In this issue of Blood, Sobh et al show that NSD2 overexpression in t(4;14) multiple myeloma (MM) diverts S-adenosylmethionine (SAM) to the epigenome, impairing SAM-dependent creatine biosynthesis, thus creating a reliance on adenylate kinase 2 (AK2)–facilitated mitochondrial adenosine triphosphate distribution (see figure).1
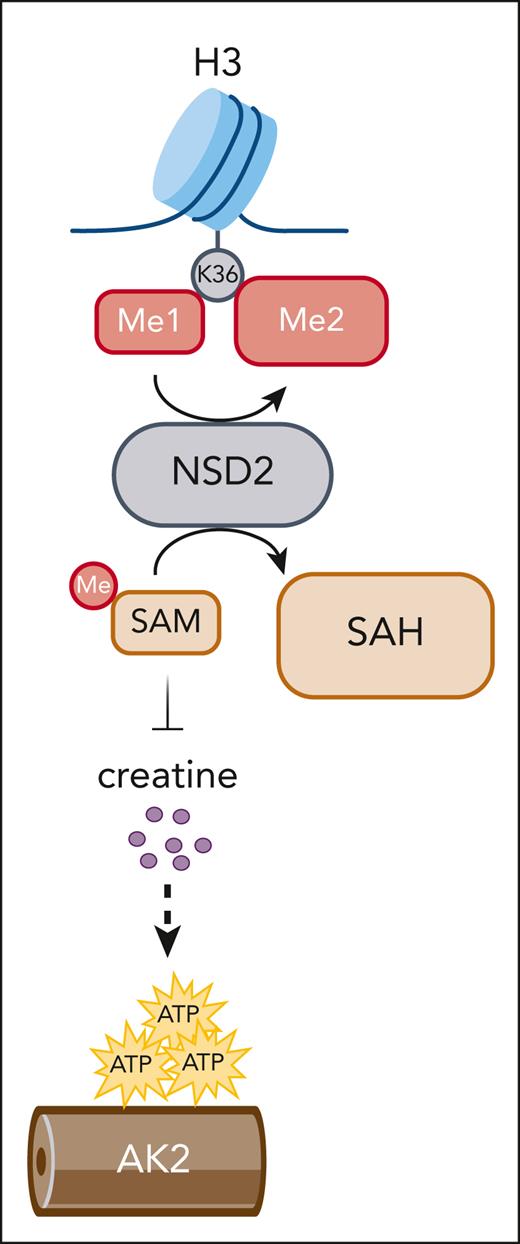
NSD2 converts SAM to SAH to globally dimethylate H3K36, impairing SAM-dependent creatine biosynthesis. Creatine deficiency, in turn, creates a reliance on AK2-facilitated mitochondrial adenosine triphosphate distribution. AK2, adenylate kinase 2; SAH, S-adenosylhomocysteine.
NSD2 converts SAM to SAH to globally dimethylate H3K36, impairing SAM-dependent creatine biosynthesis. Creatine deficiency, in turn, creates a reliance on AK2-facilitated mitochondrial adenosine triphosphate distribution. AK2, adenylate kinase 2; SAH, S-adenosylhomocysteine.
The t(4;14) chromosome translocation was identified almost 30 years ago.2 It is an unusual translocation in that 2 oncogenes are dysregulated, 1 on each of the derivative chromosomes. On der14, the powerful 3′ heavy-chain enhancer dysregulates the expression FGFR3 in three-fourths of patients, which is further activated by somatic mutations in 40% of patients with expression.3 In contrast, on der4, most patients express a hybrid fusion transcript between the immunoglobulin heavy-chain J and/or I-mu exon, and exons of NSD2, nuclear SET domain 2, initially called MMSET or WHSC1.4 In one-third of cases, the breakpoints occur between coding exons of NSD2, resulting in the translation of an amino-truncated NSD2 lacking one of the PWWP1 domains important for recognizing methylated histones, and leading to mislocalization of NDS2 from nuclei to nucleoli.5 NSD2 functions as a histone H3 lysine 36 dimethylase (H3K36me2)6 and, remarkably in MM with t(4;14), results in a global increase in H3K36me2 and a corresponding global decrease in H3K27me3.7
Sobh and colleagues used a genome-wide CRISPR screen in an isogenic pair of MM cell lines with and without NSD2 overexpression. Pathway analysis of the selective dependencies in cells with dysregulated NSD2 identified enrichment for metabolic pathways and led the authors to focus on one of the top hits, adenylate kinase 2 (AK2). Using metabolomics analyses, and various cellular assays they examined the effects of AK2 suppression on cell viability, replication stress, and sensitivity to proteasome inhibitors. They determined that NSD2 overexpression leads to a significant diversion of one-carbon metabolism toward methylating the epigenome (H3K36me2). This metabolic shift results in depletion of creatine, and a dependence on AK2, a mitochondrial membrane protein that plays a key role in maintaining the cell’s energy balance.
The study is significant for several reasons. It is paradigm shifting to consider methylation of the epigenome so extensive that it depletes the one-carbon pool of the cell. The finding of a increased dependency on AK2 in t(4;14) vs non-t(4;14) MM suggests a therapeutic window for targeting AK2. This is particularly important given the slow progress in directly targeting NSD2. The first clinical trial of an inhibitor of the methyltransferase activity of the SET domain of NSD2 using KTX-1001 is currently underway (NCT05651932), while therapeutics targeting the PWWP1 domain, both inhibitors and PROTACs, are being explored preclinically.8 Finally, it appears that AK2 is also a selective dependency for other lymphoid tumors, expanding the relevance of this study to other hematologic malignancies.
Conflict-of-interest disclosure: The authors declare no competing financial interests.