Abstract
We identified an antibody-secreting human B-cell line (HTD8), which actively replaces the production of the original λ light chain with a new λ chain (light chain shifting) at a high rate. Loss of the original rearranged λ light chain occurs by significantly reducing the amount of transcript expressed. Expression of the new λ chain, which replaces the original λ chain, occurs by rearranging new VJ segments on a previously excluded allele. V λ gene usage of these new rearrangements are biased toward Vλ4, Vλ6, and Vλ10 families, which are known to be the least frequently used. In striking contrast to the plasma cell phenotype, recombination activating genes, RAG-1 and RAG-2, were expressed in the HTD8 cells and were shown to be necessary, but insufficient for inducing expression of the new λ chain. These results suggest that human plasma cells have the potential to actively undergo light chain replacement.
ALLELIC EXCLUSION is a fundamental principle in B-cell development and is the basis for clonal selection, which limits one cell to produce a single antibody with a certain specificity. Ig gene rearrangements occur in a highly ordered fashion during B-cell development1 and is initiated by DNA rearrangement at the heavy-chain locus.2 In developing B cells, the expression of a complete Ig heavy-chain protein is accompanied by a drastic change in the targeting of V(D)J recombinase activity. Recombinase activity changes from being predominantly active at the heavy-chain locus in pro-B cells to being exclusively restricted to the light chain loci in pre-B cells. This switch in locus-specific recombinase activity results in allelic exclusion at the Ig heavy-chain locus.3 The resulting pre-B cells then can rearrange their light chain genes, although some pro-B cells have been shown to be able to rearrange their light chain genes before their heavy-chain genes.4 Expression of the light chain genes and assembly with the heavy chains leads to the formation of complete Ig molecules that are expressed on the cell surface. To ensure antibody monospecificity, it is generally believed that further light chain rearrangement is shut off as soon as a functional light chain gene rearrangement is generated, resulting in expression of the complete IgM on the cell surface of immature B cells.2,5,6 However, the mechanism for allelic exclusion at the light chain locus has not been clearly elucidated.3 Continuous rearrangement of the light chain genes has been reported in some cell surface Ig positive (sIg+) B-cell lines7-10 and in mature B cells in vivo,11,12 but not in Ig-secreting plasma cells. We have shown that long-term culture of a human B-cell hybridoma line with concanavalin A (Con A) induce at a high frequency production of various novel λ light chains, which replaced the original light chain.13,14 This differential light chain expression led to an alteration of the original antigen binding ability. We call this process light chain shifting.14 To investigate the mechanism of how this alteration occurs in antibody-secreting cells, we established a plasma cell clone (termed HTD8). Clone HTD8 had undergone secondary rearrangement after losing its ability to express the originally rearranged λ chain at a high frequency. To explain how the loss of original light chain expression occurs, the receptor editing model has been proposed. It has been demonstrated that the V genes coding for an autoreactive light chain can be replaced with V genes coding for a functional nonautoreactive light chain in mice expressing a transgenic sIg with antiself specificity.15 16 In this model, rearrangement of the new λ light chain occurs on the originally rearranged allele, which results in the deletion of the original light chain. In contrast, we see in our plasma cells that loss of the original chain production not only precedes expression of the new λ light chain, but is also independent of the new λ light chain gene rearrangement.
Components of the recombinational machinery necessary for Ig rearrangement include the recombination activating genes, 1 and 2 (RAG-1 and RAG-2).17 Mature B cells, in which RAG-1 and RAG-2 expression is downregulated,18 have been shown to undergo no further Ig gene rearrangement. However, recently RAG-1 and RAG-2 proteins have been shown to be reexpressed in mature B cells that have been stimulated in vitro with lipopolysaccharide and interleukin-4 (IL-4) and also in germinal center B cells in immunized mice. This indicates that the control of RAGs expression has yet to be fully elucidated.19-22 In striking contrast to antibody producing cells in the plasma phenotype stage, the RAG genes are expressed in our established plasma cell line. B cells leaving the bone marrow have been hypothesized to have fixed sIg that can only be altered by somatic hypermutation. We describe a possible mechanism by which expression of the original light chain is replaced with a new light chain in cells at the plasma stage of B-cell development.
MATERIALS AND METHODS
Cells.
The human hybridoma HB4C5, which secretes antibody specific to human histone H2B, was generated by fusing a B lymphocyte with the IgM secreting human plasma line NAT-30.23 Con A–resistant clones derived from HB4C5 cells were isolated as previously described.24 These cells were cultured in ERDF medium (Kyokuto Pharmacy, Tokyo, Japan) containing 5% fetal calf serum (Whittaker Bioproducts, Walkersville, MD). The human mature B-cell line, Raji, and the human T-cell line, Molt-4, were cultured as recommended by American Type Culture Collection (Manassas, VA). To identify HTD8 cells, Con A–resistant variants were seeded in 96-well culture plates, and the supernatant of each well 3 weeks after subcloning was examined for the expression of λ light chains.
Cell stimulation.
Cells were cultured in a 5% CO2 atmosphere at 37°C in the presence of 50 ng/mL of phorbol 12-myristrate 13-acetate (PMA).25 The cells and the culture supernatants were then obtained and assayed for the expression of λ light chain and RAG proteins. PMA was purchased from Sigma (St Louis, MO).
Flow cytometric analysis.
Cultured cells were washed in phosphate-buffered saline (PBS) and stained on ice with the appropriate monoclonal antibodies for 30 minutes in 100 μL of PBS and the relative fluorescence intensity was detected by flow cytometry (FACS Calibur; Becton Dickinson, Mountain View, CA). Antibodies used were mouse anti-CD20 (Southern Biotechnology, Birmingham, AL), mouse anti-CD38 (Immunotech, Cedex, France), anti-mouse IgG1 conjugated to fluorescein isothiocyanate (FITC) (Southern Biotechnology), and goat anti-human Ig heavy and light chains conjugated to FITC (Biosource, Camarillo, CA). Irreverent FITC-conjugated mouse or goat IgG was used as a negative control.
Isolation of sIg− cells.
To isolate sIg− cells, cells were incubated in a culture dish (10 cm in diameter; Becton Dickinson) coated with 100 μg/mL of anti-human IgM antibody (Biosource). The cell population left unattached to the dish was harvested, and these cells were cultured in ERDF medium containing 5% fetal calf serum.
Western blot analysis.
Cell extracts were prepared by one cycle of freeze-thaw lysis (1 × 107 cells) in PBS containing 1 mmol/L phenylmethyl sulfonyl fluoride (PMSF) as described.26 The resulting lysate was spun for 5 minutes at 4°C, and supernatant was retained as whole-cell extract. Samples from supernatant cell extract were boiled for 5 minutes in sample buffer, electrophoresed on sodium dodecyl sulfate (SDS)-polyacrylamide gels (10%) and transferred to a nitrocellulose membrane.27 The blotted RAG proteins were detected by immunoblotting with 1 μg/mL of anti-human RAG-1 or RAG-2 antibodies (Pharmingen, San Diego, CA). Immunoreactive proteins were detected by incubation with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Sigma) at 1:2,000 dilution for 1 hour at room temperature; immobilized HRP was visualized using an enhanced chemiluminescence assay (Amersham, Buckinghamshire, UK). To detect the human λ light chain, the blotted membrane was incubated in a 1:500 dilution of HRP-conjugated goat anti-human λ light chain (Biosource) for 1 hour. The membranes were washed in PBS containing 0.05% Tween 20 and developed with 1.6 mmol/L 4-chloro-1-naphthol, 0.01% H2O2 in PBS with 20% methanol.
Nucleotide sequence analysis.
Total RNA prepared using the TRIzol reagent (GIBCO-BRL, Gaithersburg, MD) was subjected to cDNA synthesis and the resultant cDNA served as a template for polymerase chain reaction (PCR) amplification using specific primers. cDNA was synthesized from total RNA using a kit (Amersham) according to the instructions provided by the manufacturer. PCR was done in 50-μL reaction volumes containing 10 mmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, 1.5 mmol/L MgCl2, 20 μg/mL gelatin, 1 μmol/L of each primer, 0.2 mmol/L of each deoxyribonucleotide triphosphate, and 1 U of Taq polymerase (Takara, Osaka, Japan). A single PCR cycle consisted of an incubation period of 0.7 minutes at 94°C, 1 minute at 54°C, and 1 minute at 72°C. The PCR primers used were as follows. The variable regions of the λ light chain genes28: P-LLF, 5′-ACTAGAATTCATG(AG)- CCTG(CG)(AT)C(CT)CCTCTC(CT)T(CT)CT(CG)(AT)(CT)CC-3′ and P-LLR, 5′-ACTAGCGGCCGCCTATGAACATTC(CT)G(CT)AGGGGC-3′; EBNA-1 gene: EBNA1F, 5′-CCGAAATTTGAGAACATTGC-3′ and EBNA1R, 5′-TCACTCCTGCCCTTCCTCAC-3′; LMP-1 gene: LMP1F, 5′-ACTAAGCTTATGGAACGCGACCTTGAGAG-3′ and LMP1R, 5′-ACTAGCGGCCGCTTAGTCATAGTAGCTTAGC-3′ and glyceraldehyde-3-phosphate dehydrogenase gene (G3PDH): P-G3PDF, 5′-CATCACCATCTTCCAGGAGC-3′ and P-G3PDR, 5′-GGATGATGTTCTGGAGAGCC-3′. The nucleotide sequences of V region genes were determined after subcloning into pGEM-T vector (Promega, Madison, WI), using the DyeDeoxy terminator kit (Perkin Elmer-Cetus, Norwalk, CT) on the DNA sequencer ABI 310 (Perkin Elmer-Cetus) according to the manufacturer’s instruction.
DNA analysis.
Genomic DNA was prepared as previously described.29 A total of 1 ng of genomic DNA was used for the PCR assay. The PCR was used to detect the VλC5-to-Jλ coding joint for the original λ light chain λC5 or the VλJλ coding joints for the new λ light chains (D8-1, D8-2, D8-3, and D8-4). The following primers: VλC5P (λC5-CDR1 region-specific) 5′-AACAGCTCCAACATTGGGAA-3′ (sense), Vλ4P (Vλ4-CDR1 region-specific) 5′-CTCTGAGCAGTGGGCACAGC-3′ (sense), Vλ6P (Vλ6-CDR1 region-specific) 5′-AGTTGCAGCATTTTCAGCAAC-3′ (sense), and JλP (conserved 3′ of Jλ) 5′-TCAGTTTAGTCCCTCCGCC-3′ (antisense) were used. PCR reactions were performed as described above for reverse transcriptase (RT)-PCR. Aliquots were withdrawn at cycles 20 and 25 for separate analysis to ensure that amplification was a linear range. Reaction products were run on 0.8% agarose gels, followed by Southern blot hybridization. The PCR products were transferred to nylon transfer membranes (Hybond-N+, Amersham) and hybridized with32P-radiolabeled probes. cDNA fragments coding for each λ light chain, which were used for analyzing the variable region sequence, was digested from their respective vectors with restriction enzymes. The resulting fragment each containing the specific V gene was labeled using Redi-Prime (Amersham) and α32P-deoxycytidine triphosphate (Amersham) for use as probes for the detection of the corresponding VλJλ coding joints. To check for possible Taq polymerase errors, all PCR products were sequenced and compared with the previously defined sequences.
Northern blots.
Total RNA, 10 μg per lane, was subjected to electrophoresis through a 1% agarose gel and transferred onto nylon membranes and hybridized with the 32P-radiolabeled probes. The membrane was air-dried and autoradiographed on x-ray film. For original light chain, μ heavy chain, and β-actin transcripts, the appropriate PCR fragments (VλC5-Jλ, μNAT-30, and β-actin) were used as probe.14
RESULTS
Isolation of a plasma clone expressing differential λ light chains.
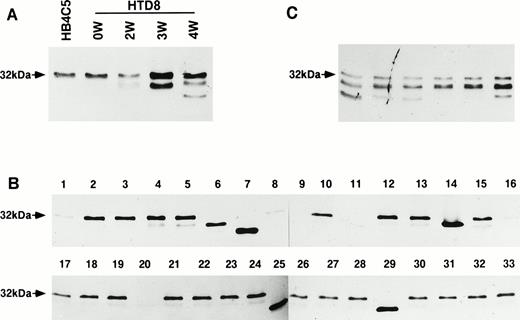
To obtain glycosylation variants of Ig, we previously isolated glycosylation mutants of Ig-secreting cells that were resistant to the cytotoxic effect of Con A from the human hybridoma line, HB4C5. HB4C5 cells secrete IgM reactive to the human histone, H2B.13,30The expressed λ light chain is relatively large (32 kD), the size of which is due to a carbohydrate chain linked to its variable region.31 Initially, some of the variant clones were able to secrete λ light chains of different apparent molecular sizes. The various antibodies secreted from these clonal mutants showed either a loss of reactivity or varied cross-reactivity with different antigens.13 When we subcloned these Con A–resistant variant cells, we isolated a single-cell variant clone (HTD8). As with the parental HB4C5 line, HTD8 initially secreted the original λ light chain species. However, as the clone was allowed to expand, other various λ light chain species were detected. At week 3, a 28-kD species and at week 4 an even smaller species were found (Fig 1A). To determine whether a single-cell clone can secrete more than one λ light chain, we further subcloned the expanded HTD8 cells using the limiting dilution method in 96-well culture plates and allowed the cloned cells to undergo only 12 cell divisions. Almost all of the subclones (22 of 33 randomly chosen clones) secreted again mainly the 32-kD species and a detectable amount of the 28-kD λ chain. Secretion of the various new λ light chains, which are determined by their difference in size from the original, was observed in five wells (Fig 1B, lanes 6, 7, 14, 25, and 29). A complete lack of λ light chain expression, or at least reduced production to undetectable amounts, was shown in six wells (Fig1B, lanes 1, 8, 9, 11, 16, and 20). Another six randomly chosen wells from the 96-well culture plate were cultured for a further 4 weeks, and as in Fig 1A, week 4, almost all of these subclones again demonstrated that the 32-kD λ light chain species was the dominant species and the 28-kD chain was the minor species produced (Fig 1C). From Fig 1B, four of the five novel light chain producers were isolated and named D8-1, D8-2, D8-3, and D8-4. In addition, a clone, which did not secrete any λ light chains (D8-11), was also isolated. The original λ chain was not detected in the cytoplasmic fractions of any of these novel λ chain-producing clones (data not shown). Although these four clones were cultured for over 6 months, they secreted the novel λ chain stably, and the original 32-kD λ chain was not detected at all. These results indicate that HTD8 subpopulations obtained by expansion are able to secrete either the original or a new λ light chain or none at all.
Immunoblot analysis of the expression of differential λ light chains by HTD8 cells. The pattern of λ light chain secretion was assayed by immunoblotting. (A) The parental HB4C5 cells are shown to secrete only the 32-kD λ light chain. During the continuous culture of HTD8 cells, culture supernatants taken from 0, 2, 3, and 4 weeks after the initial cloning were assessed using an anti-human λ light chain antibody. (B) The HTD8 cells were further cloned by the limiting dilution method in 96-well culture plates, the culture supernatants from 33 randomly chosen wells were subjected to SDS-polyacrylamide gel electrophoresis (PAGE), transferred to a nitrocellulose membrane, and the λ light chains were detected with an anti-human λ light chain antibody/HRP conjugate. (C) Cloned HTD8 cells were cultured in 96-well plates for 4 more weeks, and six randomly selected wells were assayed for λ chain production.
Immunoblot analysis of the expression of differential λ light chains by HTD8 cells. The pattern of λ light chain secretion was assayed by immunoblotting. (A) The parental HB4C5 cells are shown to secrete only the 32-kD λ light chain. During the continuous culture of HTD8 cells, culture supernatants taken from 0, 2, 3, and 4 weeks after the initial cloning were assessed using an anti-human λ light chain antibody. (B) The HTD8 cells were further cloned by the limiting dilution method in 96-well culture plates, the culture supernatants from 33 randomly chosen wells were subjected to SDS-polyacrylamide gel electrophoresis (PAGE), transferred to a nitrocellulose membrane, and the λ light chains were detected with an anti-human λ light chain antibody/HRP conjugate. (C) Cloned HTD8 cells were cultured in 96-well plates for 4 more weeks, and six randomly selected wells were assayed for λ chain production.
Vλ gene usage in the new light chain gene expression.
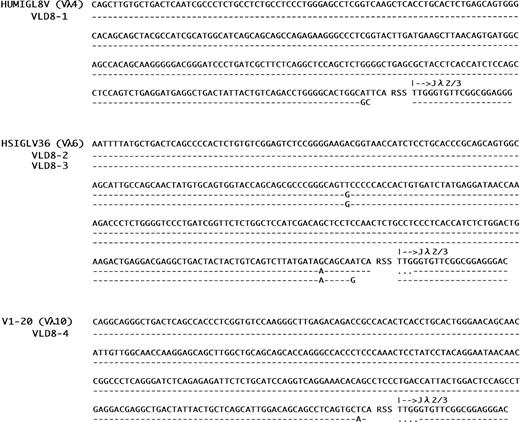
From the subclones secreting only the novel λ chain (D8-1, D8-2, D8-3, and D8-4), cDNA was synthesized and analyzed for V gene sequencing (Fig 2). The nucleotide sequences of V regions of the λ light chains produced by the four subclones are different from the original. The germ-line V gene for the original λ light chain (λC5) has been shown to belong to the Vλ1 family,31 as grouped by Chuchana et al.32 The germ-line Vλ families used in the light chain genes from the four subclones were shown to be Vλ4, Vλ6, and Vλ10. Strikingly, these Vλ gene families are the least frequently used.33 In addition to the four genes, another seven new light chains sequenced so far also used the Vλ genes from these seldom used Vλ gene families (data not shown). These results indicate that the usage of Vλ genes in HTD8 cells is biased towards these minor families. Although D8-2 and D8-3 cells express genes encoded by the same germ-line Vλ gene (HSIGLV36), they apparently arose from independent recombination events, as judged by having distinct V-J joining sequences. This was confirmed by genomic DNA analysis using Vλfamily-specific probes (see Fig 3). We found no significant differences in the growth rates of these new light chain-expressing subclones, and doubling times of all subclones were comparable to the original cells (data not shown). Therefore, we conclude that the bias in Vλ gene usage is not the result of a differential growth rate.
Nucleotide sequences of variable regions of new λ light chains expressed in HTD8 subclones. Nucleotide sequences of the variable regions of the light chains expressed in HTD8 subclones (D8-1, D8-2, D8-3, and D8-4) are shown in a 5′ to 3′ direction. VLD8-1, VLD8-2, VLD8-3, and VLD8-4 represents the λ light chain genes expressed by the subclones of HTD8. Sequences are compared with the corresponding germ-line Vλ (HUMIGL8V, HSIGLV36, and V1-20) and Jλ2/3 sequences. Designations in brackets indicate their corresponding Vλ gene family. Dashes sequence identity with the germline genes. Dots indicate where gaps have been introduced to maximize homology.
Nucleotide sequences of variable regions of new λ light chains expressed in HTD8 subclones. Nucleotide sequences of the variable regions of the light chains expressed in HTD8 subclones (D8-1, D8-2, D8-3, and D8-4) are shown in a 5′ to 3′ direction. VLD8-1, VLD8-2, VLD8-3, and VLD8-4 represents the λ light chain genes expressed by the subclones of HTD8. Sequences are compared with the corresponding germ-line Vλ (HUMIGL8V, HSIGLV36, and V1-20) and Jλ2/3 sequences. Designations in brackets indicate their corresponding Vλ gene family. Dashes sequence identity with the germline genes. Dots indicate where gaps have been introduced to maximize homology.
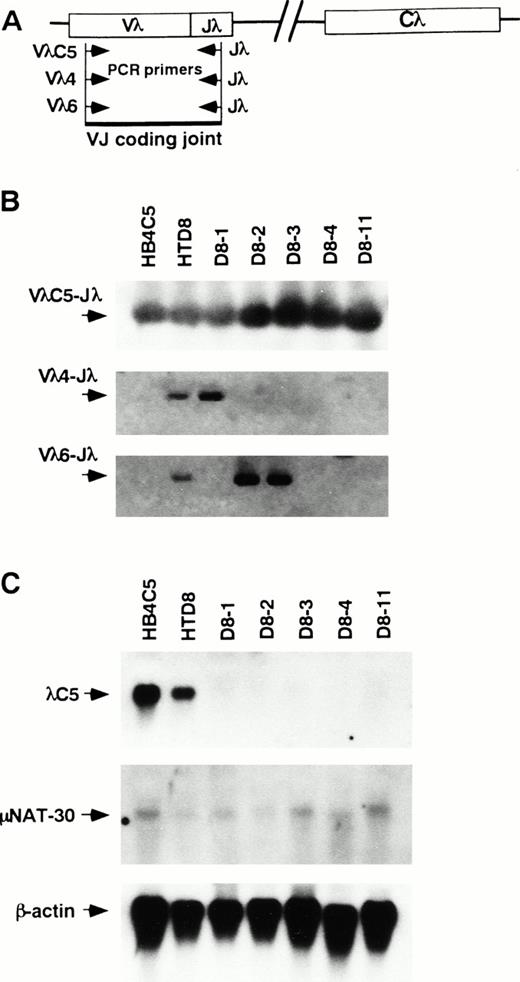
Presence of the original Vλ to Jλ coding joint and the new Vλ to Jλ rearrangement and quantification of the original λ chain transcript. PCRs were performed with primers that amplify the indicated VJ coding joint formation in the genomic DNA from HB4C5, HTD8, and its subclones. (A) A diagram of the PCR primers used to detect the VJ coding joint. The direction of primers used are indicated with arrowheads. (B) The PCR-amplified products were run on agarose gels and assayed for hybridization to a VλC5, Vλ4, and Vλ6 specific probes by Southern blotting. Bands corresponding to VJ coding joint for the original light chain λC5, Vλ4 joined to Jλ, and Vλ6 jointed to Jλ are shown (C) Expression of the original λ light chain and the μ heavy-chain mRNAs were examined by Northern blotting. Cells used as a source of mRNA are indicated above each lane.
Presence of the original Vλ to Jλ coding joint and the new Vλ to Jλ rearrangement and quantification of the original λ chain transcript. PCRs were performed with primers that amplify the indicated VJ coding joint formation in the genomic DNA from HB4C5, HTD8, and its subclones. (A) A diagram of the PCR primers used to detect the VJ coding joint. The direction of primers used are indicated with arrowheads. (B) The PCR-amplified products were run on agarose gels and assayed for hybridization to a VλC5, Vλ4, and Vλ6 specific probes by Southern blotting. Bands corresponding to VJ coding joint for the original light chain λC5, Vλ4 joined to Jλ, and Vλ6 jointed to Jλ are shown (C) Expression of the original λ light chain and the μ heavy-chain mRNAs were examined by Northern blotting. Cells used as a source of mRNA are indicated above each lane.
Rearrangement of the new λ chain on a previously excluded allele and reduction of the original λ chain transcript are found in the HTD8 subclones.
To determine the genetic event responsible for expression of a differential λ light chain in Ig-secreting cells, we analyzed λ light chain gene rearrangement in HB4C5 cells, HTD8 cells, and the four subclones secreting only the novel λ chain (D8-1, D8-2, D8-3, and D8-4). The four clones were resubcloned three times by limiting dilution before use. If rearrangement for the expression of a new λ light chain occurs on the previously rearranged λ locus, the original VλC5 to Jλ coding joint should be replaced by the novel rearrangement and thus be deleted from the chromosome. If rearrangement for the new light chain occurs on a previously excluded allele, both the original and the new VλJλcoding joint should be detected. PCR was performed on the genomic DNA from each cell line using primers appropriate to the CDR1 of λC5 and the Jλ regions to detect the original VλC5-to-Jλ coding joint formation. The combination of these primers provides the ability to detect VJ rearrangement necessary to produce λC5 in the Igλ loci. Genomic DNA was subjected to PCR and the reaction products were analyzed by Southern blot hybridization. A diagram of the PCR assay strategy is shown in Fig 3A. In DNA from all of the subclones examined, the VJ coding joint for λC5 was detected (Fig 3B). Furthermore, authenticity of all of the PCR products was confirmed by determining the partial sequence of these products (data not shown). The presence of the VJ coding joint for λC5 suggest that the original rearranged locus is retained in the chromosome thus implying that the rearrangement for a new light chain occurs on another allele. To corroborate this view, we examined for the presence of novel VλJλcoding joint formation specific for the new light chain using primers specific to the Vλ4 and Vλ6 families (Fig3B). PCR products hybridized with Vλ4 and Vλ6-specific probes were detected slightly in the DNA from HTD8 cells, suggesting that the new VJ rearrangements occurred during the cell population expansion. This result coincides with the fact that not only the original light chain, but also a smaller amount of the new light chains, was found in the supernatant of the HTD8 bulk cell population (Fig 1A). Because only rearranged λ light chain genes can be detected by this assay, each subclone is shown to contain both the originally rearranged allele plus another rearranged gene on a previously excluded Igλ allele. These results indicate that the new λ chain expression in HTD8 cells results from rearrangement on a previously excluded allele. The subclones also retain the original rearranged gene, although the original λ protein is not expressed. To address the loss of production of the original λ light chain in these subclones, we examined for the expression of the original light chain transcript by Northern blot analysis (Fig 3C). The original λ chain transcript was seen in all subclones, however the level was much lower when compared with HTD8 cells and the parental HB4C5 cells. In contrast, no difference was seen in the expression level of the μ heavy-chain transcript. The significant reduction of the level of mRNA may be a reason why the original λ chain protein is not detected in the HTD8 subclones, although we cannot rule out other possibilities.
RAG genes expression is necessary, but insufficient for inducing rearrangement of the new λ light chain gene in HTD8 cells.
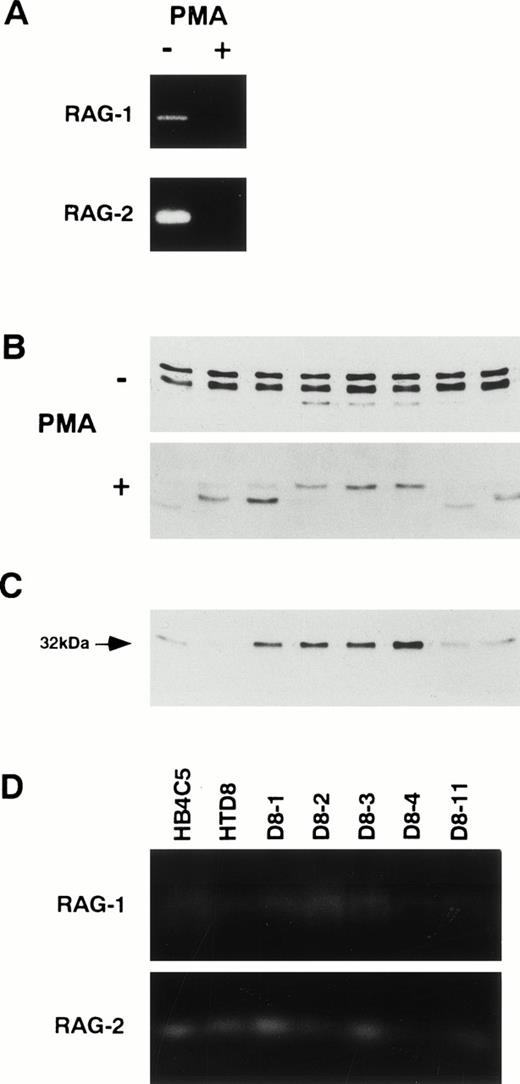
RAG-1 and RAG-2 gene products have been shown to be essential for initiating Ig gene rearrangement, and RAG expression is terminated after the formation of sIg, which may be a factor in maintaining allelic exclusion.18 To test for a relationship between the expression of the new λ light chain in the HTD8 cells and RAG expression, we examined for RAG expression in HTD8 cells (Fig 4A). We found that both proteins were expressed in the HTD8 cells. To test whether the level of RAG expression correlates with the ongoing new λ light chain expression in HTD8 cells, we treated HTD8 cells with PMA, which has been shown to decrease the RAG expression (Fig 4A).25 The level of RAG expression was significantly reduced as compared with the control. To assess the effect of the reduction of the RAG expression on the new light chain expression, HTD8 cells were cultured in 96-well culture plates in the presence of PMA for 3 weeks, and expression of the new λ light chain from eight randomly chosen wells was compared (Fig 4B). Although the new λ light chain was detected in some of the PMA-treated clones, other clones producing only the 32-kD original λ light chain were also found. When the PMA-treated clones expressing only the 32-kD light chain were further maintained in PMA-free medium for 2 weeks, only the 32-kD λ light chain continued to be expressed (Fig 4C). The failure of these cells’ ability to express the new λ light chain when treated with PMA parallels the absence of RAG expression, suggesting that the inhibition of the new λ light chain induction is a direct effect of the termination of RAG expression.
Correlation between RAG proteins expression and new light chain expression. (A) Effect of PMA that has been shown to moderate RAG expression was examined. HTD8 cells were treated with (+) or without (−) 50 ng/mL PMA. Cells were lysed in SDS lysis buffer; 50 μg/mL protein from each sample was fractionated by 10% SDS-PAGE, and RAG-1 and RAG-2 proteins were detected by immunoblotting. (B) Lambda light chain expression pattern in HTD8 cells treated with PMA. The pattern of λ light chain secretion from HTD8 cells cultured with (+) or without (−) PMA in a 96-well culture plate for 4 weeks and eight wells were randomly chosen, and the culture supernatants were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and λ light chains were detected by immunoblotting. (C) The 32-kD original λ chain producing subclones generated from the PMA treatment was cloned via the limiting dilution method, further cultured for 2 weeks in a 96-well plate, and eight randomly chosen wells were assayed for λ light chain production by immunoblotting. (D) The expression of RAG-1 and RAG-2 proteins in the cells indicated above each lane was assessed by immunoblotting.
Correlation between RAG proteins expression and new light chain expression. (A) Effect of PMA that has been shown to moderate RAG expression was examined. HTD8 cells were treated with (+) or without (−) 50 ng/mL PMA. Cells were lysed in SDS lysis buffer; 50 μg/mL protein from each sample was fractionated by 10% SDS-PAGE, and RAG-1 and RAG-2 proteins were detected by immunoblotting. (B) Lambda light chain expression pattern in HTD8 cells treated with PMA. The pattern of λ light chain secretion from HTD8 cells cultured with (+) or without (−) PMA in a 96-well culture plate for 4 weeks and eight wells were randomly chosen, and the culture supernatants were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and λ light chains were detected by immunoblotting. (C) The 32-kD original λ chain producing subclones generated from the PMA treatment was cloned via the limiting dilution method, further cultured for 2 weeks in a 96-well plate, and eight randomly chosen wells were assayed for λ light chain production by immunoblotting. (D) The expression of RAG-1 and RAG-2 proteins in the cells indicated above each lane was assessed by immunoblotting.
It has been shown that there is a strong correlation between the RAG expression levels and Ig gene rearrangement.19 Therefore, we assessed the RAG proteins level in HTD8 and the parental HB4C5 cells (Fig 4D). The expression level for the RAG-1 and RAG-2 produced in HTD8 cells did not significantly differ from HB4C5 cells. In addition, the RAG-1 and RAG-2 were expressed in the HTD8 subclones that produce only the new λ light chain or no light chain (Fig 4D). The continuous expression of RAG in the subclones indicates that new rearrangement and the following expression of new λ light chains does not terminate RAG expression in HTD8 cells. We previously showed that enhancement of RAG expression in HB4C5 cells treated with dibutyryl cyclic adenosine monophosphate (dibutyryl cAMP) did not induce light chain shifting.14 Although the subclones of HTD8 cells (D8-1, D8-2, D8-3, and D8-4) have been maintained for over 1 month in cell culture medium, other new VJ rearrangements did not occur (Fig 3B). These results suggest that constitutive and enhanced expression of the RAG genes is insufficient to rearrange new λ light chain genes.
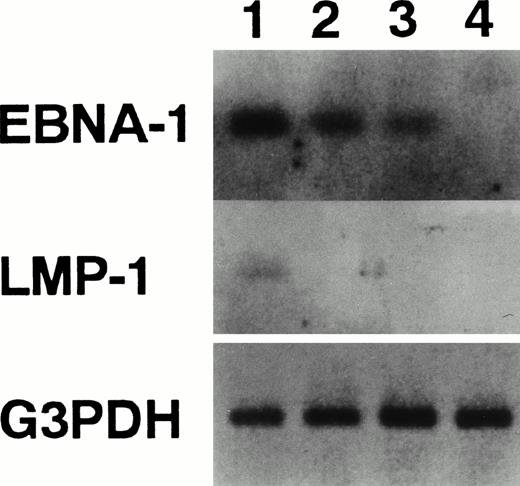
With regard to RAG expression in human mature B cells, evidence has recently been presented, which shows that expression of the Epstein-Barr virus nuclear antigen 1 (EBNA-1) protein was sufficient to induce both RAG genes.34 Conversely, expression of Epstein-Barr virus latent membrane protein (LMP)-1 downregulates RAG genes expression.22 To confirm these findings, we performed RT-PCR to detect EBNA-1 and LMP-1 RNAs (Fig5). Human Burkitt’s lymphoma line, Raji, which has been shown to lack RAG expression, expressed LMP-1 as shown previously.22 HTD8 and HB4C5 cells did not express LMP-1, but the EBNA-1 transcript was detected in both cell lines. Both genes were not detected in the human T-cell line, Molt-4. These results validate findings stating that the expression of EBNA-1 and LMP-1 are associated with RAG genes expression in human mature B cells.
Detection of mRNA for EBNA-1 and LMP-1. The expression of EBNA-1, LMP-1, and G3PDH mRNA was examined by RT-PCR. Lane 1, Raji cells; lane 2, HB4C5 cells; lane 3, HTD8 cells; and lane 4, Molt-4 cells. The PCR-amplified products were run on agarose gels and assayed for hybridization to specific internal probes (EBNA-1 probe: 5′-TGACGGAGATGAAGGAGGTG-3′; LMP-1 probe: 5′-TTGTGCTGTTCATCTTTGGC-3′) by Southern blotting.
Detection of mRNA for EBNA-1 and LMP-1. The expression of EBNA-1, LMP-1, and G3PDH mRNA was examined by RT-PCR. Lane 1, Raji cells; lane 2, HB4C5 cells; lane 3, HTD8 cells; and lane 4, Molt-4 cells. The PCR-amplified products were run on agarose gels and assayed for hybridization to specific internal probes (EBNA-1 probe: 5′-TGACGGAGATGAAGGAGGTG-3′; LMP-1 probe: 5′-TTGTGCTGTTCATCTTTGGC-3′) by Southern blotting.
Loss of sIg expression occurs at a high frequency in HTD8 cells.
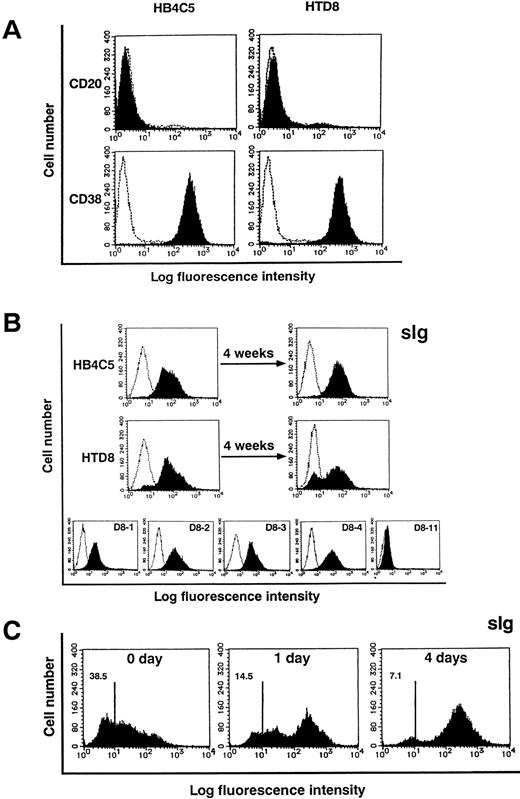
We have previously shown that HB4C5 cells display characteristics common to B cells in the plasma cell state with regard to Ig secretion and the expression pattern of surface antigens.14 HTD8 cells and all of the subclones, except D8-11 cells, secrete IgM. To elucidate the difference between HTD8 and the parental HB4C5 cells, we examined for the expression of the antigens CD20 and CD38. The expression pattern of these antigens in human plasma cells was shown to be CD20−CD38+.35Both of our cell lines are also CD20−CD38+, indicating that the HTD8 cells are in the same plasma state as wild-type plasma cells (Fig 6A). As determined by surface staining with anti-human Ig heavy and light chain antibodies, HB4C5 cells, as well as all subclones of HTD8, except for the subclone D8-11, did not contain a notable sIg− subpopulation (Fig 6B). An sIg− subpopulation was observed, albeit barely, in the HTD8 bulk population and clearly increased (≈30%) after a further 4 weeks of cell growth (Fig 6B). This result turned out to be the most significant difference seen between HB4C5 and HTD8 cells, because after 4 weeks of culture, the HB4C5 sIg−subpopulation did not increase. This is consistent with previous results showing that Con A stimulation is able to induce the appearance of an sIg− subpopulation from the light chain shifting-inducible cell line, HB4C5.14 To examine the process for the loss of sIg, we isolated the sIg−subpopulation from the HTD8 bulk population using anti-human μ light chain antibody (Fig 6C). The sIg+ population clearly increased from this isolated sIg− HTD8 population after 4 days in culture. This demonstrates that in HTD8 cells, inactivation of the original λ light chain, which manifest as the sIg− state, actively occurs before expression of a new light chain. We also found a few λ chain−clones (see Fig 1B). The subclone, D8-11, does not contain an sIg+ phenotype, indicating that the D8-11 line was probably isolated from the sIg− subpopulation. We examined the expression of a new λ light chain gene in this subclone by RT-PCR using Vλ gene-specific primers followed by DNA sequence analysis. It appears that the D8-11 clone expresses a transcript from the new λ light chain gene, but the gene contains a frame-shift mutation at the VJ junction. This suggests that the D8-11 subclone cannot produce a new λ light chain protein due to a nonproductive rearrangement (data not shown). Therefore, subclones that fail to produce viable rearrangements may also be included in the sIg− population. These results taken together suggest that the process of sIg+ to sIg−expression is initiated when the cell loses the ability to produce the original λ light chain and is sustained when a nonproductive new rearrangement occurs, which renders the cells incompetent to express a new λ chain.
Flow cytometric analysis of CD20, CD38, and surface Ig expression. HB4C5 cells, HTD8 bulk population, and the new light chain expressing HTD8 subclones (D8-1, D8-2, D8-3, D8-4, and D8-11) were stained with FITC-conjugated anti-human CD20, CD38, or Ig antibodies and analyzed by flow cytometry (shaded histogram). The negative control was established using an irrelevant FITC-conjugated antibody of the same isotype (clear histogram). The fluorescence intensity is shown on the x-axis and the cell number on the y-axis. HB4C5 and HTD8 cells were examined for CD20 and CD38 expression (A). Presence of sIg was assessed for bulk population HB4C5 and HTD8 cells, the same cells after a 4-week culture, as well as for the HTD8 subclones (B). After isolating sIg− HTD8 cells, these cells were examined for sIg expression at day 0, 1, and 4 of culture using FITC-conjugated anti-human μ antibody (C). Numbers in the upper left corners indicate the percentage of cells that are stained negative.
Flow cytometric analysis of CD20, CD38, and surface Ig expression. HB4C5 cells, HTD8 bulk population, and the new light chain expressing HTD8 subclones (D8-1, D8-2, D8-3, D8-4, and D8-11) were stained with FITC-conjugated anti-human CD20, CD38, or Ig antibodies and analyzed by flow cytometry (shaded histogram). The negative control was established using an irrelevant FITC-conjugated antibody of the same isotype (clear histogram). The fluorescence intensity is shown on the x-axis and the cell number on the y-axis. HB4C5 and HTD8 cells were examined for CD20 and CD38 expression (A). Presence of sIg was assessed for bulk population HB4C5 and HTD8 cells, the same cells after a 4-week culture, as well as for the HTD8 subclones (B). After isolating sIg− HTD8 cells, these cells were examined for sIg expression at day 0, 1, and 4 of culture using FITC-conjugated anti-human μ antibody (C). Numbers in the upper left corners indicate the percentage of cells that are stained negative.
DISCUSSION
In this study, we showed a novel process for regulating the λ light chain expression in human plasma cells. Expression of a complete functional surface Ig complex is thought to signal the overall inactivation of Ig rearrangement.6,36 This process has been shown to be completed during B-cell development, and cells at the plasma state are thought to have lost the ability to undergo Ig rearrangement.37 The striking features of the plasma clone, HTD8, shown here are: (1) production of a new λ light chain resulting from a rearrangement on another previously excluded allele, and (2) active loss of expression of the previously rearranged λ light chain gene. Although heavy-chain allelic exclusion has been confirmed by the presence of feedback inhibition culminating with the formation of membrane-bound μ chains,38,39 the mechanism of light chain allelic exclusion remains unclear. There are a number of factors that have been shown to be necessary for Ig light chain gene rearrangement.40,41 Components of the recombinational machinery necessary for Ig rearrangement include the RAG-1 and RAG-2 genes. The RAG genes are expressed precisely when B cells rearrange the Ig heavy and light chain loci during B-cell differentiation, but not in plasma cells.17 The mechanisms that underlie the regulation of RAG gene expression are poorly understood. Although for the most part, expression of both of these genes is not seen in either normal or malignant sIg+ B cells, some exceptions to the inverse correlation between sIg expression and RAG transcription have been found.42 Bone marrow immature B cells expressing a transgenic sIg with anti-self specificity increased RAG transcript expression when the cells were cross-linked with autoantigens, which led to a secondary rearrangement.15,17 Ma et al36 showed that sIg+ B-cell lines established from Eμ-N-myc transgenic mice expressed relatively high levels of the RAG genes. We show that both HTD8 cells and the parental HB4C5 cells continuously express these RAG genes. Taken together with our results, we assume that RAG expression and sIg expression or Ig secretion are not exclusive, and that a developmental stage where the two are coexpressed may exist.
With regard to RAG expression in human mature B cells, it has been postulated that loss of sIg expression not only interrupts the signal required to terminate RAG expression, but in fact, triggers the upregulation of RAG expression.42 In contrast, because our HB4C5 and HTD8 cells express the RAG genes continuously without suffering a loss of expression of the original λ chain, we can exclude the upregulation mechanism as an explanation for new rearrangement. The RAG genes are also expressed in HB4C5 cells and the parental NAT-30 cells, both of which are in the plasma cell stage.14 RAG expression in both lines likely reflects a unique feature of the NAT-30 cells. NAT-30 cells are a subclone of the Epstein-Barr virus–bearing B-cell line Namalwa. It has been shown that expression of EBNA-1 and LMP-1 proteins was associated with RAG genes expression in human mature B cells.22,34 The expression pattern of both genes in HTD8 and HB4C5 cells coincided with the findings. EBNA-1 is a multifunctional protein that controls transcription of additional EBV genes and is itself regulated in a complex manner.43 Thus, it is possible that EBNA-1 , or an as yet undetected viral product(s), could alter RAG expression.
Ig rearrangement activity and expression of both RAG genes is known to be strongly linked in vivo and in various cell lines,16,25suggesting a possible link between ongoing RAG expression and the secondary rearrangement in our plasma cells. In fact, we found when the RAG proteins were eliminated by treatment with PMA, the cells could not produce a new λ light chain, suggesting the necessity of the RAG genes for new gene rearrangements. However, the uninterrupted maintenance of a certain level of RAG expression may be insufficient to induce expression of a new λ light chain, as shown by parental HB4C5 cells, which secrete only the original λ light chain and HTD8 cells, which can secrete a new or the original λ light chain. We did not detect any difference in the levels of RAG proteins between these two cell lines, which suggests that new λ chain expression in HTD8 cells is not caused by a greater amount of expression of the RAG genes. In addition, we have found that the increased level of RAG expression caused by dibutyryl cAMP treatment did not generate any new light chain in HB4C5 cells. However, when HB4C5 cells were treated with Con A for a long period of time, secondary gene rearrangement occurred on a previously excluded allele of the Ig λ locus leading to the production of a new λ light chain.14 This indicates that the potential for recombinase activity is present in the HB4C5 cells, and that other regulatory factors are needed for the activation of gene rearrangement.
A critical difference between the HTD8 cells and the parental HB4C5 cells is the expression pattern of sIg. HTD8 cells were able to generate subclones lacking the production of the original λ light chain at a high frequency on cell proliferation, consequently leading to a subpopulation of sIg− cells. Especially noteworthy is the fact that none of the HTD8 subclones produced both the original and new light chains together, although subclones that were light chain negative were found. These results indicate that loss of the original λ light chain precedes expression of a new λ light chain, therefore secondary rearrangement would not account for triggering the loss of original λ chain production. Conversely, this result indicates that loss of the original λ light chain could play a role in providing the induction signal for new rearrangement. HTD8 cells continually express both RAG proteins without losing the expression of sIg. Moreover, expression of the new λ light chain is seen only in HTD8 cells, but not in its parental HB4C5 cells, although both cell lines express RAG proteins at the same level. This pattern is consistent with the result that loss of the production of the original λ light chain is restricted to HTD8 cells, but not in the parental HB4C5 cells. These results suggest that the loss of the original light chain is an intermediate step providing a stimulus for new rearrangement. We speculate that the expression of a new λ light chain may represent a mechanism somewhat analogous to the receptor editing model that has been described for autoantibody regulation.15 16 This receptor editing model proposes that B cells expressing an autoantibody undergoes a change through a secondary rearrangement of its light chain thus becoming a nonautoreactive cell. Compared with our HTD8 cells, significant differences are present. During the editing process, the previously active VJ coding joint is rendered inactive by a nested rearrangement. This new rearrangement results in the deletion of the original coding joint. Thus, in the receptor editing model, loss of the original coding joint is a passive process. In contrast, HTD8 subclones, which do not produce the original λ light chain, nonetheless retain the originally formed VJ coding joint for the original λ chain. This confirms that a new rearrangement occurred on a previously excluded allele. We show that the original λ chain mRNA level is significantly lower in cells producing the new light chain compared with cells producing only the original chain. This shows that the loss of production of the original λ light chain is a result of the low expression level for the original λ transcript, not the elimination of the VJ coding joint formation on the chromosome. In contrast to the low amount of original λ chain produced, the production level of the new λ chain was high. If the significantly reduced level of the original λ chain transcript was caused by a defect in the transcriptional regulatory machinery, production of the new λ chain should be also low. Analysis of the differences between the HTD8 cells and the parental HB4C5 cells will be useful in obtaining insights into the mechanism responsible for the highly frequent loss of rearranged λ light chain production in plasma cells.
We found that V λ genes used in secondary rearrangements in the HTD8 cells were biased towards the Vλ4, Vλ6, and Vλ10 families. It is intriguing that these Vλ genes belong to the least frequently expressed families.33Recently, it has been shown that differences in the promoter of the V gene segment is strongly associated with germline transcriptional activity and the frequency of rearrangement, thus consequently, it may affect the V gene usage in the expressed repertoire.44 This finding suggests that the frequency of Vλ gene usage is fixed by the promoter sequence and is therefore unable to change. Our results suggest that not only structural differences in the promoter region of V gene segments, but also the presence of other regulatory factors, can modulate usage of the Vλ gene, although the mechanism of this bias usage remains unclear. HTD8 cells will be useful in investigating the regulatory factors concerning Vλ gene usage.
Based on these results, we have formulated the following hypothesis for the light chain shifting in our HTD8 cells. First, the expression level of the original λ chain transcript is significantly reduced. A low level of the original λ light chain transcript and/or loss of the original λ light chain protein caused by some impairment may trigger the activation of VJ recombination in another λ locus. In the other systems, extinction of surface IgM expression is thought to induce secondary rearrangement.43 Thus in HTD8 cells, the loss of original light chain protein, which fails to make a functional Ig receptor, might be responsible for directly signaling a locus-specific recombinase activation event. When recombinase-accessible Vλ genes, but not recombinase-accessible heavy-chain V genes, are induced by this signal together with the continuously expressed RAG genes, a new rearrangement occurs in the λ locus. If the secondary rearrangement is successful, a new λ chain protein is produced replacing the original light chain. Still maintaining allelic exclusion, this process confirms production of a single λ light chain and maintains single antibody specificity, although the original specificity might be altered. Furthermore, although expression of the RAG genes is not terminated by production of a new light chain and sIg expression, further rearrangements leading to the production of another new λ chain have not been found. If the expression of a new λ light chain signals a modulation of recombinase activity, this finding may shed light on the recombination event, even though the RAG genes are continuously expressed in the cells.
ACKNOWLEDGMENT
The authors thank Perry Seto for proofreading the manuscript and Kyoko Ueda and Kousuke Morizumi for their excellent technical help.
Supported in part by grants from the Program for Promotion of Basic Research Activities for Innovative Biosciences (to H. T.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hirofumi Tachibana, PhD, Department of Food Science and Technology, Faculty of Agriculture, Kyushu University, 6-10-1, Hakozaki, Higashi-ku, Fukuoka, 812-8581, Japan; e-mail:tatibana@agr.kyushu-u.ac.jp.