Recent studies have provided evidence for associations between common polymorphic markers in the coagulation factor VII (FVII) gene and plasma FVII levels. Here we describe two common, nonrelated, functional polymorphisms in the promoter region of the FVII gene, a G to T substitution at position −401 and a novel G to A substitution at position −402. Both polymorphisms strongly influence the binding properties of nuclear protein(s). The rare −401T allele is associated with a reduced basal rate of transcription of the FVII gene in human hepatoblastoma cells and with reduced plasma concentrations of total FVII (VIIag) and fully activated FVII molecules (VIIa). In contrast, the rare −402A allele confers increased transcriptional activity and is associated with increased plasma FVII levels. Together, the two polymorphisms explained 18% and 28% of the variation in VIIag and VIIa, respectively, in a group of 183 healthy, middle-aged men. It is concluded that these polymorphisms are important for the regulation of the plasma levels of FVII and that they are likely to be useful genetic markers to resolve the issue of whether a causal relationship exists between FVII levels and risk of coronary heart disease.
COAGULATION FACTOR VII (FVII) is a vitamin K– dependent protease that plays an important role in the extrinsic pathway of blood coagulation. FVII is synthesized principally in the liver and is secreted as an inactive single-chain glycoprotein. In the presence of tissue factor, inactive FVII is converted by limited proteolysis to its fully activated two-chain form, VIIa. Activation can be effected by a number of activated coagulation factors, including Xa, IXa, XIIa, and thrombin. After activation, VIIa rapidly converts FIX and FX into their active forms, thus initiating the generation of thrombin and fibrin clot formation.
The coagulant activity of FVII has been identified as a potential risk indicator for coronary heart disease (CHD). In the Northwick Park Heart Study, FVII coagulant activity was an independent predictor of myocardial infarction in initially healthy middle-aged men,1,2 and particularly of fatal coronary events.3 However, two recent large-scale prospective studies, the Prospective Cardiovascular Münster Study4 and the Edinburgh Artery Study,5 failed to identify the FVII level as a predictor of future CHD. Results from cross-sectional studies are also contradictory. Some studies have reported increased plasma FVII levels in groups with manifest CHD or at risk of CHD,6-10 whereas others did not.11-13
Recent studies have provided evidence for associations between common polymorphic markers in the FVII gene and plasma FVII levels. Two common polymorphisms, a G to A substitution in the codon for amino acid 353 (R353Q) causing a substitution of arginine by glutamine in the FVII protein14 and a decanucleotide insertion/deletion at position −323 in the promoter region (−323P0/1015), have been extensively studied in relation to FVII levels in the past few years.10,16-25Heterozygotes for the rare alleles have 20% to 40% lower FVII coagulant activity than homozygotes for the wild-type alleles,17,21,22 and allele frequencies differ among populations at different risk of CHD.25 FVII gene polymorphisms may explain up to about one third of the FVII level variation in plasma.21 This indicates that plasma FVII levels are to a major extent determined by genetic influences. However, there is considerable controversy over whether these genetically determined variations in plasma FVII levels influence the risk of CHD.10,23 25-27 It is also notable that there is no distinct evidence at present that any of the FVII gene polymorphisms described to date are directly related to changes in FVII metabolism. It is therefore likely that some of these polymorphisms are in linkage disequilibrium with other polymorphisms in the FVII gene, which are of physiological relevance for FVII metabolism. In view of the proposed role of FVII as a risk factor for CHD, it is important to define the nature of these functional polymorphisms.
The rate of FVII gene transcription is indicated to be a critical factor in the regulation of plasma FVII levels. Three common mutations in the promoter of the FVII gene have been described: the −323P0/10 polymorphism, a T to C substitution at position −122 (−122T/C), and a G to T substitution at position −401 (−401G/T).15,28 It has been proposed that the −323P0/10 polymorphism may influence the rate of transcription of FVII.28 However, in preliminary studies using electromobility shift assay (EMSA), we found no evidence that either the −323P0/10 polymorphism or the −122T/C polymorphism affected the interaction with hepatic nuclear proteins, suggesting that these mutations may be irrelevant for FVII gene transcription. We therefore made a more extensive screening of the promoter of the FVII gene to uncover additional polymorphisms and performed functional studies in vitro of all promoter polymorphisms encountered. Here we describe a new, common polymorphism at position −402 that is unrelated to the previously described FVII polymorphisms. It is also shown that the rare alleles of the polymorphisms at positions −401 and −402 are associated with marked changes in the rate of FVII gene transcription in vitro in human hepatoblastoma (HepG2) cells and independently contribute to altered plasma levels of FVII in vivo in healthy middle-aged men.
MATERIALS AND METHODS
Subjects.
A total of 183 men aged 35 to 50 years (40.1 ± 2.8 years, mean ± SD) were investigated in this project. They were recruited at random from the general population of the greater Stockholm area using a registry containing all permanent residents in Stockholm County. Of those initially invited, 81% agreed to participate in the research program. All the men were interviewed to exclude individuals with a history of cardiovascular disease. Additional exclusion criteria were the presence of concomitant disorders, like severely impaired renal function, arteritis, collagenosis, and diabetes mellitus, a history of alcohol abuse or other forms of addiction, and non-Swedish origin of the subject.
Blood sampling, DNA procedures, and biochemical methods.
Blood sampling and preparation of plasma for FVII analyses were as described.10 For DNA procedures, nucleated cells from frozen whole blood were prepared according to Sambrook et al29 and DNA was extracted by a salting-out method.30 The FVII protein concentration was determined as FVII antigen (VIIag), using an enzyme immunoassay (Factor VII EIA kit, Dako A/S, Glostrup, Denmark, a kind gift from Dr Mirella Ezban, Novo Nordisk A/S, Gentofte, Denmark). Plasma levels of activated FVII (VIIa) were determined with a clotting assay31,32 using soluble recombinant truncated tissue factor (a kind gift from Prof James H. Morrissey, Oklahoma Medical Research Foundation, Oklahoma City, OK). Coagulation times were converted to VIIa concentration by comparison with a standard curve constructed from varying concentrations of purified recombinant factor VIIa (a kind gift from Dr Mirella Ezban, Novo Nordisk A/S). Data were analyzed using the Windows Research Software supplied with the ACL-300 coagulometer (Instrumentation Laboratories, Milano, Italy), as described.17 The within and between run coefficients of variation, respectively, were 1.8% and 2.9% for VIIag and 3.9% and 9.1% for VIIa.
Gene sequencing.
For the nucleotide sequencing of the promoter of the FVII gene, a 705 base pair (bp) section spanning from position −491 to +210 was amplified by polymerase chain reaction (PCR) using the forward primer 5′-GGCTCACCTAAGAAACCAGC and the reverse primer 5′-AAGAAATTGAACAGGAGCCG. This PCR-fragment was used as template for further amplifications as part of the Taq DyeDeoxy Terminator Cycle sequencing system (Perkin Elmer, Applied Biosystems Division, Foster City, CA). Nested primers, designed on the basis of the published sequences of the promoter of the FVII gene, were used for the analysis of overlapping sections of 150 to 250 bp in both directions.
Genotyping.
Genotyping for the −401G/T and the −402G/A polymorphisms was performed using a PCR fragment amplified by the forward primer 5′-GGCTCACCTAAGAAACCAGC and the reverse primer 5′-GTTGACATTCCCCATGGGAC, followed by digestion with the restriction enzymes TaiI (for the −401G/T polymorphism) and BslI (for the −402G/A polymorphism). TaiI recognizes the nucleotide sequence ACGT and therefore cleaves the −401T allele. In contrast, the restriction enzyme BslI recognizes the nucleotide sequence CC(N7)GG, and will therefore only cleave the wild-type allele. Because it was observed that the −401G/T and the −402G/A polymorphisms are not related (see Results), it was possible to deduce the genotype of the −402 G/A polymorphism by subtraction of the genotype for the −401G/T polymorphism from the genotype obtained by digestion withBslI. The conditions for the genotyping were: PCR in a 25 μL reaction mixture containing 50 to 500 ng of genomic DNA, 1.2 μmol/L of the primers, 50 mmol/L KCl, 1 mmol/L MgCl2, 10 mmol/L Tris-HCl pH 9.0, 0.1% Triton X-100, 0.2 mmol/L of each dNTP, and 1 unit Termostable Taq polymerase. The reaction mixtures were incubated for 3 minutes at 94°C, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 59°C for 1 minute, and extension at 72°C for 2 minutes. Digestion with the appropriate restriction enzyme was performed in 25 μL of the PCR product using 2.7 units of TaiI at 65°C for 3 hours or 2.7 units ofBslI at 55°C for 4 hours.
Additional assays were developed to genotype for the −401G/T and −402 G/A polymorphisms using primers containing nucleotide mismatches, which made it possible to use restriction enzymes specific for the two polymorphisms. The forward primers were 5′-CAAATATTTACATCCACACCCAGAATAC (−401G/T polymorphism) and 5′-CAAATATTTACATCCACACCCAAGCTGC (−402G/A polymorphism), whereas the same reverse primer 5′-GTTAAGGTGTGTGTGGCACC was used for both PCR amplifications (mismatches in primers are underlined). Because it was found in preliminary experiments that the forward primers influenced the cleavage efficiency of the restriction enzymes, a second PCR reaction was introduced using the forward primer 5′-CAAATATTTACATCCACACCC and the reverse primer 5′-AGGAGAAAGGTCAGGTGACC. The −401G/T and −402G/A polymorphisms were subsequently assayed with the restriction enzymes XmnI and PstI, respectively. Complete agreement was observed between the different assays for genotyping the −401G/T and −402G/A polymorphisms in all DNA samples analyzed for this study.
Genotyping for the −122T/C and −323P0/10 polymorphisms was performed using the forward primer 5′-TCGCATGATTGCTATGGGAC and the reverse primer 5′-GTTGACATTCCCCATGGGAC. The −122T/C polymorphism was analyzed using the restriction enzyme Bsp1286 I. For the analysis of the −323P0/10 polymorphism, the PCR fragment was cleaved by the restriction enzyme EcoRI, followed by electrophoresis using 3% MetaPhor (FMC Bioproducts, Rockland, ME). In addition to the four promoter polymorphisms, the MspI polymorphism in exon 8 of the FVII gene was analyzed as described by Green et al.14 All restriction enzymes were purchased either from New England Biolabs (Beverly, MA) or from Boehringer Mannheim (Mannheim, Germany).
EMSA.
Nuclear extracts were prepared according to Alksnis et al.33 All buffers were freshly supplemented with leupeptin (0.7 μg/mL), aprotinin (16.6 μg/mL), phenylmethyl sulfonyl fluoride (PMSF) (0.2 mmol/L) and 2-mercaptoethanol (0.33 μL/mL). The protein concentration in the extracts were estimated by the method of Kalb and Bernlohr.34 Incubation for EMSA was conducted as described35 and the reaction products were applied to 7% polyacrylamide gel (80:1 acrylamide/N,N′-methylene-bisacrylamide weight ratio), whereafter electrophoresis was performed in 22.5 mmol/L Tris/22.5 mmol/L boric acid/0.5 mmol/L EDTA buffer for 2.5 hours at 200 V. Nonradioactive competitor DNAs, either identical, or of the opposite allelic variant, or of nonspecific origin, were added in 100-fold excess of the labeled DNA.
DNA constructs.
Two sets of double-stranded oligonucleotides were constructed, constituting the 30 bp sequence around the polymorphic region of the −401G/T and −402G/A polymorphisms, flanked by BamHI and BglII ends. The double-stranded oligonucleotides were ligated head to tail into BamHI-digested HCAT vector.36 The correct sequence and orientation of the inserts were tested by DNA sequencing. Promoter-CAT plasmids were constructed using 607 bp promoter fragments (position −490 to +117) ligated into a SplI- and SalI-restricted pCAT-Basic vector as described by the supplier (Promega, Madison, WI). The promoter fragments were obtained by PCR amplification of DNA samples from subjects homozygous for either the −402A or the −401T alleles, or homozygous for the wild-type condition, using the forward primer 5′-ACATGCATGCGCTCACCTAAGAAACCAGCC and the reverse primer 5′-ACGCGTCGACGTTTATGGAGAAAACCTCCCC.
Transient transfection assay.
HepG2 cells were cultured in 90-mm dishes in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. Confluent cells were transfected using the calcium-phosphate DNA coprecipitation method, essentially as described by Sambrook et al.29 pSV-β-galactosidase (Promega) was cotransfected as an internal control. In all experiments, 5 μg of CAT-construct and 5 μg of β-galactosidase plasmid were added to the medium. CAT activity was analyzed using the method described by Sambrook et al29 and quantified using a phosphorimager (Bio-imaging Analyzer BAS-2500, Fuji Photo Film Co, Tokyo, Japan). β-galactosidase activity was determined as described by the supplier (Promega). CAT levels were expressed in arbitrary units after standardization for β-galactosidase activity. All constructs were tested in triplicates in four independent transfection experiments.
Statistical methods.
Distributions of continuous variables in groups were expressed as means ± standard deviation. Logarithmic transformation was performed on all skewed variables to obtain a normal distribution before statistical computations and significance testing were undertaken. Allele frequencies were estimated by gene counting. A χ2 test was used to compare the observed numbers of each FVII genotype with those expected for a population in Hardy-Weinberg equilibrium. The normalized linkage disequilibrium coefficient (D’) for all pairs of FVII polymorphisms was calculated according to Ott.37One-way analyses of covariance (with age or age and plasma triglycerides as covariates) and two-way analyses of variance performed by the general linear model procedure were performed to test whether genetic variation within the FVII locus was associated with differences in VIIag or VIIa. The Scheffé multiple comparison test was used as a post-hoc test. The percentage of genotype-based variation in FVII levels was calculated according to Sing and Davignon.38Differences in binding affinity of nuclear proteins for the polymorphic sites and differences in transcriptional activity between promoter constructs were evaluated by paired Student’s t-test.
RESULTS
Detection of a new, common polymorphism at position −402 of the FVII promoter.
A 705 bp section of the proximal promoter of the FVII gene was sequenced in both directions using DNA samples from 10 subjects with a broad range of plasma FVII levels. Minor differences were observed compared with the sequence reported by O’Hara et al39: an extra C was observed at −149, and an extra G was observed at −459. No differences were found compared with the sequences published by Pollak et al28 and Erdmann and Heim.40 The nomenclature introduced by the latter authors will be used throughout this report. Three previously described mutations were observed in several subjects: the decanucleotide sequence insert (5′-CCTATATCCT-3′) at position −323 (−323P0/10), the T to C substitution at position −122 (−122T/C), and the G to T substitution at position −401 (−401G/T). In addition, a G to A substitution at position −402 was found in several subjects.
Assays were developed for the detection of all four promoter mutations, and the nucleotide sequence of the promoter of the FVII gene was determined in DNA samples from additional subjects who were homozygous for the rare haplotype −122C/−323P10/−401T. No additional polymorphisms were detected when in total nine alleles with this haplotype were analyzed. The nucleotide sequence was also determined in DNA samples from subjects who were homozygous for the rare −402A allele. Again, no additional polymorphisms were detected when in total 11 alleles with the −402A allele were analyzed.
Allele-specific binding of two nuclear proteins to the −401G/T and −402G/A polymorphic sites.
The possibility that the decanucleotide insertion at −323 and/or the nucleotide substitutions at positions −122, −401, and −402 might affect the interaction with nuclear proteins was analyzed with EMSA using nuclear extracts derived from HepG2 cells. No evidence was found for differences in binding characteristics of a 30 bp DNA fragment containing either the −122T or the −122C site of the FVII promoter (data not shown). Furthermore, no differences were found in the binding characteristics of a 35 bp DNA fragment containing the decanucleotide insertion and a comparable 25 bp DNA fragment without the insertion (data not shown).
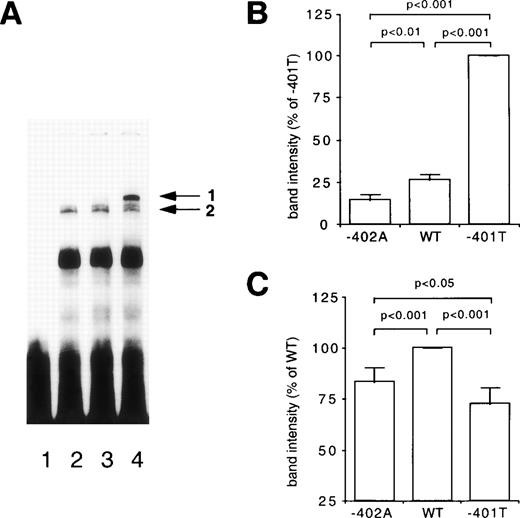
The binding characteristics of the −401G/T and −402G/A polymorphisms were analyzed using three different 30 bp DNA fragments. The wild-type fragment was a 30 bp fragment containing the common −401G and −402G sites. For the analysis of the −401G/T polymorphism, a 30 bp fragment containing the rare −401T site and the common −402G site was used. This fragment is referred to as the −401T fragment. The 30 bp fragment termed −402A contained the rare −402A site and the common −401G site. Considerable quantitative differences were observed for two protein-DNA complexes when the binding characteristics of the three DNA fragments were analyzed in detail. As shown in Fig 1A, substantial quantities of protein-DNA complex 1 (indicated with arrow 1) were observed for the −401T fragment. In contrast, only minor quantities of complex 1 were noted for the wild-type and the −402A fragments. Lower quantities of protein-DNA complex 2 (indicated with arrow 2) were observed with the −402A and the −401T fragments than with the wild-type fragment. Occasionally, we noted that the protein-DNA complex 2 was split in two bands with slightly different mobility. The observed differences between the three fragments were quantified in five independent experiments, using different nuclear extracts from HepG2 cells. In agreement with the data presented in Fig 1A, significant quantitative differences were observed between the three fragments for the two protein-DNA complexes (Fig 1B and 1C). These quantitative differences were substantiated by the results from EMSA studies in which the binding characteristics of the three fragments were analyzed using different concentrations of nuclear extract (data not shown). Competition experiments showed that all three fragments are able to compete effectively for the two protein-DNA complexes when either the −401T fragment (Fig 2), the −402A fragment, or the wild-type fragment (data not shown) were used as radiolabeled probe, indicating specific protein-DNA interactions. Taken together, the EMSA studies provided evidence for differences in the binding characteristics of the three fragments with regard to two nuclear proteins.
Differential binding of nuclear proteins to the −401 and −402 polymorphic sites. (A) EMSA of nuclear extract derived from HepG2 cells bound to a 30 base pair DNA fragment containing either the −402A site (lanes 1 and 2), the wild-type (WT) site (lane 3), or the −401T site (lane 4) of the FVII promoter. Arrows 1 and 2 denote the allele specific factors. Lane 1, without extract; lanes 2 to 4, 0.20 mg/mL HepG2 extract. (B and C) The relative intensity of the protein-DNA complexes 1 and 2 obtained with the three DNA fragments were quantified in five independent experiments. Bars indicate mean values and standard deviations. The statistical significance of differences was determined by Student’s paired t-test.
Differential binding of nuclear proteins to the −401 and −402 polymorphic sites. (A) EMSA of nuclear extract derived from HepG2 cells bound to a 30 base pair DNA fragment containing either the −402A site (lanes 1 and 2), the wild-type (WT) site (lane 3), or the −401T site (lane 4) of the FVII promoter. Arrows 1 and 2 denote the allele specific factors. Lane 1, without extract; lanes 2 to 4, 0.20 mg/mL HepG2 extract. (B and C) The relative intensity of the protein-DNA complexes 1 and 2 obtained with the three DNA fragments were quantified in five independent experiments. Bars indicate mean values and standard deviations. The statistical significance of differences was determined by Student’s paired t-test.
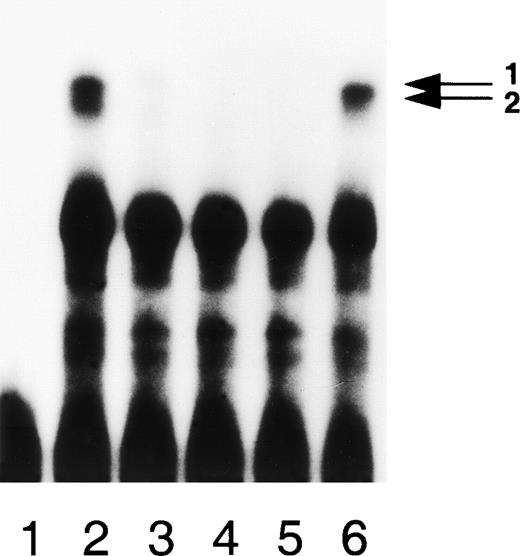
Binding of nuclear proteins is specific. EMSA of nuclear extract derived from HepG2 cells bound to the −401T fragment in the presence of unlabeled DNA as competitor. Arrows 1 and 2 refer to the allele-specific factors. Lane 1, without extract; lane 2, 0.20 mg/mL of HepG2 extract in the absence of competitor; lanes 3 to 6, 0.20 mg/mL of HepG2 extract in the presence of 200-fold excess of unlabeled DNA as competitor. Competitors used were: −402A site (lane 3), WT site (lane 4), −401T site (lane 5), and nonrelated 30 bp fragment (lane 6).
Binding of nuclear proteins is specific. EMSA of nuclear extract derived from HepG2 cells bound to the −401T fragment in the presence of unlabeled DNA as competitor. Arrows 1 and 2 refer to the allele-specific factors. Lane 1, without extract; lane 2, 0.20 mg/mL of HepG2 extract in the absence of competitor; lanes 3 to 6, 0.20 mg/mL of HepG2 extract in the presence of 200-fold excess of unlabeled DNA as competitor. Competitors used were: −402A site (lane 3), WT site (lane 4), −401T site (lane 5), and nonrelated 30 bp fragment (lane 6).
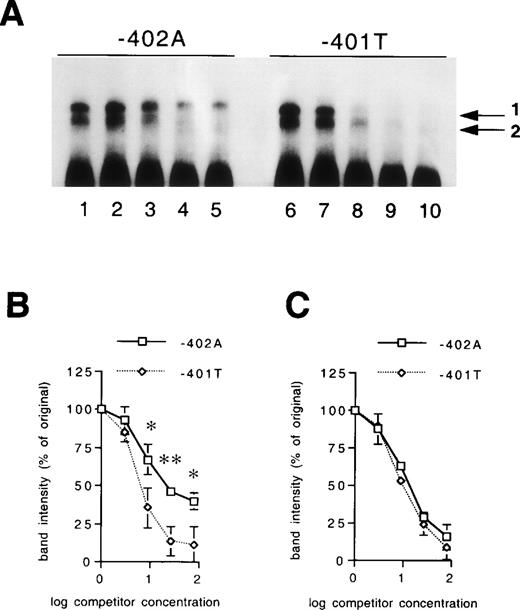
The differences in the binding characteristics of the three fragments were further explored by quantitative competition experiments. In Fig 3, the abilities of unlabeled −401T and −402A fragments to compete with a labeled −401T fragment are compared. As expected, the unlabeled −401T fragment was a more efficient competitor for the protein-DNA complex 1, as compared with the unlabeled −402A fragment, whereas no significant differences were observed between unlabeled −401T and −402A fragments regarding competition for protein-DNA complex 2 (Fig 3A). These differences were quantified in three independent experiments (Fig 3B and 3C). Similar experiments were performed comparing the abilities of unlabeled −402A and wild-type fragments to compete with the labeled wild-type fragment. These studies showed that the unlabeled −402A fragment is less effective than the unlabeled wild-type fragment in competing for protein-DNA complex 1 (data not shown). In all, the results from the EMSA studies indicate that both the −401G/T mutation and the −402G/A mutation influence the specific binding properties of two nuclear proteins.
Differences in binding affinity of the nuclear proteins for the polymorphic sites. (A) EMSA of nuclear extract derived from HepG2 cells bound to the −401T fragment in the presence of increasing concentrations of unlabeled −402A fragment (lanes 2 to 5) or −401T fragment (lanes 7 to 10) as competitor. Arrows 1 and 2 refer to the allele-specific factors. Lanes 1 and 6, 0.20 mg/mL of HepG2 extract in the absence of competitor; lanes 2 to 5 and 7 to 10, 0.20 mg/mL of HepG2 extract in the presence of threefold (lanes 2 and 7), ninefold (lanes 3 and 8), 27-fold (lanes 4 and 9) and 81-fold (lanes 5 and 10) excess DNA as competitor. The intensity of the protein-DNA complexes 1 and 2 were quantified in three independent experiments. The relative intensities of the protein-DNA complexes 1 and 2 in the presence of increasing concentrations of unlabeled DNA fragments are shown in B and C, respectively. Line plots indicate mean values and standard deviations. The statistical significance of differences was determined by Student’s paired t-test. *P < .05; **P< .01.
Differences in binding affinity of the nuclear proteins for the polymorphic sites. (A) EMSA of nuclear extract derived from HepG2 cells bound to the −401T fragment in the presence of increasing concentrations of unlabeled −402A fragment (lanes 2 to 5) or −401T fragment (lanes 7 to 10) as competitor. Arrows 1 and 2 refer to the allele-specific factors. Lanes 1 and 6, 0.20 mg/mL of HepG2 extract in the absence of competitor; lanes 2 to 5 and 7 to 10, 0.20 mg/mL of HepG2 extract in the presence of threefold (lanes 2 and 7), ninefold (lanes 3 and 8), 27-fold (lanes 4 and 9) and 81-fold (lanes 5 and 10) excess DNA as competitor. The intensity of the protein-DNA complexes 1 and 2 were quantified in three independent experiments. The relative intensities of the protein-DNA complexes 1 and 2 in the presence of increasing concentrations of unlabeled DNA fragments are shown in B and C, respectively. Line plots indicate mean values and standard deviations. The statistical significance of differences was determined by Student’s paired t-test. *P < .05; **P< .01.
The −401G/T and −402G/A polymorphisms modulate the transcription of the FVII gene.
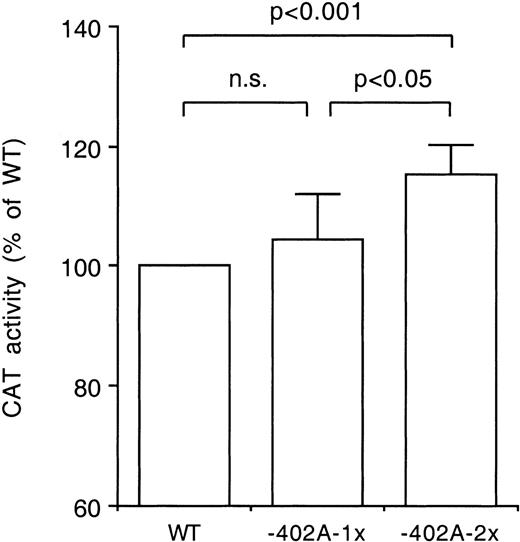
Transfection studies in HepG2 cells were conducted to evaluate whether the −401G/T and the −402G/A polymorphisms influence the rate of transcription of the FVII gene. CAT activities were compared between constructs harboring either a single or two tandemly arranged 30 bp fragments of the FVII promoter. These constructs contained either the −401T, the −402A or the wild-type sites. As shown in Fig 4, higher CAT activities were observed with the −402A than with the wild-type constructs. In contrast, significantly lower CAT activities were observed with the −401T than with the wild-type constructs (Fig 5). It is notable that the effects of the tandemly arranged constructs were greater than those of the single-fragment constructs.
Difference in transcriptional activity of the −402A allele. The CAT activities of constructs harboring one or two tandemly arranged 30 base pair fragments with either the wild-type (WT) site or the −402A site were compared in transfection studies using HepG2 cells. The constructs were tested in triplicate in four independent experiments. The CAT activities of the −402A-1x and −402A-2x constructs were expressed relative to the activities of the WT-1x and WT-2x constructs, respectively, the latter jointly indicated as WT. Bars indicate mean values and standard deviations. The statistical significance of differences was determined by Student’s pairedt-test.
Difference in transcriptional activity of the −402A allele. The CAT activities of constructs harboring one or two tandemly arranged 30 base pair fragments with either the wild-type (WT) site or the −402A site were compared in transfection studies using HepG2 cells. The constructs were tested in triplicate in four independent experiments. The CAT activities of the −402A-1x and −402A-2x constructs were expressed relative to the activities of the WT-1x and WT-2x constructs, respectively, the latter jointly indicated as WT. Bars indicate mean values and standard deviations. The statistical significance of differences was determined by Student’s pairedt-test.
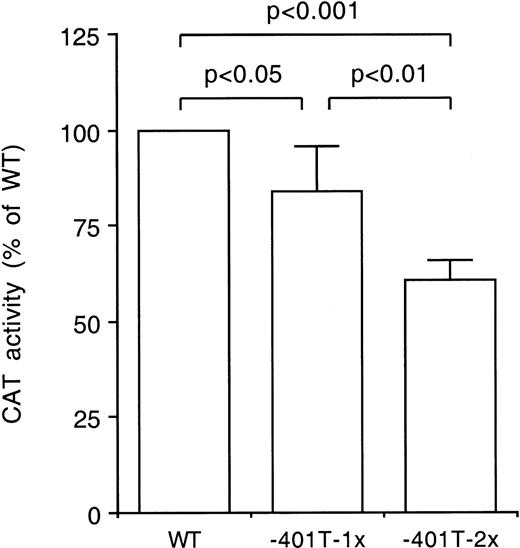
Difference in transcriptional activity of the −401T allele. The CAT activities of constructs harboring one or two tandemly arranged 30 base pair fragments with either the wild-type (WT) site or the −401T site were compared in transfection studies using HepG2 cells. The constructs were tested in triplicate in four independent experiments. The CAT activities of the −401T-1x and −401T-2x constructs were expressed relative to the activities of the WT-1x and WT-2x constructs, respectively, the latter jointly indicated as WT. Bars indicate mean values and standard deviations. The statistical significance of differences was determined by Student’s pairedt-test.
Difference in transcriptional activity of the −401T allele. The CAT activities of constructs harboring one or two tandemly arranged 30 base pair fragments with either the wild-type (WT) site or the −401T site were compared in transfection studies using HepG2 cells. The constructs were tested in triplicate in four independent experiments. The CAT activities of the −401T-1x and −401T-2x constructs were expressed relative to the activities of the WT-1x and WT-2x constructs, respectively, the latter jointly indicated as WT. Bars indicate mean values and standard deviations. The statistical significance of differences was determined by Student’s pairedt-test.
Additional transfection experiments were performed using 609 bp FVII promoter constructs. In agreement with other reports,36 it was found that the activity of the FVII promoter is remarkably weak. The expression of the FVII promoter in HepG2 cells was only two- to threefold higher than the control plasmid without promoter, whereas in the same experiments the SV40 promoter gave 25- to 50-fold higher CAT values. This phenomenon considerably hampered the detection of differences in CAT activity between the long FVII promoter constructs. In four experiments a significant reduction (–34% ± 17%,P < .05) in CAT activity was observed for the −401T construct compared with the wild-type construct. A small increase (+13% ± 21%) in CAT activity was noted for the −402A construct, but this increase did not reach the conventional level of statistical significance.
Allele frequencies and degree of linkage disequilibrium.
Genotyping was performed in 183 healthy, population-based men aged 35 to 50 years. All FVII polymorphisms were found to be in Hardy-Weinberg equilibrium. Allele frequencies were as follows: −122T/C (0.91/0.09), −323P0/10 (0.91/0.09), −401G/T (0.91/0.09), −402G/A (0.71/0.29), and R353Q (0.92/0.08). The −122T/C, −323P0/10, and −401G/T polymorphisms exhibited complete allelic association. The normalized linkage disequilibrium coefficient (D’) was −0.4123 (P < .0009) for the −401G/T and −402G/A polymorphisms, 0.9985 (P < .0001) for the −401G/T and R353Q polymorphisms, and −0.4076 (P<0.0026) for the −402G/A and R353Q polymorphisms.
Associations between the −401G/T and −402G/A polymorphisms and the plasma concentrations of activated FVII and FVII mass.
The relationships between the −401G/T and −402G/A polymorphisms and FVII plasma levels were analyzed in detail in the 183 apparently healthy, population-based men. As shown in Table1, the rare −401T allele was associated with significantly lower plasma concentrations of FVII protein (VIIag) and fully activated FVII molecules (VIIa), respectively, than the common −401G allele. In contrast, the rare −402A allele was associated with significantly higher FVII levels than the common −402G allele. Furthermore, subjects who were homozygous for a rare allele had a greater change in FVII levels than subjects who were heterozygous for a rare allele. The influences of the −401G/T and −402G/A polymorphisms on the VIIag concentration were about equally strong, accounting for 11.0% and 10.1% of the variation in VIIag, respectively (Table 1). In contrast, the impact of the −401G/T polymorphism on the VIIa concentration was stronger (% attributable phenotypic variation 19.1%) than the corresponding effect of the −402G/A polymorphism (% attributable phenotypic variation 4.6%). By comparison, the R353Q polymorphism in exon 8 accounted for 11.1% of the variation in VIIag (P < .0001) and 23.7% of the variation in VIIa (P < .0001).
Associations Between the −401G/T and −402G/A Polymorphisms in the Promoter Region of the Factor VII Gene Locus and Plasma Concentrations of Factor VII Protein and Activated Factor VII in Healthy Men
| −401 Genotype . | −401G/G . | −401G/T . | −401T/T . | Covariance Analysis . | % of Phenotypic Variation . |
|---|---|---|---|---|---|
| N | 151 | 31 | 1 | ||
| VIIag (ng/mL) | 503 ± 110 | 408 ± 90 | 331 | <.0001 | 11.0 |
| VIIa (ng/mL) | 5.0 ± 1.2 | 3.1 ± 0.7 | 2.6 | <.0001 | 19.1 |
| −402 Genotype | −402G/G | −402G/A | −402A/A | Covariance Analysis | % of Phenotypic Variation |
| N | 89 | 81 | 13 | ||
| VIIag (ng/mL) | 453 ± 100 | 510 ± 119 | 565 ± 83 | <.001 | 10.1 |
| VIIa (ng/mL) | 4.5 ± 1.6 | 4.7 ± 1.6 | 5.8 ± 1.6 | <.001 | 4.6 |
| −401 Genotype . | −401G/G . | −401G/T . | −401T/T . | Covariance Analysis . | % of Phenotypic Variation . |
|---|---|---|---|---|---|
| N | 151 | 31 | 1 | ||
| VIIag (ng/mL) | 503 ± 110 | 408 ± 90 | 331 | <.0001 | 11.0 |
| VIIa (ng/mL) | 5.0 ± 1.2 | 3.1 ± 0.7 | 2.6 | <.0001 | 19.1 |
| −402 Genotype | −402G/G | −402G/A | −402A/A | Covariance Analysis | % of Phenotypic Variation |
| N | 89 | 81 | 13 | ||
| VIIag (ng/mL) | 453 ± 100 | 510 ± 119 | 565 ± 83 | <.001 | 10.1 |
| VIIa (ng/mL) | 4.5 ± 1.6 | 4.7 ± 1.6 | 5.8 ± 1.6 | <.001 | 4.6 |
Values are mean ± SD. One-way analysis of covariance (with age as covariate) was performed to test whether genetic variation within the factor VII promoter was associated with differences in factor VII mass concentration (VIIag) and plasma levels of activated factor VII (VIIa). The percentage of genotype-based variation in factor VII levels was calculated according to Sing and Davignon.34
To further determine the impact of genetic variation on FVII levels in plasma, genotypes for each pair of polymorphisms were defined and ranked in order of FVII mean values (Table2). In addition to the two functional −401G/T and −402G/A polymorphisms in the FVII promoter, the R353Q polymorphism in exon 8 was included in this analysis. The highest VIIag and VIIa concentrations were encountered in subjects who were homozygous for the −401G, −402A, and 353R alleles. Covariance analysis controlling for the influence of age on VIIag and VIIa (Pearson correlation coefficients: r = 0.06, ns, and r = 0.23,P < 0.01, respectively) showed that the mean FVII level differences between combined genotypes were highly significant (Table2). Of note, the genetic contribution to the variation in VIIa concentration was much higher than the genetic contribution to the variation in VIIag. Whereas genetic variation associated with genotypes for each pair of polymorphisms accounted for 12% to 18% of the total variation in VIIag, the corresponding figures for VIIa were 26% to 28%. The joint effect on VIIag of the −401G/T and R353Q polymorphisms, which are in strong linkage disequilibrium, was as expected significantly smaller (11.6%) than the joint effects of the other two pairs of weakly linked FVII polymorphisms (17.8% for both the −401G/T and −402G/A polymorphisms and the −402G/A and R353Q polymorphisms). In contrast, the three pairs of FVII polymorphisms had a similar impact on the plasma concentration of VIIa.
Mean Values of Plasma Concentrations of Factor VII Protein and Activated Factor VII in Groups of Healthy Men With Different Combinations of Factor VII Genotypes
| Genotypes . | Factor VII Protein . | Covariance Analysis . | % of Phenotypic Variation . | ||
|---|---|---|---|---|---|
| No. . | % . | VIIag (ng/mL) . | |||
| [−401G/T]/[−402G/A] | |||||
| GG/AA | 13 | 7.1 | 565 ± 83 | 7.56 | 17.8 |
| GG/GA | 71 | 38.8 | 524 ± 114 | <.0001 | |
| GG/GG | 67 | 36.6 | 469 ± 101 | ||
| GT/GA | 10 | 5.5 | 409 ± 106 | ||
| GT/GG | 21 | 11.5 | 408 ± 84 | ||
| TT/GG | 1 | 0.5 | 331 | ||
| [−401G/T]/[R353Q] | |||||
| GG/RR | 151 | 82.5 | 503 ± 110 | 7.58 | 11.6 |
| GT/RR | 4 | 2.2 | 462 ± 87 | <.0001 | |
| GT/RQ | 27 | 14.8 | 400 ± 89 | ||
| TT/RQ | 1 | 0.5 | 331 | ||
| [−402G/A]/[R353Q] | |||||
| AA/RR | 13 | 7.1 | 565 ± 83 | 9.45 | 17.8 |
| GG/RR | 73 | 39.9 | 522 ± 114 | <.0001 | |
| GA/RR | 69 | 37.7 | 469 ± 100 | ||
| GG/RQ | 20 | 10.9 | 398 ± 81 | ||
| GA/RQ | 8 | 4.4 | 396 ± 110 | ||
| Genotypes | Activated Factor VII | Covariance Analysis | % of Phenotypic Variation | ||
| No. | % | VIIa (ng/mL) | |||
| [−401G/T]/[−402G/A] | |||||
| GG/AA | 13 | 7.1 | 5.8 ± 1.6 | 13.89 | 27.6 |
| GG/GG | 67 | 36.6 | 5.0 ± 1.6 | <.0001 | |
| GG/GA | 71 | 38.8 | 4.9 ± 1.5 | ||
| GT/GA | 10 | 5.5 | 3.2 ± 1.0 | ||
| GT/GG | 21 | 11.5 | 3.1 ± 0.7 | ||
| TT/GG | 1 | 0.5 | 2.6 | ||
| [−401G/T]/[R353Q] | |||||
| GG/RR | 151 | 82.5 | 5.0 ± 1.6 | 20.63 | 26.3 |
| GT/RR | 4 | 2.2 | 3.4 ± 1.0 | <.0001 | |
| GT/RQ | 27 | 14.8 | 3.1 ± 0.8 | ||
| TT/RQ | 1 | 0.5 | 2.6 | ||
| [−402G/T]/[R353Q] | |||||
| AA/RR | 13 | 7.1 | 5.8 ± 1.6 | 14.79 | 25.5 |
| GG/RR | 73 | 39.9 | 4.9 ± 1.6 | <.0001 | |
| GA/RR | 69 | 37.7 | 4.9 ± 1.5 | ||
| GG/RQ | 20 | 10.9 | 3.1 ± 0.7 | ||
| GA/RQ | 8 | 4.4 | 2.9 ± 0.9 | ||
| Genotypes . | Factor VII Protein . | Covariance Analysis . | % of Phenotypic Variation . | ||
|---|---|---|---|---|---|
| No. . | % . | VIIag (ng/mL) . | |||
| [−401G/T]/[−402G/A] | |||||
| GG/AA | 13 | 7.1 | 565 ± 83 | 7.56 | 17.8 |
| GG/GA | 71 | 38.8 | 524 ± 114 | <.0001 | |
| GG/GG | 67 | 36.6 | 469 ± 101 | ||
| GT/GA | 10 | 5.5 | 409 ± 106 | ||
| GT/GG | 21 | 11.5 | 408 ± 84 | ||
| TT/GG | 1 | 0.5 | 331 | ||
| [−401G/T]/[R353Q] | |||||
| GG/RR | 151 | 82.5 | 503 ± 110 | 7.58 | 11.6 |
| GT/RR | 4 | 2.2 | 462 ± 87 | <.0001 | |
| GT/RQ | 27 | 14.8 | 400 ± 89 | ||
| TT/RQ | 1 | 0.5 | 331 | ||
| [−402G/A]/[R353Q] | |||||
| AA/RR | 13 | 7.1 | 565 ± 83 | 9.45 | 17.8 |
| GG/RR | 73 | 39.9 | 522 ± 114 | <.0001 | |
| GA/RR | 69 | 37.7 | 469 ± 100 | ||
| GG/RQ | 20 | 10.9 | 398 ± 81 | ||
| GA/RQ | 8 | 4.4 | 396 ± 110 | ||
| Genotypes | Activated Factor VII | Covariance Analysis | % of Phenotypic Variation | ||
| No. | % | VIIa (ng/mL) | |||
| [−401G/T]/[−402G/A] | |||||
| GG/AA | 13 | 7.1 | 5.8 ± 1.6 | 13.89 | 27.6 |
| GG/GG | 67 | 36.6 | 5.0 ± 1.6 | <.0001 | |
| GG/GA | 71 | 38.8 | 4.9 ± 1.5 | ||
| GT/GA | 10 | 5.5 | 3.2 ± 1.0 | ||
| GT/GG | 21 | 11.5 | 3.1 ± 0.7 | ||
| TT/GG | 1 | 0.5 | 2.6 | ||
| [−401G/T]/[R353Q] | |||||
| GG/RR | 151 | 82.5 | 5.0 ± 1.6 | 20.63 | 26.3 |
| GT/RR | 4 | 2.2 | 3.4 ± 1.0 | <.0001 | |
| GT/RQ | 27 | 14.8 | 3.1 ± 0.8 | ||
| TT/RQ | 1 | 0.5 | 2.6 | ||
| [−402G/T]/[R353Q] | |||||
| AA/RR | 13 | 7.1 | 5.8 ± 1.6 | 14.79 | 25.5 |
| GG/RR | 73 | 39.9 | 4.9 ± 1.6 | <.0001 | |
| GA/RR | 69 | 37.7 | 4.9 ± 1.5 | ||
| GG/RQ | 20 | 10.9 | 3.1 ± 0.7 | ||
| GA/RQ | 8 | 4.4 | 2.9 ± 0.9 | ||
Values are mean ± SD. One-way analysis of covariance (with age as covariate) was performed to test whether genetic variation within the factor VII promoter was associated with differences in factor VII mass concentration (VIIag) and plasma levels of activated factor VII (VIIa).
Because there is evidence that the plasma FVII levels are influenced by factors like age, body mass index (BMI), and plasma lipid concentrations (reviewed in41), we tested whether these factors differed according to −401G/T and −402G/A genotypes. However, no relationships were observed between the two polymorphisms and age, BMI, and plasma concentrations of cholesterol and triglycerides in the major lipoprotein classes VLDL, LDL, and HDL (data not shown).
DISCUSSION
In this study we conducted an extensive screening of the promoter region of the FVII gene to detect new polymorphisms that might influence the plasma levels of FVII and the ensuing risk of CHD. One novel common polymorphism was found, a G to A substitution at position −402. This polymorphism was evaluated with respect to effects on transcriptional regulation in vitro, along with previously detected polymorphisms in the promoter region of the FVII gene locus. The −401G/T and −402G/A polymorphisms proved to be functional in vitro in HepG2 cells and associated with altered plasma concentrations of FVII in vivo in population-based healthy human volunteers. Together the −401G/T and −402G/A polymorphisms accounted for 18% and 28% of the variation in the plasma concentrations of total FVII and fully activated factor FVII molecules, respectively.
Three basic observations were made in relation to both the −401G/T and −402G/A polymorphisms. Firstly, results from the EMSA studies using nuclear extracts from HepG2 cells indicated that the two mutations influence the specific binding properties of either both (−401T) or one (−402A) of two nuclear protein complexes. Secondly, transfection studies provided evidence that the rare −401T allele is associated with a decreased basal transcription rate, whereas the rare −402A allele appeared to confer increased transcriptional activity compared with constructs harboring the wild-type allele. Thirdly, significant associations were found between the two polymorphisms and both the FVII mass concentration and the plasma level of the fully activated FVII molecule. The hypothesis that ensues from these observations is that they are interrelated and part of a sequence of events starting at the level of the FVII promoter and ultimately leading to modulation of the FVII levels in plasma. Thus, it is envisaged that the −401G/T and −402G/A polymorphisms affect the binding of two hepatic nuclear proteins to the promoter of the FVII gene, with accompanying changes of FVII expression in hepatocytes and FVII secretion by the liver. This, in turn, results in decreased (−401T allele) or increased (−402A allele) plasma levels of FVII.
The molecular mechanisms remain to be determined in detail. The present data could be interpreted to suggest the hypothesis that the nuclear protein contained in complex 1 in the EMSA studies functions as a transcriptional repressor by interfering with the binding/activation of the protein contained in complex 2 that would act as a transcriptional activator. The repressor protein, which binds strongly to the rare −401T allele but only to a limited extent to the rare −402A allele, thus seems to determine the differences in the activity of the FVII promoter related to the −401G/T and −402G/A polymorphisms. However, further experiments using for example methylation interference assays are needed to show that the proteins contained in complexes 1 and 2 compete for binding at the same or overlapping sites.
The physiological significance of the −401G/T and −402G/A polymorphisms was analyzed in transfection studies in HepG2 cells. Initially, we evaluated the effects of these polymorphisms using long promoter constructs. In agreement with Pollak et al,28 a reduction in CAT activity was observed for the construct containing the −122C/−323P10/−401T alleles when compared with a construct harboring the wild-type alleles. As was also observed by these authors, the activity of the FVII promoter was found to be remarkably weak. This phenomenon seriously limits the application of long promoter constructs for the analysis of the basal rate of transcription of the FVII gene. We therefore evaluated the effects of the −401G/T and −402G/A mutations using single or tandemly arranged 30 bp fragments of the FVII promoter. The use of these constructs made it possible to evaluate more accurately the impact of the two mutations on the basal rate of transcription of the FVII gene. These studies clearly showed that the −401T allele is associated with a reduced rate of transcription, whereas the −402A allele is related to an increased rate of transcription of the FVII gene.
In agreement with the report of Dell’Acqua et al,42 the −401G/T polymorphism showed complete allelic association with the −323P0/10 and −122T/C polymorphisms. Because neither of these latter polymorphisms were found to exhibit allele-specific interactions with hepatic nuclear proteins, it could be argued that the −401G/T site accounts for the association of the −323P10 allele with significantly lower plasma FVII concentrations, which has been shown in several populations.21,22,24,25 It should be emphasized in this context that failure to detect allele-specific interactions with nuclear proteins on EMSA does not completely rule out the potential role of a promoter polymorphism. Indeed, Pollak et al28 proposed that the −323P0/10 polymorphism is of functional significance. However, in agreement with our results, no protein-binding sites were found by these authors in the vicinity of the −323 site, and the −401G/T site appears to have been contained in the constructs used in the transfection studies.
The impact of the −401G/T and −402G/A polymorphisms on the plasma FVII levels was evaluated in apparently healthy, middle-aged Swedish men. When the two functional promoter polymorphisms were analyzed in conjunction, they contributed to 18% of the variation in the total FVII protein concentration (VIIag) in plasma and to 28% of the variation in the plasma concentration of fully activated FVII molecules (VIIa). Furthermore, the impact of the −401G/T site on VIIa was much stronger than the corresponding effect of the −402G/A site, whereas the two promoter polymorphisms had similar effects on VIIag. It seems conceivable that the −401G/T and −402G/A polymorphisms would only affect the synthesis of FVII. The R353Q site in exon 8 reduces the secretion of FVII43and is in strong linkage disequilibrium with the −401G/T site in most populations studied so far. The question then arises of whether the R353Q site contributes to the high proportion of the variation in VIIa associated with the −401G/T polymorphism or whether the −401G/T locus is linked to other mutations in coding regions of the FVII gene. It is notable in this context that the charge change ensuing from the substitution of a positively charged arginine with a neutral glutamine at amino acid position 353 may affect the interaction of FVII with lipid surfaces with ensuing effects on the activity state of the FVII molecule.44 It is noteworthy that the in vivo data are in good agreement with the results from the in vitro transfection studies. Indeed, in both transfection experiments and clinical studies it was found that the −401T allele was associated with a major reduction in transcriptional activity and plasma FVII mass concentration, whereas the enhancing effect of the −402A allele on these parameters was relatively small. However, the overall effects of the −401G/T and −402G/A polymorphisms at the population level were comparable, due to the three-fold higher frequency of the −402A allele as compared with the −401T allele.
There is some epidemiological evidence that FVII coagulant activity is a predictor of future CHD.1-4 However, data is contradictory both with respect to plasma FVII level relations to CHD1-13 and regarding the potential relationship between genetic markers associated with FVII coagulant activity and risk of CHD.10,23,25-27 It is noteworthy in this context that none of the genetic markers used to date to evaluate the relations of FVII with CHD are known to be related to changes in FVII metabolism. The results presented in this report indicate that the −401G/T and −402G/A polymorphisms are physiologically relevant for FVII metabolism and suggest that they may be useful genetic markers for resolving the issue of whether a causal relationship exists between FVII levels and CHD. Needless to say, the predictive power of individual polymorphisms may vary between populations depending on differences in the overall risk factor burden, environmental influences, and gene-environment interactions. It is interesting to note that the frequencies of the protective Q353 and −401T alleles were considerably lower amongst the healthy 50-year old Swedish men participating in the present study (0.08 and 0.09, respectively) compared with healthy Italian populations (0.22 to 0.25 and 0.13, respectively)21,26 43 in which the incidence of myocardial infarction is much lower.
ACKNOWLEDGMENT
The authors thank Tobias Burt for help with the genotyping.
Supported by grants from the Swedish Medical Research Council (8691), the Swedish Heart-Lung Foundation, the Marianne and Marcus Wallenberg Foundation, the European Commission (BMH4-CT96-0272), the Petrus and Augusta Hedlund Foundation, the King Gustaf V 80th Birthday Foundation, the Professor Nanna Svartz Foundation, and the Foundation for Old Servants.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Ferdinand M. van ’t Hooft, MD, King Gustaf V Research Institute, Karolinska Hospital, S-171 76 Stockholm, Sweden; e-mail: gerd@instmed.ks.se.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal