Cytokines manifest their function through regulation of gene expression. We searched for immediate-early cytokine responsive genes by the mRNA differential display technique using interleukin-3 (IL-3)–dependent OTT-1 cells, and have isolated a novel cDNA which encodes 210 amino acids and shows 87% amino acid identity to human SNAP-23 (synaptosomal-associated protein of 23 kD). The message for this protein (mouse SNAP-23) was induced in OTT-1 cells by IL-3, granulocyte-macrophage colony-stimulating factor (GM-CSF), and IL-5. The experiment using C-terminal deletion mutants of the common β subunit (βc) of IL-3/GM-CSF/IL-5 receptors showed that expression of SNAP-23 was associated with the Ras-Raf-MAPK pathway, but not with the JAK-STAT pathway. Moreover, SNAP-23 was induced in response to a wide variety of cytokines, including IL-2, IL-3, IL-5, IL-10, stem cell factor, G-CSF, GM-CSF, leukemia inhibitory factor, and erythropoietin. Constitutive expression of SNAP-23 was seen in various tissues, including heart, lung, kidney, liver, spleen, and small intestine. Possible involvement of SNAP-23 in cytokine signal transduction is discussed.
CYTOKINES, including interleukins (ILs) and colony-stimulating factors (CSFs) regulate proliferation, differentiation, and cellular functions of various lineages of immune and hematopoietic cells.1 These cytokines exert their biological functions through specific receptors expressed on their target cells.
Although the cytoplasmic regions of these cytokine receptors lack any intrinsic enzymatic activity such as protein tyrosine kinases (PTK), most of these cytokines rapidly induce tyrosine phosphorylation of cellular proteins, indicating a role for PTKs in their signaling.2-6 Several shared signaling events activated by different cytokines have been identified; most cytokines including IL-2, IL-3, IL-5, IL-6, granulocyte-macrophage CSF (GM-CSF), and erythropoietin (EPO) activate p21Ras and the Raf-MAP kinase cascade that leads to the induction of c-fos and c-jun. In addition to this well-defined pathway, a distinct subfamily of PTKs known as Janus kinases (JAKs) have recently been found to play important roles in cytokine signaling.3,4 This JAK-mediated signaling pathway, originally found in the interferon (IFN) system, is now believed to be shared by various cytokines. Deletion analysis of the common β (βc) subunit of the IL-3, GM-CSF, and IL-5 receptors has established that the distal and the proximal regions are required for activation of the Ras-Raf-MAP kinase cascade and the JAK-STAT pathway, respectively.7
There are several techniques for identification of differentially expressed genes, including differential and subtractive hybridization.8 We have previously identified some differentially expressed genes by the cDNA library subtraction technique.9-11 In this study we attempted to obtain genes induced by IL-3, GM-CSF, and IL-5 using the mRNA differential display technique, and we isolated some novel inducible genes (one of which we show in this reprot is a mouse homologue of the human synaptosomal-associated protein-23 kD [SNAP-23]).
MATERIALS AND METHODS
Cells.
IL-3–dependent murine myelomonocytic cell line OTT-1,12pro-B cell line Ba/F3, mast cell line MC9, and myeloid progenitor cell lines L-G3 and L-GM313 were maintained in RPMI 1640 supplemented with 10% fetal calf serum (FCS) and 2 ng/mL murine IL-3. The murine erythroid cell line SKT614 and myeloid leukemia cell line M1 were cultured in RPMI 1640 medium containing 10% FCS. The IL-2–dependent T-cell line CTLL-2 was cultured in RPMI 1640 medium containing 10% FCS and 1 nmol/L IL-2.
Generation of deletion mutants and transfectants.
Differential display.
mRNA differential display was performed according to the method described by Liang and Pardee,17 with some modification. Briefly, RNA preparations were subjected to a DNase I (GenHunter, Brookline, MA) digestion step to eliminate any remaining genomic DNA, and 0.2 μg of total RNA was used for reverse transcription and polymerase chain reaction (PCR) in a 20-μL reaction volume. The cycling parameters are as follows: 94°C for 30 seconds, 40°C for 2 minutes, 72°C for 30 seconds for 40 cycles, followed by 72°C for 5 minutes. The amplified cDNAs were then separated on a 6% DNA sequencing gel. AmpliTaq DNA polymerase was purchased from Perkin-Elmer Cetus Corp (Foster City, CA). [α-35S]dATP (1,200 Ci/mmol) was obtained from Amersham (Arlington Heights, IL). Differentially induced bands were recovered from dried denaturing polyacrylamide gels and reamplified in a 40-cycle PCR using corresponding primers.
Cloning and sequencing of cDNA probes.
Reamplified cDNA fragments were cloned into the pCRII vector using the TA cloning system from Invitrogen (San Diego, CA). Sequencing of the cloned cDNA fragments with either T7 or SP6 primer was performed by using a Dye Terminator Cycle Sequencing Ready Reaction kit from Perkin-Elmer Cetus Corp.
Isolation of the full-length cDNA.
A partial cDNA fragment of mouse SNAP-23 obtained by differential display was labeled with digoxigenin (DIG) by using a DIG-DNA labeling kit (Boehringer Mannheim, Indianapolis, IN) and used for colony hybridization screening of the cDNA library from OTT-1 cells stimulated with IL-3 according to the manufacturer's instructions. The longest clone (A2-2) was sequenced by using synthetic oligonucleotide primers.
Northern blot analysis.
The cells were factor-depleted for 6 hours in RPMI 1640 medium containing 1% bovine serum albumin (BSA), then stimulated with cytokines at the following concentrations for various periods: murine (m) IL-2, 10 ng/mL; mIL-3, 10 ng/mL; mIL-5, 10 ng/mL; mIL-10, 10 ng/mL; mGM-CSF, 10 ng/mL; human (h) GM-CSF, 5 ng/mL; hG-CSF, 50 ng/mL; hEPO, 5 U/mL; murine stem cell factor (mSCF), 50 ng/mL; murine leukemia inhibitory factor (mLIF), 50 ng/mL. Cytokines except for mIL-2 were purchased from R & D systems (Minneapolis, MN). mIL-2 was kindly provided by DNAX Research Institute (Palo Alto, CA). For Northern blotting, poly(A)+RNA was separated on 1.2% agarose gels containing 2.4% formaldehyde, and transferred to positively charged nylon membranes (Boehringer Mannheim). After fixing RNA onto the membranes under calibrated UV irradiation, the membranes were hybridized with 32P-labeled SNAP-23 probes by using the quick hybridization solution (Stratagene, La Jolla, CA). After 2 hours of hybridization, membranes were washed in 2× SSC (1× SSC = 44.6 mmol/L sodium chloride, 5 mmol/L trisodium citrate, pH 7.0), 0.1% sodium dodecyl sulfate (SDS) solution at 68°C for 15 minutes twice, 0.1× SSC, 0.1% SDS at 68°C for 20 minutes, and exposed to x-ray films for an appropriate period. An EcoRI-digested 0.7-kbp fragment of the clone A2-1 was used as a probe.
RESULTS
Isolation of genes induced by IL-3/GM-CSF/IL-5.
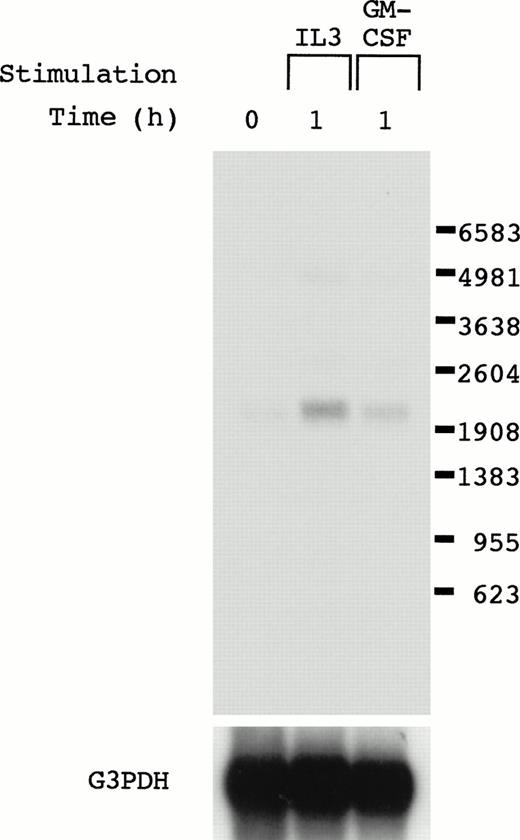
To obtain genes whose induction is mediated by IL-3, GM-CSF, and IL-5, we used a cytokine-dependent cell line, OTT-1,12 which responds to IL-3, GM-CSF, and IL-5, and the mRNA differential display technique. We obtained mRNA from OTT-1 cells before and 1 hour after stimulation with IL-3, GM-CSF, and IL-5, and performed mRNA differential display using them. cDNA bands detected in OTT-1 cells stimulated with IL-3, GM-CSF, and IL-5 were excised from the display gel and cloned into the pCRII TA vector. To eliminate false-positive clones, the induction of the mRNA expression was examined in the OTT-1 cells stimulated with IL-3 and GM-CSF by Northern blot analysis using the EcoRI fragment of these clones as a probe. When we used the clone 2.1 as a probe, 2.2 kb of mRNA was detected in OTT-1 cells stimulated with IL-3 and GM-CSF, but not in them without stimulation (Fig 1), indicating that clone 2.1 mRNA was induced after stimulation with IL-3 and GM-CSF.
Northern blot analysis of the cDNA clone 2.1. OTT-1 cells were deprived of serum and mIL-3 for 6 hours, and then stimulated with mIL-3 and mGM-CSF for 1 hour. Poly(A)+ RNA (4 μg/lane) was hybridized with the cDNA (clone 2.1) and G3PDH probes.
Northern blot analysis of the cDNA clone 2.1. OTT-1 cells were deprived of serum and mIL-3 for 6 hours, and then stimulated with mIL-3 and mGM-CSF for 1 hour. Poly(A)+ RNA (4 μg/lane) was hybridized with the cDNA (clone 2.1) and G3PDH probes.
Structure of protein encoded by the 2.1 cDNA.
The initial 2.1 cDNA clone obtained from the differential display contained about 250 bp of coding region of the mRNA. To obtain a full-length cDNA, we screened 1 × 105 independent clones from a cDNA library of IL-3–stimulated OTT-1 cells with randomly DIG-labeled cDNA for clone 2.1 and obtained 12 positive clones. Four clones among them were sequenced (Fig 2). Three clones, A2-2, A6-1, and A6-2, contained an ATG codon followed by a single open reading frame, and had an in-frame stop codon upstream of the ATG, but clones A6-1 and A6-2 had some deletion at the end of 3′-untranslated region. The sequence of clone A2-1 matched to a fragment between nucleotides 350 to 1890 of the longest clone A2-2. The cDNA sequence of A2-2 encodes a predicted protein of 210 amino acids, and the cystein-rich region between residues 79 and 87, to which palmitate is linked by thioester bonds,18 was found (Fig 2A). A search for the nucleotide data base, DDBJ, showed that clone A2-2 is a mouse homologue of SNAP-2319 and shows 98.6% similarity and 86.7% identity to human SNAP-23 at the amino acid level (Fig 2B).
(A) Nucleotide and predicted protein sequences of mouse SNAP-23. In amino acid sequences, the asterisk indicates the stop codon, and cystein-rich region is underlined. (B) Comparison of the predicted amino acid sequences with mouse and human SNAP-23. Vertical lines indicate amino acid identity.
(A) Nucleotide and predicted protein sequences of mouse SNAP-23. In amino acid sequences, the asterisk indicates the stop codon, and cystein-rich region is underlined. (B) Comparison of the predicted amino acid sequences with mouse and human SNAP-23. Vertical lines indicate amino acid identity.
Induction of the SNAP-23 message by stimulation with IL-3, GM-CSF, and IL-5.
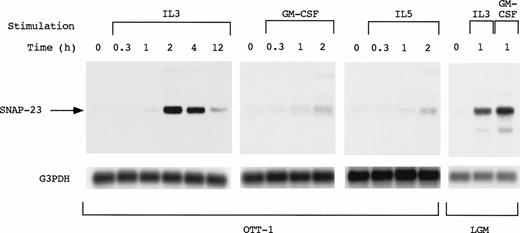
SNAP-23 induction by stimulation with IL-3, GM-CSF, and IL-5 was examined by Northern blotting using OTT-1 cells and L-GM3 cells (Fig 3). In OTT-1 cells, induction of SNAP-23 by IL-3 began 1 hour after stimulation, reached a maximal level at 2 hours, and was detected at 12 hours. SNAP-23 was also induced after stimulation with GM-CSF and IL-5 in the same manner. In L-GM3 cells, which proliferate in response to IL-3 and differentiate in response to GM-CSF, a high level of SNAP-23 induction was observed 1 hour after stimulation with both IL-3 and GM-CSF.
Induction of SNAP-23 message by stimulation with IL-3/GM-CSF/IL-5 in factor-dependent cells. OTT-1 and L-GM3 cells were deprived of serum and mIL-3 for 6 hours, then stimulated with IL-3/GM-CSF/IL-5 for the indicated periods (h). Poly(A)+RNA (2 μg/lane) was separated from the cells and blotted with SNAP-23 and G3PDH probes.
Induction of SNAP-23 message by stimulation with IL-3/GM-CSF/IL-5 in factor-dependent cells. OTT-1 and L-GM3 cells were deprived of serum and mIL-3 for 6 hours, then stimulated with IL-3/GM-CSF/IL-5 for the indicated periods (h). Poly(A)+RNA (2 μg/lane) was separated from the cells and blotted with SNAP-23 and G3PDH probes.
Induction of the SNAP-23 message mediated by the membrane-distal region of the common β (βc) subunit of IL-3/GM-CSF/IL-5 receptor.
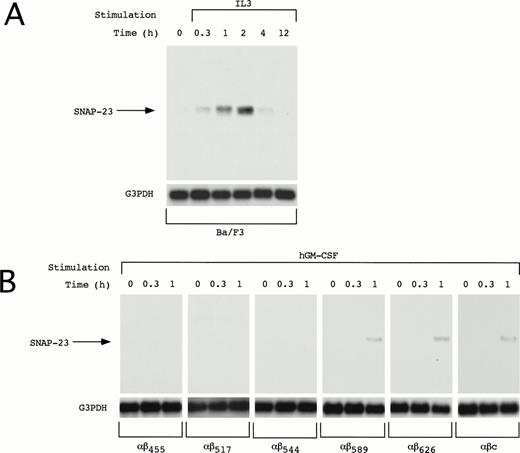
We also examined induction of SNAP-23 in other IL-3–dependent cell line Ba/F3. In Ba/F3 cells, SNAP-23 mRNA was induced by IL-3 at earlier time points than in OTT-1 cells; its induction began 20 minutes after stimulation, reached the maximal level at 1 to 2 hours, and thereafter decreased to the basal level in 12 hours (Fig 4A).
Induction of SNAP-23 in Ba/F3 cells and Ba/F3 transfectants. Factor- and serum-starved Ba/F3 cells and Ba/F3 transfectants were stimulated with mIL-3 and hGM-CSF for the indicated time (h), respectively. Poly(A)+ RNA (1 μg/lane) was extracted from the cells and blotted with SNAP-23 and G3PDH probes.
Induction of SNAP-23 in Ba/F3 cells and Ba/F3 transfectants. Factor- and serum-starved Ba/F3 cells and Ba/F3 transfectants were stimulated with mIL-3 and hGM-CSF for the indicated time (h), respectively. Poly(A)+ RNA (1 μg/lane) was extracted from the cells and blotted with SNAP-23 and G3PDH probes.
The common β (βc) subunit of the IL-3, GM-CSF, and IL-5 receptors is responsible for signal transduction of these cytokines; a membrane-proximal region of the cytoplasmic domain is responsible for activation of the JAK-STAT pathway and the distal region is required for activation of the Ras-Raf-MAP kinase cascade. To address whether the induction of SNAP-23 is mediated by the membrane-proximal or -distal region, deletion mutants of the βc subunit of the IL-3, GM-CSF, and IL-5 receptors were tested for their ability to induce the SNAP-23 mRNA (Fig 4B). We previously generated a series of deletion mutants of the common β subunit of hGM-CSF receptor (R) and established the Ba/F3 stable transfectants expressing the wild-type hGM-CSFR α subunit and a series of mutant β subunits.7,15 16 The induction of SNAP-23 was detected in BaF3/αβ589, BaF3/αβ626, and BaF3/αβc (wild type) 1 hour after stimulation with hGM-CSF, but not in BaF/αβ544, BaF/αβ517, and BaF/αβ415 (Fig 4B). This result shows that the membrane-distal region (544-589), which is required for activation of the Ras-Raf-MAP kinase cascade, is also required for the induction of SNAP-23.
Induction of SNAP-23 message by stimulation with other cytokines inducing proliferation and/or differentiation.
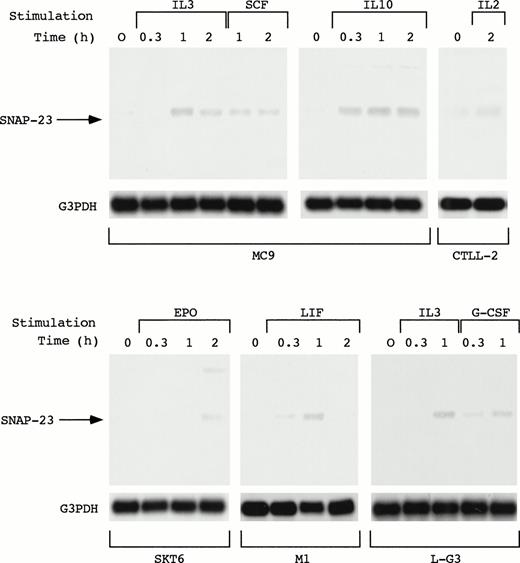
As described above, SNAP-23 was induced by stimulation with IL-3, GM-CSF, and IL-5. We also examined the induction of SNAP-23 by stimulation with SCF and IL-10 in an IL-3–dependent mast cell line MC9, G-CSF in myeloid progenitor cell line L-G3, leukemia inhibitory factor (LIF) in a LIF-responsive myeloid leukemia cell line M1, EPO in erythroid cell line SKT6, and IL-2 in IL-2–dependent T-cell line CTLL-2 (Fig 5). When MC9 was stimulated with IL-3, SCF, and IL-10, which lead to cell proliferation, SNAP-23 was induced in the same manner; induction began 20 minutes after the stimulation and high level of the expression was kept until 2 hours after the stimulation. Furthermore, we also detected the induction of SNAP-23 in CTLL-2 cells stimulated with IL-2.
Induction of SNAP-23 by the stimulation with various cytokines. Factor-dependent cell lines MC9, CTLL-2, and L-G3 were deprived of serum and growth factors, then stimulated with mIL-3, mSCF, mIL-10, mIL-2,or hG-CSF for the indicated time (h). SKT6 cells and M1 cells were stimulated with hEPO and mLIF for the indicated time (h), respectively. Poly(A)+ RNA (1 μg/lane) was separated from the cells and blotted with SNAP-23 and G3PDH probes.
Induction of SNAP-23 by the stimulation with various cytokines. Factor-dependent cell lines MC9, CTLL-2, and L-G3 were deprived of serum and growth factors, then stimulated with mIL-3, mSCF, mIL-10, mIL-2,or hG-CSF for the indicated time (h). SKT6 cells and M1 cells were stimulated with hEPO and mLIF for the indicated time (h), respectively. Poly(A)+ RNA (1 μg/lane) was separated from the cells and blotted with SNAP-23 and G3PDH probes.
When SKT6 cells, which proliferate without EPO and undergo limited erythroid differentiation in response to EPO,14 were stimulated with EPO, SNAP-23 was induced 2 hours after stimulation. In addition, SNAP-23 was induced by stimulation with LIF in M1 cells.
In L-G3 cells, which differentiate into neutrophils in response to G-CSF,13 G-CSF as well as IL-3 induced SNAP-23 1 hour after the stimulation. Thus, SNAP-23 was induced by a wide range of cytokines regardless of biological consequences induced by cytokines.
Expression of SNAP-23 message in tissues.
SNAP-23 expression was examined in various mouse tissues. While SNAP-23 expression requires the stimulation of cytokines in hematopoietic cell lines, a high level of SNAP-23 mRNA was detected without stimulation in various tissues including heart, lung, liver, skeletal muscle, kidney, skin, stomach, and small intestine (Fig 6). The SNAP-23 message was also detected in spleen, smooth muscle, and pancreas at lower intensity. In the brain and the testis, expression of SNAP-23 message was marginal. Such a wide distribution of SNAP-23 suggests important roles of SNAP-23 in various tissues as well.
Distribution of SNAP-23 mRNA in various mouse tissues. Poly(A)+ RNA (2 μg/lane) from various mouse tissues was hybridized with the SNAP-23 probe.
Distribution of SNAP-23 mRNA in various mouse tissues. Poly(A)+ RNA (2 μg/lane) from various mouse tissues was hybridized with the SNAP-23 probe.
DISCUSSION
We previously identified two novel genes, CIS and OSM, induced by various cytokines through the JAK-STAT pathway by using a cDNA library subtraction technique.9-11 In this study we searched for the genes differentially or commonly induced by IL-3, GM-CSF, and IL-5 by using the mRNA differential display technique and obtained several novel genes. Here we describe a gene, SNAP-23, which is commonly induced by IL-3, GM-CSF, and IL-5. SNAP-23 is also induced by several other cytokines, including IL-2, IL-10, EPO, SCF, G-CSF, and LIF. Despite the inducible expression of SNAP-23 by various cytokines in vitro, SNAP-23 is widely expressed in various tissues without any stimulation. The constitutive expression of SNAP-23 message in vivo may be due to various cytokines existing in the body.
SNAP-23 is an isoform of SNAP-25 which is one of the SNARE proteins.19 Docking and fusion of the cytoplasmic vesicles to the cell membrane involves the formation of a trimeric core complex of three proteins, vesicle-associated membrane protein (VAMP), syntaxin, and SNAP-25,20 in transport of proteins along the secretory pathway of eukaryotic cells. Although different isoforms of VAMP and syntaxin have been reported in many cell types,20-24 SNAP-25 has been exclusively detected in neuronal tissues.25 On the other hand, human SNAP-23 is ubiquitously expressed.19 A mouse homolog of SNAP-23 we cloned in this study is widely expressed in most tissues. Thus, it is likely that SNAP-23 is functionally equivalent to SNAP-25 and replaces SNAP-25 in nonneuronal tissues.
Cytokine receptors transmit their signals through multiple signaling pathways. Among them, Ras activation pathway is well characterized.26-28 Ras is activated after formation of Shc-Grb-SOS complex or tyrosine phosphorylation of Vav. Activation of Ras then leads to induction of c-fos and c-jun via activation of Raf and MAPK. Activation of the Ras-Raf-MAPK cascade is induced by most cytokines in all hematopoietin receptors with some exception, such as IL-4. Another well-characterized signaling pathway is the JAK-STAT pathway, which is activated by virtually all cytokines. In the IL-3/GM-CSF/IL-5 receptor systems, the binding of the ligand to the receptor results in rapid tyrosine phosphorylation of Jak2, which leads to the induction of some genes, including OSM and CIS, via activation of STAT5. These two pathways require the distinct region of the βc subunit of IL-3/GM-CSF/IL-5 receptors; membrane-proximal and -distal regions of βc subunit activate JAK-STAT pathway and Ras-Raf-MAPK cascade, respectively. Deletion analysis using truncated βc subunit of the IL-3/GM-CSF/IL-5 receptors suggested that SNAP-23 is one of the target genes downstream of the Ras-Raf-MAP kinase cascade. Alternatively, induction of SNAP-23 is mediated by a yet uncharacterized pathway that interacts with the distal region of the βc subunit which is also required for activation of the Ras-Raf-MAPK pathway.
Shimazaki et al29 reported protein kinase C (PKC)-mediated phosphorylation of SNAP-25. In addition, recent reports suggest that PI3 kinase, whose activation also requires the membrane-distal region of the βc subunit, activates the PKC.30 31 After induction of the SNAP-23 by cytokines, SNAP-23 may also be phosphorylated by PKC downstream of cytokine receptors to become functionally active.
In summary, we have shown that mouse SNAP-23 is induced by stimulation with various cytokines, implicating important roles of SNAP-23 in cytokine-mediated function. However, it remains to be investigated whether SNAP-23 simply functions as a protein for the fusion of vesicle and plasma membrane, or if it also participates in signal transduction. Some investigators reported that the SNARE proteins participate in the cytokinesis and the resealing of the plasma membrane as well.32,33 Therefore, one may speculate that SNAP-23 also plays a role in cellular functions other than exocytosis such as cell proliferation or differentiation. In fact, SNAP-23 message was induced in the process of both proliferation and differentiation. The generation of the antibody against SNAP-23 and mice lacking the SNAP-23 gene will provide a clear insight into the role of SNAP-23 induced by cytokines in hematopoietic cells. After we completed this work, Araki et al34 reported that the mouse SNAP-23, which is the same as the one we cloned, interacts with syntaxin 4 for the translocation of GLUT4 vesicles to the plasma membrane in adipocytes.
ACKNOWLEDGMENT
The nucleotide sequence data reported in this paper will appear in the DNA Data Bank of Japan, European Molecular Biology Laboratory, and GenBank nucleotide sequence databases with the following accession number: AB007444.
Supported by a Grant-in-Aid for Encouragement of Young Scientists from the Ministry of Education, Science and Culture, Japan. The Department of Hemopoietic Factors is partly supported by Chugai Pharmaceutical Company Ltd.
Address reprint requests to Yoshihiro Morikawa, MD, Department of Anatomy and Neurobiology, Wakayama Medical School, Kyubancho-27, Wakayama 640, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.