Abstract
Recombinant adeno-associated virus (rAAV) vectors are being evaluated for gene therapy applications. Using purified rAAV containing a mutationally marked globin gene (Aγ*) and sites 2, 3, and 4 from the locus control region (rHS432Aγ*), but lacking a drug-resistance gene, we investigated the relationship between multiplicity of infection (MOI), gene expression, and unselected genome integration in erythroid cells. Most primary erythroid progenitors were transduced as reflected by Aγ* mRNA in mature colonies but only at an MOI of greater than 5 × 107. Using immortalized erythroleukemia cells as a model, we found that fewer than one half of the colonies that contained the Aγ* transcript had an integrated, intact rHS432Aγ* genome. rHS432Aγ* integrated as a single copy with expression at approximately 50% the level of an endogenous γ globin gene. A second vector, rHS32Aγ*3′RE, containing the regulatory element (RE) from 3′ to the chromosomal Aγ globin gene, integrated as an intact, tandem head to tail concatamer with a median copy number of 6 with variable expression per copy ranging from approximately onefold to threefold that of an endogenous γ globin gene. These results establish that purified rAAV can be used to achieve integration and functional expression of a globin gene in erythroid cells, but only when high MOIs are used.
THE ABILITY TO TRANSFER a globin gene into repopulating hematopoietic stem cells and to achieve its expression in differentiating erythroid cells would create the potential for gene therapy of severe β-thalassemia and sickle cell anemia. Attempts to develop retroviral vectors for this purpose have been hampered by the relative inefficiency of gene transfer by such vectors into stem cells.1-5 Furthermore, the regulatory elements derived from the locus control region (LCR) that are required to achieve high-level expression of a linked globin gene cause proviral genome instability during virus production and transfer of the viral genome.3,6,7 Only recently have stable vectors been derived that are capable of transferring a globin gene with regulatory elements into primary hematopoietic cells.8-11 Unfortunately, the long terminal repeats of the integrated retroviral genome may cause suppression of globin gene expression in erythroid cells.12
An alternative vector system that is potentially useful for therapeutic globin gene transfer is that based on adeno-associated virus (AAV).13-16 This single-stranded DNA virus with a 4675 nucleotide genome depends on coinfection with a helper DNA virus, eg, adenovirus or a Herpes virus, which provides functions needed in trans for AAV genome replication and encapsidation. The wild-type AAV genome preferentially integrates into a specific region of chromosome 1917,18 by a mechanism that depends on its nonstructural (Rep) proteins.19-22 Only the 145 nucleotide inverted terminal repeats (ITRs) are required in cis to generate a rAAV vector; the Rep and capsid proteins can be provided in trans in packaging cells that are also expressing adenoviral proteins.13-16 Recent advances have provided packaging systems that yield purified recombinant AAV (rAAV) preparations of relatively high titer.23-25
rAAV vectors have been shown to transfer, integrate, and express globin genes in erythroleukemia cells when linked to a drug selection marker that facilitates recovery of clonal cell lines.26-28 However, the frequency of rAAV transduction and genomic integration was not established with this experimental design, because drug selection allows recovery of resistant clones even at low efficiencies of transduction and integration. Drug selection also demands that the rAAV genome be integrated into a transcriptionally active region of chromatin that is also likely to be permissive for globin gene expression. Expression of a globin gene has been observed in primary, progenitor-derived erythroid colonies without drug selection.29 However, available data have shown that an episomal rAAV genome may express its encoded genes for several days,30-33 raising the possibility that the globin mRNA found in erythroid colonies 14 days after transduction of purified hematopoietic progenitors was derived from a nonintegrated genome. Supporting this interpretation is our finding that the rAAV genome, although present initially after engraftment of transduced bone marrow cells, may disappear during long-term reconstitution in a nonhuman primate, bone marrow transplantation model (A.W. Nienhuis and R. Donahue, unpublished observations).
Our experiments have been designed to determine the fate of an unselected rAAV genome containing a globin gene and linked regulatory elements from the LCR in hematopoietic cells. We found that integration of an unrearranged genome permits stable expression of a globin gene in many but not all clonal erythroleukemia cell lines, all of which had initially expressed the gene after rAAV transduction. Integration frequency appeared to depend on vector design and multiplicity of infection.
MATERIALS AND METHODS
Vector preparation.Two rAAV vectors were used. Each contained a human γ globin gene that had been mutationally marked by a 6-bp deletion in its 5′ untranslated region to allow its transcript to be measured in the presence of normal human γ globin mRNA.34 The first vector, rHS432Aγ*, which contains three core hypersensitive sites from the LCR, has been previously described.29 The second vector, rHS32Aγ*3′RE was derived from rHS432Aγ* by removing the site 4 LCR fragment and adding a 750-bp EcoRI-HindIII fragment from downstream of the human Aγ globin gene that contains a regulatory element (RE) that associates with the nuclear matrix.35 Preparation of each of these rAAV vectors, using the cognate plasmids as substrates, was according to previously published methods.23,36 Purification was accomplished by isopynic density centrifugation in CsCl. The rAAV particles were isolated from fractions with an average density of approximately 1.38 g/mL and resuspended in HEPES-buffered saline containing 50 mmol/L HEPES, pH 7.4, and 150 mmol/L NaCl. Residual adenovirus was inactivated by heating the preparation at 56°C for 1 hour. The vector preparations had rAAV particle titers of 3 to 5 × 1013/mL as determined by slot blot hybridization.29,33 Slot blot analysis with a probe for AAV sequences33 confirmed that the titer for wild-type AAV was ≤10% that of rAAV. Assessment of contaminating helper adenovirus was performed by exposing 293 cells to vector stocks, serially passaging the cells for 4 weeks, and scoring as positive any degree of cytopathic effect. Replication competent adenovirus was not detected by this assay with a limit of detection of ≤100 pfu/mL. Electron microscopy was used to verify that the preparations were free of adenovirus or cellular contaminants.33
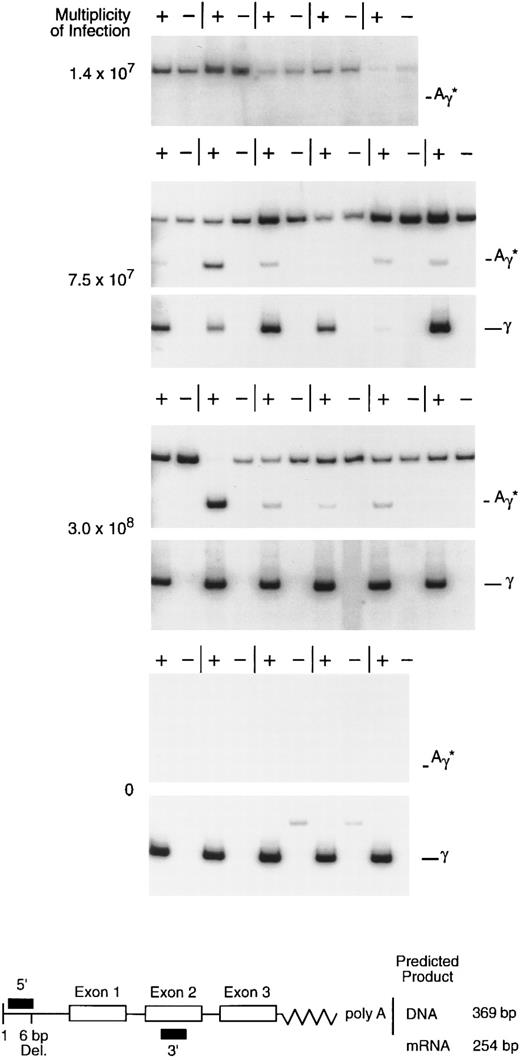
rHS432Aγ*-mediated gene transfer and expression in primary, progenitor-derived, human erythroid colonies. CD34-selected bone marrow cells were exposed to rAAV at various multiplicities of infection and cultured, and individual colonies were analyzed for mRNA at 14 to 16 days. The diagram below shows the design of the RT-PCR analysis; the upper band (369 bp), when present, is derived from genomic or vector DNA regardless of the use of reverse transcriptase (+ or −), whereas the lower (254 bp) reverse transcriptase-dependent band (+) is derived from processed globin mRNA. The 5′ primer designed to detect the Aγ* mRNA spans a 6-bp deletion in the 5′ untranslated region (diagram below), whereas the 5′ primer designed to detect the wild-type γ mRNA includes the deleted sequences (not shown). The upper band with the Aγ* primers in several of the figures is derived from rAAV vector particles, episomal DNA, or possibly integrated rAAV genomic DNA; its presence in the absense of Aγ* gene expression (upper panel, 1.4 × 107 MOI) suggests that this signal may be derived from particle DNA or intracellular, single-stranded DNA.
rHS432Aγ*-mediated gene transfer and expression in primary, progenitor-derived, human erythroid colonies. CD34-selected bone marrow cells were exposed to rAAV at various multiplicities of infection and cultured, and individual colonies were analyzed for mRNA at 14 to 16 days. The diagram below shows the design of the RT-PCR analysis; the upper band (369 bp), when present, is derived from genomic or vector DNA regardless of the use of reverse transcriptase (+ or −), whereas the lower (254 bp) reverse transcriptase-dependent band (+) is derived from processed globin mRNA. The 5′ primer designed to detect the Aγ* mRNA spans a 6-bp deletion in the 5′ untranslated region (diagram below), whereas the 5′ primer designed to detect the wild-type γ mRNA includes the deleted sequences (not shown). The upper band with the Aγ* primers in several of the figures is derived from rAAV vector particles, episomal DNA, or possibly integrated rAAV genomic DNA; its presence in the absense of Aγ* gene expression (upper panel, 1.4 × 107 MOI) suggests that this signal may be derived from particle DNA or intracellular, single-stranded DNA.
Detection of the rAAV-encoded transcript in erythroleukemia (K562) cell colonies. Erythroleukemia cells were transduced with rHS432Aγ* at an MOI of 3 × 108; cultured in semisolid medium for 14 days, after which individual colonies were plucked; and expanded for 2 to 4 days in liquid medium. An aliquot of the cells was used for RNA extraction. Lanes 1 through 12, RT-PCR analysis to detect the Aγ* transcript; (+) is with reverse transcriptase and (−) is the control without reverse transcriptase. Lane 13, water blank for the PCR reaction. Lane 14, RNA extracted from a cell line containing a single integrated copy of the Aγ* gene linked to HS2. Lane 15, RNA from control K562 cells that had not been transduced.
Detection of the rAAV-encoded transcript in erythroleukemia (K562) cell colonies. Erythroleukemia cells were transduced with rHS432Aγ* at an MOI of 3 × 108; cultured in semisolid medium for 14 days, after which individual colonies were plucked; and expanded for 2 to 4 days in liquid medium. An aliquot of the cells was used for RNA extraction. Lanes 1 through 12, RT-PCR analysis to detect the Aγ* transcript; (+) is with reverse transcriptase and (−) is the control without reverse transcriptase. Lane 13, water blank for the PCR reaction. Lane 14, RNA extracted from a cell line containing a single integrated copy of the Aγ* gene linked to HS2. Lane 15, RNA from control K562 cells that had not been transduced.
Cell culture.Human erythroleukemia (K562) cells37 were cultured in Improved Modified Eagles Medium (IMEM) containing 10% fetal calf serum (FCS). Human hematopoietic progenitors were purified by positive selection for CD34 expression by antibody affinity purification38 (Ceprate Stem Cell Concentrator; Cell Pro, Inc, Bothwell, WA). Transduction was performed by exposing 105 cells to rAAV in 2 mL of serum-free IMEM for 14 to 16 hours at 37°C in 5% CO2 with gentle, continuous rocking.29 Stem cell factor (100 ng/mL), interleukin-3 (IL-3; 20 ng/mL), and IL-6 (50 ng/mL) were added during transduction of CD34+ cells. Stem cell factor was obtained from Pepro Tech (Rocky Hill, NJ), and IL-3 and IL-6 were obtained from Life Technologies (Gaithersburg, MD). After transduction, the cells were washed in IMEM containing 10% FCS and plated in semisolid medium containing methylcellulose (MethoCult GF; Terry Fox Laboratories, Vancouver, British Columbia, Canada). After 14 to 16 days, individual erythroid cell colonies were plucked from the cultures with a micropipette and placed in suspension culture in Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS. RNA or DNA was extracted from an aliquot after culture for periods ranging from 2 days to several weeks. K562 cells were induced to undergo erythroid differentiation over 3 days by the addition of 40 mmol/L hemin as monitored by benzidine staining.
Nucleic acid analysis.RNA was recovered from individual primary erythroid colonies or K562 cells using RNA STAT-60 (Tel-Test Inc, Friendswood, TX) and concentrated by ethanol precipitation. Detection and semiquantitation of mRNA species was by reverse transcriptase-polymerase chain reaction (RT-PCR) methodology using reagents provided by the manufacturer (Promega, Madison, WI) and primers specific for β-actin, wild-type γ globin mRNA, or the mutationally marked human γ globin mRNA species. The conditions for RT-PCR analysis were as follows: (1) for β-actin, 25 cycles of 1 minute at 94°C, 1 minute at 60°C, and 1 minute at 72°C; or (2) for globin mRNAs, 25 to 30 cycles of 1 minute at 94°C, 1 minute at 60°C, and 1 minute at 72°C. The sequences of the β-actin primers were as follows: 5′ GATGATATCGCCGCGCTCGT and 5′ GGTCATCTTCTCGCGGTTGG. RNA samples were tested for β-actin sequences to verify that each sample was intact and capable of reverse transcription and PCR amplification. The sequences of the globin mRNA specific primers and the methodology for analysis of the PCR products have been described.29 The PCR products were resolved on a polyacrylamide gel (Protogel; National Diagnostics, Atlanta, GA) and quantitated using a phosphoimager (Molecular Dynamics, Sunnyvale, CA). DNA was extracted from K562 cells using standard methods and analyzed by Southern blot methodology39 using a 938-bp BamHI-EcoRI fragment containing intron II of the human Aγ globin gene as a probe.
Fluorescence in situ hybridization (FISH).Colcemid-arrested K562 cells were used as a source of metaphase chromosomes. Recombinant AAV DNA (rHS32Aγ*3′RE) was nick-translated with digoxygenin-11-UTP and hybridized overnight at 37°C to fixed metaphase chromosomes with the inclusion of 500 μg/mL of Cot1 competitor DNA (Life Technologies, Bethesda, MD) according to previously established methods.40 Signals were detected by incubating the slides with fluorescein-conjugated sheep antidigoxygenin antibodies (Boehringer Mannheim, Indianapolis, IN) followed by 4,6-diamidino-2-phenylindole (DAPI) counterstaining. Individual fluorescein- and DAPI-labeled signals were collected with a Nikon epifluorescence microscope coupled to a CCD camera and merged using CytoVision software (Applied Imaging, Pittsburgh, PA).
RESULTS
Multiplicity of infection required for transduction of erythroid progenitors.The availability of concentrated, highly purified rHS432Aγ* permitted titration of the ratio of vector particles to target cells required for introduction of the vector genome into primary clonogenic progenitors. Enriched CD34+ bone marrow cells (50% to 80% pure) were exposed to various multiplicities of infection (MOI) and cultured in semisolid medium. Individual erythroid colonies were plucked from the culture after 14 to 16 days of incubation and RNA recovered for analysis. At an MOI of 1.4 × 107, 0 of 11 colonies contained the Aγ* transcript, whereas at MOIs of 7.5 × 107 and 3.0 × 108, 9 of 11 and 17 of 22 colonies contained the Aγ* transcript, respectively (Fig 1 and data not shown). These data substantiate our earlier observations regarding the ability of rAAV to introduce and express a globin gene in primary erythroid progenitors29 and establish that, with highly purified preparations, a very high MOI is required for this purpose.
Prolonged expression of the rHS432Aγ* globin gene without genomic integration.The limited proliferative potential of primary clonogenic erythroid progenitors precludes attempts to investigate the relationship between viral uptake and initial gene expression and subsequent genomic integration. To study this relationship, we used human erythroleukemia (K562) cells37 as a model. A preliminary titration was performed to determine the MOI required to transduce a majority of clonogenic K562 cells. After transduction, the cells were plated in semisolid medium and incubated for 16 days, after which individual clones were plucked and transferred to liquid culture. Twenty-four hours later, RNA was extracted from an aliquot of the cells. Twenty colonies were analyzed from cells transduced at four different MOIs, with the following frequencies of detection of the Aγ* transcript: 105 = 1 of 20; 106 = 3 of 20; 107 = 11 of 20; and 108 = 16 of 20. In another experiment, all 12 colonies derived from cells transduced at an MOI of 3 × 108 that were analyzed were found to contain the Aγ* globin mRNA transcript (Fig 2 and Table 1). Seventeen days later, a second aliquot from these 12 clones and an aliquot from 11 others were analyzed; the cells from 19 of the 23 clones studied contain the Aγ* globin mRNA (Table 1). DNA from 16 of these clones was analyzed by Southern blot analysis. Nine were found to contain an integrated vector genome, but in only 5 of these was the genome unrearranged. Among the 11 clones that lacked an intact, unrearranged rHS432Aγ* genome were 7 that contained Aγ* mRNA, showing that this transcript could persist for 35 days without rAAV genome integration.
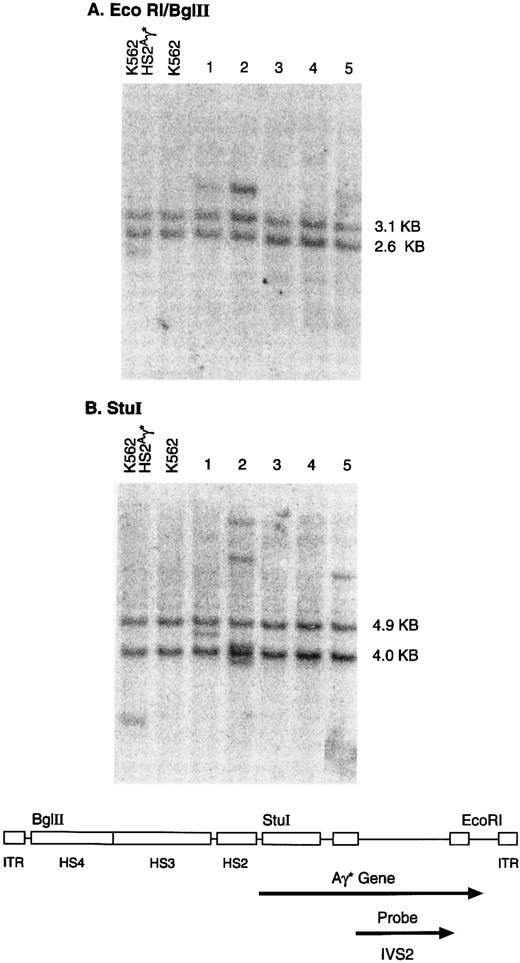
Representative results for clones containing an intact genome are shown in Fig 3. Restriction with EcoRI and Bgl II released the predicted 4.4-kb unrearranged genomic fragment in DNA from 3 of 5 clones; in 1 of these clones (no. 2), the intensity of the vector band suggested the presence of multiple integrated copies. Digestion with Stu I, which cuts once in the vector genome releases a junction fragment, the length of which is a characteristic of the integration site. Clones 1 and 5 contain a single integrated provirus, whereas clone 2 contains at least 4 genomic copies, each of which appears to be integrated at a separate chromosomal location (Fig 3).
Genomic integration of the rHS432Aγ* genome. Individual clones, derived from K562 cells transduced at an MOI of 3 × 108, were cultured in semisolid medium for 14 days and then expanded in liquid culture for 3 to 4 weeks. DNA was extracted and analyzed by the Southern blot methodology after restriction with EcoRI and Bgl II (A) or Stu I (B). DNA extracted from a cell line containing a single integrated copy of an rAAV genome that includes the Aγ* gene (K562 HS2Aγ*) served as a positive control (left lane), whereas K562 cells provided DNA that served as a negative control (second lane from left). The bands of 2.6 and 3.1 kb (A) or the 4.0- and 4.9-kb bands (B) present in each lane contain the endogenous γ globin genes. The diagram below indicates the organization of the rAAV vector genome and the location of specific restriction endonuclease sites. EcoRI and Bgl II (A) release an internal rAAV band of 4.4 kb, whereas Stu I releases a junction fragment of variable length that includes a portion of the rAAV genome and human chromosomal DNA.
Genomic integration of the rHS432Aγ* genome. Individual clones, derived from K562 cells transduced at an MOI of 3 × 108, were cultured in semisolid medium for 14 days and then expanded in liquid culture for 3 to 4 weeks. DNA was extracted and analyzed by the Southern blot methodology after restriction with EcoRI and Bgl II (A) or Stu I (B). DNA extracted from a cell line containing a single integrated copy of an rAAV genome that includes the Aγ* gene (K562 HS2Aγ*) served as a positive control (left lane), whereas K562 cells provided DNA that served as a negative control (second lane from left). The bands of 2.6 and 3.1 kb (A) or the 4.0- and 4.9-kb bands (B) present in each lane contain the endogenous γ globin genes. The diagram below indicates the organization of the rAAV vector genome and the location of specific restriction endonuclease sites. EcoRI and Bgl II (A) release an internal rAAV band of 4.4 kb, whereas Stu I releases a junction fragment of variable length that includes a portion of the rAAV genome and human chromosomal DNA.
RNA extracted from clones 1 and 5 after an additional 4 weeks of exponential growth in culture was found to contain the Aγ* transcript at concentrations approximately equivalent to 7% or 9%, respectively, of the mRNA species encoded by the endogenous γ globin genes. After correction for relative copy number (6 endogenous v 1 transduced), the rAAV-encoded globin gene appeared to be expressed at 42% or 54%, respectively, of the level of each endogenous gene. Clone 2 contained a high level of the Aγ* transcript, consistent with presence of the multiple integrated copies of the rAAV genome in this cell line. Further analysis of expression from this vector genome was not performed because of the high frequency of genome rearrangement.
Integration of rHS32Aγ* 3′RE as a tandem array of head to tail copies.A second vector was created to further evaluate the potential of rAAV to mediate genomic integration and persistent globin gene expression. Again, all 20 K562 clones studied 18 days posttransduction at two different MOIs contained the Aγ* globin mRNA species (Table 1). These two sets of 10 clones as well as several others derived from cells transduced at the two different MOIs were studied by Southern blot analysis. Eight of 18 clones derived from cells transduced at an MOI of 4 × 108 contained an intact vector genome, whereas 23 of 24 clones derived from cells transduced at an MOI of 4 × 109 contained an intact vector genome (Fig 4 and Table 1).
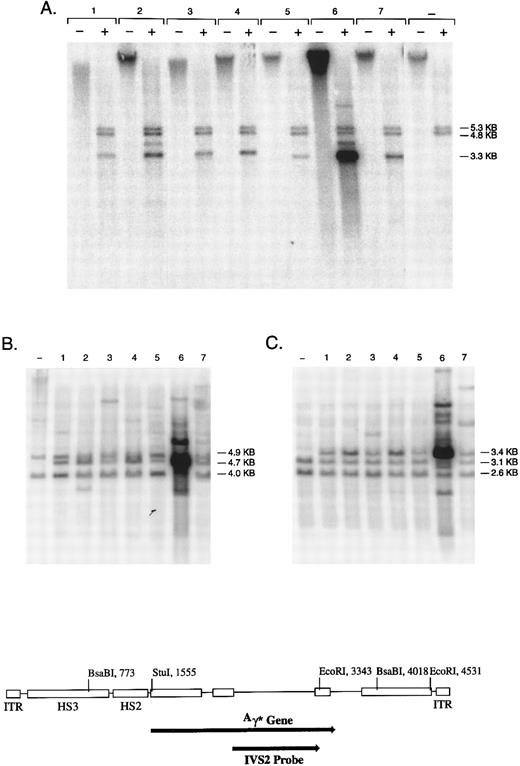
Genomic integration of the rHS32Aγ*3′RE genome. Individual colonies of K562 cells, transduced at an MOI of 4 × 109, were cultured in semisolid medium for 14 days and then expanded in liquid culture for 2 to 4 weeks. (A) DNA from 7 individual colonies analyzed without restriction (−) or cut with BsaBI (+). The bands at 4.8 and 5.3 kb contain the endogenous globin genes, whereas the band at 3.3 kb is an internal fragment derived from the vector genome (see diagram below). The negative control lanes contain DNA derived from nontransduced K562 cells. (B and C) DNA from the same 7 colonies and the negative control restricted with Stu I or EcoRI, respectively. Stu I releases variable length junction fragments, a 4.7-kb band derived from head to tail concatamers of the rAAV genome, and 4.0- and 4.9-kb fragments containing the endogenous globin genes. EcoRI releases a 3.4-kb fragment derived from tandem repeats of the rAAV genome, variable length junction fragments, and two fragments of 2.6 and 3.1 kb containing the endogenous γ globin genes.
Genomic integration of the rHS32Aγ*3′RE genome. Individual colonies of K562 cells, transduced at an MOI of 4 × 109, were cultured in semisolid medium for 14 days and then expanded in liquid culture for 2 to 4 weeks. (A) DNA from 7 individual colonies analyzed without restriction (−) or cut with BsaBI (+). The bands at 4.8 and 5.3 kb contain the endogenous globin genes, whereas the band at 3.3 kb is an internal fragment derived from the vector genome (see diagram below). The negative control lanes contain DNA derived from nontransduced K562 cells. (B and C) DNA from the same 7 colonies and the negative control restricted with Stu I or EcoRI, respectively. Stu I releases variable length junction fragments, a 4.7-kb band derived from head to tail concatamers of the rAAV genome, and 4.0- and 4.9-kb fragments containing the endogenous globin genes. EcoRI releases a 3.4-kb fragment derived from tandem repeats of the rAAV genome, variable length junction fragments, and two fragments of 2.6 and 3.1 kb containing the endogenous γ globin genes.
Southern blot analysis confirmed that the vector genome was integrated in all clones in which it was present (Fig 4 and data not shown). Analysis of uncut genomic DNA indicated that all hybridizing sequences migrated with high molecular weight DNA. Restriction with an enzyme (BsaBI) that cuts twice in the vector genome released the internal band of predicted length in every case; the signal intensity of this band suggested variable copy number among the clones. Seven of 29 clones also contained a rearranged genome as reflected by a BsaBI band that deviated in length from that predicted (Fig 4 and data not shown). Restriction with Stu I, an enzyme that cuts once in the vector genome, released a genomic length band of 4.7 kb in DNA from the vast majority of clones that contained an integrated vector genome (Table 1). This band suggests the presence of head to tail concatamers arranged in a tandem array. Table 1 summarizes all of the data obtained with respect to genome integration, the status of the vector genome, and the presence of tandem arrays.
The copy number of the tandem array was estimated by comparing the signal of the internal BsaBI band derived from the rAAV genome to the combined signal of the two BsaBI bands containing the endogenous γ globin genes (Fig 4). Three clones had very high estimated copy numbers of 25, 55, and 124, whereas in the remaining 19 clones, the copy number ranged from 3 to 14, with a median of 6. Restriction with EcoRI, an enzyme that cuts twice in the vector genome, yielded the 3.4-kb fragment predicted from the presence of such a tandem array. In addition, Stu I and EcoRI yielded one or more hybridizing fragments of variable length in each clone; these are the junction fragments between genomic DNA and integrated copies of the vector genome.
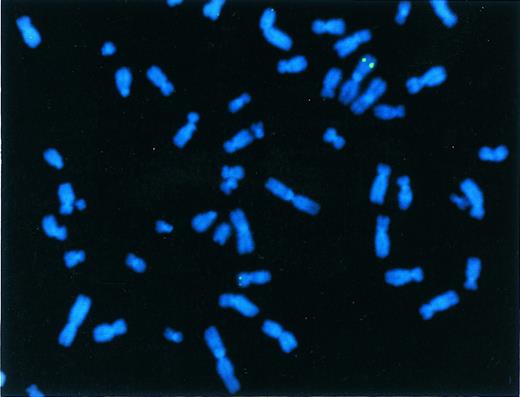
FISH was performed on several clones to verify genomic integration of the vector sequences. The genomic locus of the γ globin gene was detected at 11p15 in all clones studied. In addition, one cell line contained a clonal integration site at 1q (Fig 5), a second cell line contained a clonal integration site at 2q37 as well as several other nonclonal sites, and a third cell line contained several random, nonclonal sites of integration. The presence of nonclonal sites in cells from a single colony is consistent with delayed integration of the vector genome after the initiation of proliferation of a transduced cell. To test this hypothesis, several subclones of cell line 14 were studied. Three contained a clonal integration site on 1q, one contained a clonal integration site at the centromere of an unidentified chromosome, and another contained a clonal integration site at 2q37 (data not shown).
FISH analysis of a clone transduced with rHS32Aγ*3′RE. A signal is present on chromosome 11p15 (small chromosome below), the position of the normal β-like globin gene locus, and on chromosome 1a (large chromosome above), the position of the integrated rAAV genome in this cell line.
FISH analysis of a clone transduced with rHS32Aγ*3′RE. A signal is present on chromosome 11p15 (small chromosome below), the position of the normal β-like globin gene locus, and on chromosome 1a (large chromosome above), the position of the integrated rAAV genome in this cell line.
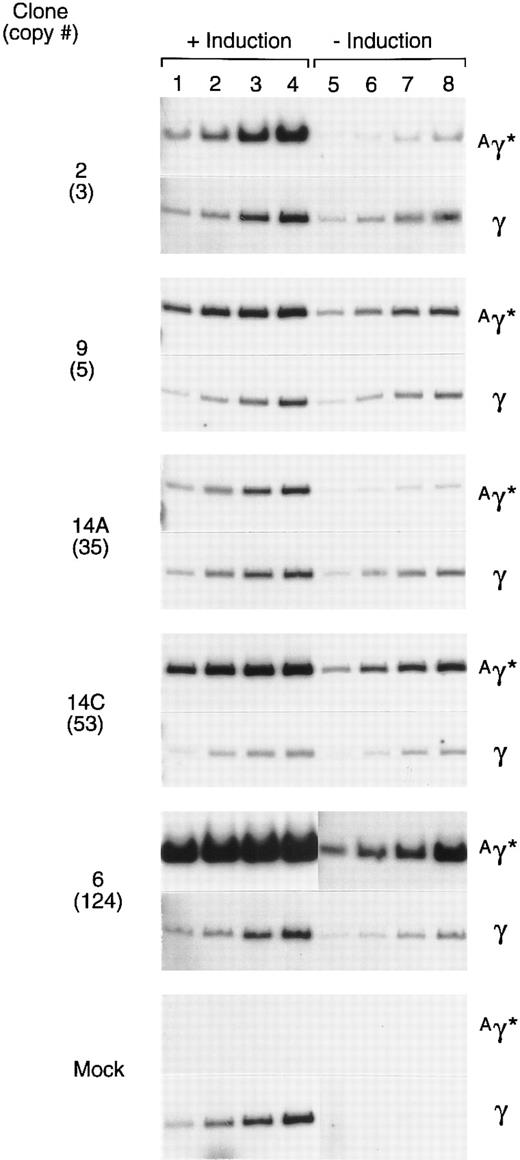
Expression of the rAAV-encoded globin genes was evaluated by semiquantitative RT-PCR. Linearity of the RT-PCR assay was established by simultaneously analyzing several different amounts of template RNA for their content of γ and Aγ* mRNAs (Fig 6). The 10 ng point was on the linear range for each sample, as determined by phosphoimager analysis. In a separate experiment, 10 ng of RNA from induced and uninduced cells of each clone was simultaneously analyzed for Aγ* and total γ mRNA under identical conditions (Fig 7). The expression per copy of the rAAV-encoded Aγ* gene ranged from 115% to 327% that of a single copy of the γ globin gene among induced cells of the 5 clones, with an average expression level of 197%. The range of expression per copy of the Aγ* gene in uninduced cells was 33% to 410% that of an endogenous gene, with an average of 135%. The 5 clones studied (Fig 6) had been in continuous culture for 6 months since transduction, indicating that the tandem arrays of the vector genome and expression levels are relatively stable.
Expression of the Aγ* gene in cells containing a tandem array of the rHS32 Aγ*3′RE genome. RNA was extracted from 5 clones after passage for 6 months in culture. Samples were extracted from uninduced (−) cells and those induced (+) by culture for 3 days in 40 mmol/L hemin. The copy number of the unrearranged vector genome, as estimated from the Southern blot analysis of BsaBI-restricted DNA is given (#) on the left. The amounts of template RNA analyzed were 10 ng (lanes 1 and 5), 25 ng (lanes 2 and 6), 50 ng (lanes 3 and 7), and 125 ng (lanes 4 and 8) for clones 2 and 6 and 5, 10, 25, and 50 ng, respectively, for the others.
Expression of the Aγ* gene in cells containing a tandem array of the rHS32 Aγ*3′RE genome. RNA was extracted from 5 clones after passage for 6 months in culture. Samples were extracted from uninduced (−) cells and those induced (+) by culture for 3 days in 40 mmol/L hemin. The copy number of the unrearranged vector genome, as estimated from the Southern blot analysis of BsaBI-restricted DNA is given (#) on the left. The amounts of template RNA analyzed were 10 ng (lanes 1 and 5), 25 ng (lanes 2 and 6), 50 ng (lanes 3 and 7), and 125 ng (lanes 4 and 8) for clones 2 and 6 and 5, 10, 25, and 50 ng, respectively, for the others.
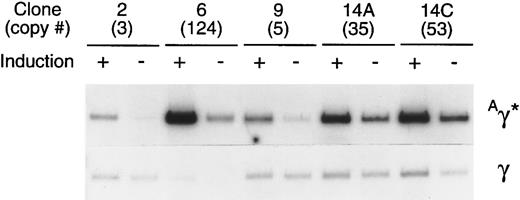
Comparison of the level of expression of the Aγ* gene in cells containing a tandem array of rHS32Aγ*3′RE genome. After 180 days in culture, 5 individual clones were either induced (+), using 40 mmol/L hemin, or not induced (−). Total RNA was isolated from each of these samples and 10 ng of the RNA was used in a semiquantitative RT-PCR assay after titration of the template amount (Fig 6). The copy number of the unrearranged vector genome, as estimated by genomic Southern analysis of BsaBI-digested DNA, is given (#) under the clone number.
Comparison of the level of expression of the Aγ* gene in cells containing a tandem array of rHS32Aγ*3′RE genome. After 180 days in culture, 5 individual clones were either induced (+), using 40 mmol/L hemin, or not induced (−). Total RNA was isolated from each of these samples and 10 ng of the RNA was used in a semiquantitative RT-PCR assay after titration of the template amount (Fig 6). The copy number of the unrearranged vector genome, as estimated by genomic Southern analysis of BsaBI-digested DNA, is given (#) under the clone number.
DISCUSSION
Our results establish that an rAAV containing a γ globin gene and linked regulatory elements can transduce human hematopoietic cells with high efficiency, leading to genomic integration and persistent gene expression. Successful transduction of primary cells with highly purified vector particles required a very high MOI. In erythroleukemia cells, the globin mRNA transcript derived from the rAAV-encoded globin gene could be detected for several weeks even in clones in which the rAAV genome had not integrated. Genomic integration appeared to occur at variable times after transduction and appeared to occur more frequently at higher MOIs. A vector containing hypersensitive sites 2 and 3 from the LCR and the regulatory element from 3′ to the Aγ globin gene integrated with highest efficiency, creating tandem head to tail arrays usually composed of 3 to 14 copies of the intact rAAV genome in each clonal cell population, which remained stable during 6 months of passage in culture. Expression of the rAAV encoded globin gene was variable, ranging from approximately onefold to threefold that of an endogenous γ globin gene at its native chromosomal location.
Recent evidence has established that rAAV can be expressed from an episomal genome without subsequent genome integration. We used an rAAV encoding the murine ecotropic receptor to evaluate the relationship between initial gene expression and subsequent genome integration.33 Ecotropic retroviruses infect mouse but not human cells because of interspecies polymorphism in the cationic amino acid transporter that acts as a receptor for this class of viruses.41 We found that human HeLa cells could be transduced at a ratio of rAAV vector particles of approximately 300 to 30,000, leading to expression of the retroviral receptor protein, permitting subsequent infection by an ecotropic retrovirus.33 However, under these conditions, integration of the rAAV genome occurred infrequently and only at higher MOI.
Expression of the γ globin gene from an episomal rAAV genome has now been documented in that we found consistent expression in all transduced clones, only some of which were subsequently found to contain the vector genome on Southern blot analysis of genomic DNA (Fig 3 and Table 1). Only at high MOIs was integration of the vector genome a consistent finding. Inclusion of the regulatory element from 3′ to the chromosomal Aγ globin gene in the rAAV vector was associated with integration of a tandem array of head to tail genomes. This element is known to associate with the nuclear matrix35 and might extend the persistence of the episomal rAAV genome and/or facilitate its entry into chromosomal DNA.
Future use of rAAV for therapeutic purposes may depend on gaining a better understanding of the mechanisms that control transduction, gene expression, and genomic integration. Viral uptake may depend on receptor number and it could account in part for the 104 higher MOI that we have found is required for transduction of hematopoietic cells compared with HeLa cells.33 Once in the cell, the single-stranded rAAV genome must be converted into a double-stranded, transcriptional template.31,32 There appears to be a correlation between the level of rAAV-encoded gene expression during the first 1 to 3 days after exposure of target cells to rAAV and the amount of double-stranded episomal genome. The adenoviral gene products encoded by the E1 and E4 (open reading frame 6) enhance the conversion of single-stranded rAAV DNA into its double-stranded form.31,32 Expression of cellular genes that facilitate DNA synthesis may be increased by these adenoviral proteins. Exposure of cells to DNA synthesis inhibitors and DNA damaging agents42 may also enhance transcription of a rAAV genome, apparently by triggering DNA repair mechanisms that facilitate its conversion to a double-stranded form.32 The higher transduction frequencies observed in cycling versus quiescent cells43 and immortalized versus primary cells44 might also be accounted for by the relative efficiency of the conversion of the rAAV genome into its double-stranded form. Formation of double-stranded DNA could also occur by annealing once the genome is uncoated, because both strands of viral DNA are packaged, but in separate virions.13 Presumably such annealing would be promoted by higher MOIs. Integration of the wild-type AAV genome appears to be mediated by the Rep proteins. Rep p78 and p68 bind to the ITRs as well as to a specific site on chromosome 19 with sequence specificity, thereby initiating integration of the AAV genome.19-22 In the absence of Rep, AAV appears to integrate at random.26 Factors that influence the integration frequency of rAAV, other than the MOI, remain undefined.
In our experiments, the high MOI required to achieve consistent integration of the rAAV genome may reflect several factors. First, viral entry into cells with low receptor numbers may be enhanced at high MOI. Second, the number of viral particles that actually enter the cell may be increased as a function of MOI and enhance the probability of annealing of the single-stranded forms to create a double-stranded template. Third, the amount of double-stranded template may be relevant to the frequency of genome integration. Finally, we cannot exclude the presence of trace contaminants of adenovirus or AAV Rep proteins, even in these highly purified rAAV preparations. Such contaminants could influence the behavior of vectors by facilitating viral uptake or influencing the fate of the rAAV genome once within the target cell. Indeed, the MOI of greater than 107 required for globin gene expression in transduced progenitor-derived colonies in these experiments compared with the MOI of 103, at which a proportion of progenitors were transduced with crude preparations29 might reflect variable amounts of functional AAV and/or adenoviral proteins in the two preparations. Our future work will focus on evaluating the effect of AAV Rep proteins and adenoviral gene products on the behavior of the rAAV genome.
Stem cell targeted gene therapy for hemoglobin disorders is likely to require integration of a transferred gene without selection of the cells in which an integration event has occurred. Therefore, it is imperative that transduction and integration occur with high frequency and that the integrated gene be expressed consistently and at a high level. Purified rAAV appears to meet these requirements when tested in erythroleukemia cells, provided that a very high multiplicity of infection is achieved. Persistence of the viral genome in an episomal state for variable periods of time after cell transduction temporally extends the opportunity for genomic integration. Repopulating stem cells are usually quiescent when purified from hematopoietic tissues but are thought to begin to proliferate when reinfused into a myeloablated recipient. Transduction of stem cells with rAAV might be accomplished ex vivo, creating the opportunity for subsequent integration as bone marrow regeneration is initiated. The ability to test this hypothesis in animal models will depend on the generation of adequate amounts of purified rAAV to achieve the required high ratio of vector particles to target stem cells. Use of immunodeficient mice in which relatively small numbers of human repopulating cells can be studied45 46 may facilitate the achievement of this objective.
ACKNOWLEDGMENT
We thank J. Johnson for outstanding assistance in the preparation of this manuscript and V. Valentine and D.N. Shapiro for performing the FISH.
Supported in part by National Heart, Lung and Blood Institute Program Project Grant No. P01 HL 53749, Cancer Center Support CORE Grant No. P30 CA 21765, and the American Lebanese Syrian Associated Charities (ALSAC).
Address reprint requests to Arthur W. Nienhuis, MD, Division of Experimental Hematology, St Jude Children's Research Hospital, 332 N Lauderdale, Memphis, TN 38105.