of endothelial cell mRNA and specificity of hybridization conditions. (D) Expression of novel cDNA CG12_1 is restricted to endothelial cells lining the vessel and the capillaries of the adventitia, identical to von Willebrand factor (C). (E) Masson Trichrome staining showing an overview through the aorta with the lumen on top, endothelial cells lining the vessel wall, a fibrous neo-intima (blue), the internal elastic lamina, the smooth muscle cell-containing media (purple), and the spongy-appearing adventitia. The boxed area is represented in (F) through (H). (F) Immunohistochemical staining using a monocyte/macrophage-specific antibody (HAM-56), showing the presence of extensive infiltration of the neointimal layer of the vessel wall, indicative of an inflammatory lesion. (G) Specific expression of MCP-1 by both macrophages and endothelial cells in this inflamed vessel wall. (H) Specific expression of ferritin by monocytes/macrophages and endothelial cells. (I) Masson Trichrome staining showing an overview through the aorta with the lumen on top, endothelial cells lining the vessel wall, a fibrous neo-intima (gray), the internal elastic lamina, the smooth muscle cell-containing media (blue/purple), and the spongy-appearing adventitia. The boxed area is represented in (J). (J) Immunostaining for macrophages (HAM-56) in this aorta section showing inflammation of the intima; the boxed area is represented in the in situ hybridizations shown in (K) through (N). (K) Expression pattern of MCP-1 in endothelial cells and macrophage/foam cells. (L) Specific expression of CA2_1 (hIAP-1) by the endothelial cells. (M) Expression of novel cDNA GG2_1 by endothelial cells. (M) and (N) are a darkfield representation of the autoradiographic images for greater clarity of hybridization of radiolabeled probes for sense mRNA for novel cDNA GG2_1 (M) and, as control for specificity, for antisense GG2_1 (N). (O) Masson Trichrome staining showing an overview through the aorta with the lumen on top, endothelial cells lining the vessel wall, a fibrous neo-intima (gray), the internal elastic lamina, the smooth muscle cell-containing media (blue/purple), and the spongy-appearing adventitia. (P) The integrity of the endothelial cell lining is shown with the specific lectin from Ulex europaeus, and specific expression of novel cDNA AG8_1 (stannin) is detected in endothelial cells (Q, darkfield representation). A more heavily inflamed area of the same aorta is shown by Masson Trichrome (R) and in the boxed area by immunostaining for macrophages (S) and expression of AG8_1 (T) in both endothelial cells and macrophages.
Pattern of in situ hybridization of mRNA expression in human vascular tissue. In situ hybridization and immuno-histochemical staining was performed on serial sections (5 μm) of formaline-fixed and paraffin-embedded vascular tissue as described in Materials and Methods. Tissues were derived from the normal iliac artery of a 12-year-old male (A through D), 0.5 cm past the aortic bifurcation and the abdominal aorta of a 49-year-old woman (E through H), or a 39-year-old woman (I through T), both 1 cm before the bifurcation. (A) Masson Trichrome staining showing an overview through the iliac artery with the lumen on top, endothelial cells lining the vessel wall, the internal elastic lamina, the smooth muscle cell-containing media, and the spongy-appearing adventitia. (B) Specific staining of smooth muscle cells, using an antibody directed against SMC-specific -actin (1A4). (C) Autoradiographic image of the specific hybridization of an antisense probe for the endothelial cell specific protein von Willebrand factor, showing the integrity of endothelial cell mRNA and specificity of hybridization conditions. (D) Expression of novel cDNA CG12_1 is restricted to endothelial cells lining the vessel and the capillaries of the adventitia, identical to von Willebrand factor (C). (E) Masson Trichrome staining showing an overview through the aorta with the lumen on top, endothelial cells lining the vessel wall, a fibrous neo-intima (blue), the internal elastic lamina, the smooth muscle cell-containing media (purple), and the spongy-appearing adventitia. The boxed area is represented in (F) through (H). (F) Immunohistochemical staining using a monocyte/macrophage-specific antibody (HAM-56), showing the presence of extensive infiltration of the neointimal layer of the vessel wall, indicative of an inflammatory lesion. (G) Specific expression of MCP-1 by both macrophages and endothelial cells in this inflamed vessel wall. (H) Specific expression of ferritin by monocytes/macrophages and endothelial cells. (I) Masson Trichrome staining showing an overview through the aorta with the lumen on top, endothelial cells lining the vessel wall, a fibrous neo-intima (gray), the internal elastic lamina, the smooth muscle cell-containing media (blue/purple), and the spongy-appearing adventitia. The boxed area is represented in (J). (J) Immunostaining for macrophages (HAM-56) in this aorta section showing inflammation of the intima; the boxed area is represented in the in situ hybridizations shown in (K) through (N). (K) Expression pattern of MCP-1 in endothelial cells and macrophage/foam cells. (L) Specific expression of CA2_1 (hIAP-1) by the endothelial cells. (M) Expression of novel cDNA GG2_1 by endothelial cells. (M) and (N) are a darkfield representation of the autoradiographic images for greater clarity of hybridization of radiolabeled probes for sense mRNA for novel cDNA GG2_1 (M) and, as control for specificity, for antisense GG2_1 (N). (O) Masson Trichrome staining showing an overview through the aorta with the lumen on top, endothelial cells lining the vessel wall, a fibrous neo-intima (gray), the internal elastic lamina, the smooth muscle cell-containing media (blue/purple), and the spongy-appearing adventitia. (P) The integrity of the endothelial cell lining is shown with the specific lectin from Ulex europaeus, and specific expression of novel cDNA AG8_1 (stannin) is detected in endothelial cells (Q, darkfield representation). A more heavily inflamed area of the same aorta is shown by Masson Trichrome (R) and in the boxed area by immunostaining for macrophages (S) and expression of AG8_1 (T) in both endothelial cells and macrophages.
DISCUSSION
We have identified 106 nonredundant HUVEC gene fragments that are differentially expressed upon cytokine stimulation of these cells. In a substantial number of cases, differential expression was observed with TNF-α (193 fragments) but not with monocyte supernatant (119 fragments), as exemplified in Fig 2. These results strongly suggest the presence of counteracting cytokines in the more complex mixture, eg, interleukin-4 (IL-4), IL-10, and IL-13 are well known to attenuate gene induction by TNF-α, whereas interferon-γ (IFN-γ) can act synergistically.29 Even for a single function as prostacyclin production by endothelial cells, different combinations of cytokines have differential effects on cells of different vascular origin.30 The minority of our panel of cytokine-responsive genes (22/106) represents mRNAs for known proteins, most of which have been shown previously to be induced by cytokines. Many of the corresponding proteins have been implicated in atherogenesis,2 showing the specificity of our approach. It seems remarkable that the transcripts for adhesion molecules VCAM-1 and ICAM-1, purported for their role in atherogenesis, were not among the 106 genes identified (Table 1), although FACScan and Northern blotting analysis showed them to be induced by both monocyte CM and TNF-α (Fig1 and data not shown). However, it should be noted that, by definition, DD/RT-PCR uses a statistically defined set of arbitrary primers, and the identification of specific transcripts is merely a chance process.15 This is in fact the main asset of this approach, because it ensures an unbiased sampling of induced and repressed known and novel genes of both low and high abundance in any given situation. Therefore, we limited our primer set to 144 combinations to ensure an unbiased sampling of activation-sensitive genes especially to find novel genes, although statistics predict that a portion of transcripts will then be missed. Nevertheless, many expected transcripts for known genes were identified, as shown in Table 1. The corresponding gene products are involved in a variety of intercellular and intracellular processes (Table 1). As expected, we find induced expression of a number of proinflammatory (proatherogenesis) genes, such as PTGS2 (Cox2) and several genes involved in leukocyte trafficking, such as IL-8, RANTES, granulocyte-monocyte colony-stimulating factor (GM-CSF), and the recently described galectin PCTA-1. In addition, the endothelial cell shows induced expression of a number of protective (antiatherogenesis) genes. Protection against oxidative stress, which accompanies many inflammatory processes as a result of monocyte/macrophage activation,28 is accomplished by the increased expression of ferritin and manganese superoxide dismutase, an enzyme essential in withstanding the intracellular oxidative burst that accompanies TNF-α activation.31,32 Identification of ferritin as a TNF-α responsive gene shows the sensitivity of our DD/RT-PCR approach, which did not identify only strongly induced genes. Like PTGS-2, ferritin expression is easily detected in unstimulated cells and is increased only threefold after stimulation. The significance of identifying moderately regulated genes is underscored by the fact that increased ferritin expression has been documented for both human and rabbit aortic atherosclerotic lesions,33 as confirmed in Fig 5. TNF-α is known for its ability to induce apoptosis in many cell types under certain conditions through activation of its cognate receptor subtype I.34,35Apparently, the endothelial cell protects itself very efficiently against TNF-α–induced apoptosis35,36 by expressing hIAP-1, a protein that is able to block cellular apoptosis.22,35,37 In addition, the antiapoptotic A20 protein, which was previously identified from a similar screen of differential endothelial cell gene expression in response to TNF-α, is expressed.38 The latter protein has recently been shown to attenuate TNF-α–induced endothelial cell activation by blocking NF-κB–dependent mechanisms.39 In our study, yet another gene (AG8_1) was identified, encoding an 88 amino acid residue protein that is the human homolog of rat stannin. Stannin is a protein that is essential for a sensitive apoptotic response by neuronal cells to organo-tin compounds. The exact functional role of this protein in this cytotoxic process has not yet been established.25,26Although apoptosis seems to be counteracted efficiently, proliferation appears to be shut down. This is apparently accomplished by repressing two genes involved in cell cycle initiation (GSPT1, RGS5) and the upregulation of an antiproliferative protein (BTG1). Endothelial cells function as an important selective barrier to separate the blood in the lumen of the vessel from the surrounding tissue. Transport of nutrients and other plasma constituents is actively regulated by the endothelium. Changes in cell shape, one of the affected functions described in Table1, could greatly compromise this barrier function, similar to observations in lung tissue, where lung fluid balance is greatly disturbed by actin rearrangements in the endothelial cells.40 Indeed, the most striking visual effect of TNF-α activation of HUVEC is the gross change of cell morphology from cobblestone to spindle shape, a well-documented phenomenon41 that we also observed upon incubation with monocyte-conditioned medium. Furthermore, several proteins involved in vesicular transport show cytokine responsive regulation, including the novel human cDNA GG10_2. As shown in Fig 4, GG10_2 represents the human homologue for the recently described murine protein Rabkinesin-6 (Y09632), a kinesin-like protein involved in intra-Golgi vesicle transport via micro-tubuli, that is regulated by the GTPase Rab-6.23
The remaining 84 of the DD/RT-PCR fragments represent genes of presently unknown function, but in 33 cases (including our 5 novel transcripts) their identity as genuine human transcripts is verified by their presence in Expressed Sequence Tag (EST) libraries.9These EST libraries contain at present more than 1,000,000 partial sequences of human cDNA clones, mostly from the IMAGE consortium48 or TIGR,9 together with information about tissue- or disease-specific expression data and possible functional data based on sequence homologies with known genes (Unigene; NCBI). In addition, a chromosomal location has been determined to aid genetic analysis, complementary to the human genome project (SCIENCE Map of the Human Genome; NCBI). Most of the ESTs that we have identified using DD/RT-PCR have been described in activated or fetal tissues: DD/RT-PCR fragment CA2_1 was identified by ESTs from adult parathyroid tumor (GB: W32947), adenoma-transformed lung cell-line (GB: U54711), and ulcerative Colitis Mucosa (GB: AA190195). The full-length cDNA CA2_1 (5.2 kb) is shown to encode hIAP1, a protein that blocks TNF-α receptor-mediated cellular apoptosis,35 37 a function that is indeed expected to be found preferentially in tumor or inflamed tissues. Similarly, expression of GG2_1 is found in EST libraries from adult multiple sclerosis lesions and HeLa carcinoma, CG12_1 in fetal tissues and ovarian cancer, and AG8_1 in neuroepithelium, fetal heart, and multiple sclerosis lesions. Similar expression profiles were found for the remaining DD-fragments corresponding to ESTs; only rarely has expression been observed in resting tissue-specific libraries such as pancreas, brain, or retina. This is remarkable given the fact that the large majority of ESTs has been obtained from resting tissue, although this is rapidly changing. This bias towards activated tissue expression in vivo, in combination with our in vitro Northern blots, strongly suggests an in vivo role in EC activation of the ESTs that remain to be studied in more detail.
Excessively long untranslated regions were found in the 4 TNF-α–induced novel transcripts. This is in remarkable contrast to the relatively short UTRs of the constitutively expressed GG10_2 (Rabkinesin-6). Long untranslated regions are frequently found in the 3′-UTR of induced transcripts, as exemplified by our novel human cDNA AG8_1, which, like the rat mRNA for stannin, contains almost 3 kb of 3′-UTR. Interestingly, the 4-kb message for PCTA1 (GB:L78132), identified as a differentially expressed mRNA in prostate carcinoma (and Table 1), has the same coding sequence as galactin-8 (GB: X91790), an mRNA from normal tissue of only 1.1 kb. Remarkably, the untranslated region of the CA2_1 (hIAP-1) transcript was located in the 5′-UTR rather than in the 3′-UTR. The functional significance of this unusually long 5′-UTR (2,750 bp) remains to be established. The DD/RT-PCR initially identified these as novel transcripts, whereas the coding sequence has already been described. At present, we cannot exclude the possibility that some of our novel sequences will turn out to represent novel transcripts for known genes, because many alternate mRNAs are already appearing in dbEST and Unigene, showing that the sequences deposited in Genbank for known mRNAs usually represent but one of several (possibly cell-type specific) transcripts.
The in vivo significance of any one of the candidate genes we identified in vitro obviously relies on in situ expression in the endothelial lining of the vascular bed. Using aorta and iliac arteries obtained during organ transplantations, our in situ hybridization approach confirms that expression of MCP-1 is a reliable marker for inflamed vascular lesions (Fig 5), as has been described.3,11,27 28 Furthermore, to our knowledge, we are the first to show in vivo expression of the antiapoptotic hIAP-1 (CA2_1), which we detected in endothelial cells overlying lesions heavily infiltrated by monocytes and foam cells (Fig 5). So, despite the fact that hIAP-1 (CA2_1) is an immediate early gene in culture, it can be detected during a chronic disease such as atherosclerosis, stressing the fact that its molecular processes are of an episodal nature. Expression of AG8_1 (stannin), expressed at low levels in cultured cells, could be confirmed in vivo, but does not seem to be absolutely specific for the endothelial cells, because occasionally monocytes/macrophages of the atherosclerotic lesions express this gene (Fig 5). Expression in vivo of GG10_2 (Rabkinesin-6), being one of the few constitutively expressed and cytokine-repressed genes in vitro has not yet been established, probably due to a low steady-state mRNA level, and awaits confirmation by an immuno-histochemical approach.
Two interesting novel genes are not yet assigned to a gene panel in Table 1. The novel cDNA fragment GG2_1 encodes an 188 amino acid protein that shows no sequence or structural homology with any protein in the EMBL databases. No structural or functional protein motifs/domains could be determined using PROSITE and Blocks, indicating that it seems to represent a totally novel protein. Although this greatly complicates its functional study, exactly these kind of proteins will be the most significant outcome of unbiased approaches such as DD/RT-PCR, because they will lead the way to more fundamental knowledge on basic processes. Interestingly, expression of GG2_1, which is expressed at relatively high levels both in unstimulated and cytokine-activated cultured cells, could be detected in the arterial vascular material screened so far only with great difficulty, although it seems restricted to arterial lesion endothelial cells (Fig 5). This might indicate that its expression level in vivo is quite low and is markedly induced by culturing the cells in vitro, something that has been documented for other genes as well.42 We cannot speculate about the significance of this at present, because its protein sequence does not yield clues about a possible function of this novel protein. Equally significant is the novel cDNA CG12_1, which represents an endothelial cell-specific, 331 amino acid protein. We determined in vivo expression of CG12_1, for which we detected low expression in unstimulated cultured cells (Fig 3), that was upregulated approximately 20-fold. The mRNA for this apolipoprotein-L–like protein can be detected readily in the normal endothelial lining of nonatherosclerotic iliac artery and aorta (Fig 5). In addition, expression is seen in capillary endothelial cells in the adventitia, but not in smooth muscle cells. Also, in atherosclerotic vascular material, expression was restricted to endothelial cells, indicating that the expression of this novel gene seems limited to endothelial cells in all tissues examined so far. Interestingly, the deduced amino acid sequence for CG12_1 is 70% homologous (50% identical) to the recently described human apolipoprotein-L, an HDL-associated lipoprotein produced by the pancreas.24 The fact that CG12_1 is highly homologous to an HDL-associated protein and seems to be expressed in the vessel wall in an endothelial cell-specific way might indicate a role for the vessel wall in lipoprotein homeostasis.
The next stage of our studies will embody the analysis of the expression of our panel of 106 EC genes in a quantitative way in healthy and diseased vascular tissue to confirm which gene or gene panels play a dominant in vivo role during atherosclerosis. A thorough mapping of gene expression patterns in various (patho-)physiological situations will give indispensable information concerning alterations in functional properties related to altered gene expression patterns. Prescreening of the total human gene repertoire for potential candidate genes will greatly accelerate gene expression mapping of cellular anomalies that are characteristic for a specific disease.8,9 The results from the present study on cytokine-responsive genes may then be combined with data from other differential screening procedures on vascular endothelial cells in a variety of processes, such as hemodynamic forces,43hyperhomocysteinemia,44 endothelial differentiation,42 endothelial cell proliferation,45 and IL-146 or angiotensin II47 stimulation of endothelium. This will yield detailed information on the exact nature of what distinguishes an activated or dysfunctional endothelial cell at the gene expression level from a healthy or resting cell, information needed to describe the patho-physiology of the atherosclerotic vessel wall at the molecular level. The 5 novel cDNAs that we described in this report illustrate that many genes potentially related to these molecular processes remain to be identified.
Supported by the Molecular Cardiology Program of the Netherlands Heart Foundation (The Hague) (Grant No. M93.007).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Anton J.G. Horrevoets, PhD, Department of Biochemistry, Academic Medical Center Room K1-160, Meibergdreef 15, 1105 AZ Amsterdam, The Netherlands; e-mail: a.j.horrevoets@amc.uva.nl.