In this issue of Blood, Wienecke et al1 study the sensitive detection of measurable residual disease (MRD) in peripheral blood in patients with acute myeloid leukemia (AML) and post–allogeneic transplant. The work underscores the potential of sensitive genetic assays in the context of mutation context and kinetics to better understand response and relapse and could have important impact on studies.
The detection of MRD among patients with AML is an important predictor of outcome. This is true for patients with AML undergoing chemotherapy or allogeneic transplant and holds across age groups, AML subtypes, time of MRD assessment, specimen source, and type of assays used.1-6 The clinical challenge is that patients with AML with MRD following chemotherapy have a high rate of relapse compared with those without MRD. Patients with MRD at the time of or following allogeneic transplantation also do poorly. In each situation, the hazard ratio of relapse in MRD+ compared with negative cases runs approximately two- to threefold range. Approximately one-third of MRD+ cases will not relapse, and approximately one-third of MRD− cases will relapse. How should we use MRD? How can we improve it use to better predict outcome and inform interventional therapy?
The study by Wienecke et al is a retrospective analysis of 74 patients with AML who received an allogeneic transplant and later relapsed. They used a sensitive error-corrected next-generation sequencing (NGS) panel of 46 genes/hot spots with an estimated limit of detection of ∼0.01%. Cases were selected by having serial peripheral blood samples, including a relapse sample. This allowed the team to evaluate the “stability” (defined as mutations found at diagnosis and relapse) of different mutations and the kinetics of MRD over time. The study only included relapse cases, so the authors cannot comment on the cases of MRD positivity without relapse.
Often the potential of MRD studies are handcuffed by using an arbitrary cutoff of MRD positivity and negativity, and thus lumping all MRD into a dichotomous, nonquantitative variable. This study illustrates how much interesting nuance is lost with this approach. The authors find that at least 1 MRD marker became detectable in a time-dependent manner, predictably more common the closer to clinical relapse. Only 10% of cases had MRD detected a full 6 months prior to relapse, and 64% of patients had MRD detected by 1 month prior to relapse (alas, the latter finding may not be particularly helpful for strategies meant to preempt relapse). The relapse kinetics after transplant were influenced by the “functional class” of mutations and their stability during molecular progression. Mutations in epigenetic modifier and splicosome genes had a higher prevalence of MRD positivity, greater stability before relapse (mutation present a diagnosis and at relapse), and a slower tempo from first detection to relapse, and mutations in signaling genes (eg, FLT3-ITD) demonstrated less stability (eg, not detectable at diagnosis, but detectable at relapse) and a shorter time to relapse. These differences in mutation-dependent kinetics mirrors that found in AML cases after receiving chemotherapy.7 Approximately 80% of all MRD mutations found at relapse were present at diagnosis. Thus, a future potential application could be a cheaper, faster, targeted approach strategy for mutation testing posttransplant, focusing on only mutations present at the time of transplantation (especially if coupled with flow cytometry as another way to detect MRD). Both DMT3A, TET2, and ASXL1 (DTA) and non-DTA mutations displayed similar relapse kinetics during the follow-up period after allogenic hematopoietic cell transplantation. This adds to a growing mixed picture of what DTA mutations mean in the context of treated AML cases, as opposed to their presence in clonal hematopoesis of indeterminate potential (CHIP) in normal individuals. In the posttransplant setting, the study of DTA mutations is complicated, such that mutations might arise from donor CHIP. Perhaps the best way to understand DTA in the allogeneic setting is by use of single-cell genomics where donor and recipient mutations can be more easily discriminated.8
There are some limitations of the study. There were a variety of ages, AML history, transplant preparative regimens, and so on. Potential selection bias makes broad conclusions difficult given it took 200 patients screened to arrive at 74 patients to study who relapsed. The study did not have concurrent bone marrow samples to compare with peripheral blood, nor are there data using flow cytometry as a measure of MRD (a loss since many centers use flow as a fast and convenient measure of MRD2,4).
Relapse is the main cause of treatment failure in AML following transplantation. The detection of MRD is associated with relapse and survival in AML, but it is quite imperfect, as some patients without MRD relapse, and some with MRD do not. This study did not look at MRD cases that did not relapse, but they found many MRD− cases (36%) that went on to relapse. The findings suggest that if we want to detect MRD and act to prevent relapse, we are going to need a monitoring strategy driven by aggressive monitoring (with more frequent use of peripheral blood?), with potential tailoring based on mutation subtype. Better detection might also require the addition of other modalities (flow cytometry) and even more sensitive NGS techniques, which may improve MRD detection.9
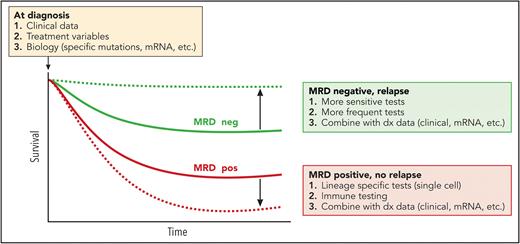
A goal of MRD research is shown schematically in the figure. MRD− patients nonetheless relapse and die (thick red line), and MRD+ cases often do not relapse (thick green line). For MRD to guide therapy and become an early “surrogate” of disease response for clinical trials, we need to separate these curves so that MRD status more fully predicts outcome. Better and/or multiple methods of MRD detection may help. Of tantamount importance is prospective studies that can model MRD and outcomes in the context of pretreatment clinical and biology studies, as well as sensitive, multimodality MRD detection (such as the MEASURE NCT05224661 study).
The association of MRD and relapse/survival. The association of MRD status during treatment and survival are shown in thick green (MRD neg) and red (MRD pos) curves. Potential solutions to improve the predictive power of MRD status are boxed. Many MRD– cases still relapse. This may be sampling error, limitations of the assay sensitivity, rapid kinetics (with missed detection given the former 2 variables), the evolution or loss of MRD markers, and so on. On the other hand, some MRD+ cases do not relapse. These could be from clones with very slow kinetics (in check from immune mechanisms?), mutations occurring in other lineages (lymphoid), among others. The box next to the blue arrow suggests predictive models that add clinical variables, therapy, and other genetic/biological parameters that could be combined with MRD kinetics to improve the predictive capabilities of MRD monitoring. Professional illustration by Patrick Lane, ScEYEnce Studios.
The association of MRD and relapse/survival. The association of MRD status during treatment and survival are shown in thick green (MRD neg) and red (MRD pos) curves. Potential solutions to improve the predictive power of MRD status are boxed. Many MRD– cases still relapse. This may be sampling error, limitations of the assay sensitivity, rapid kinetics (with missed detection given the former 2 variables), the evolution or loss of MRD markers, and so on. On the other hand, some MRD+ cases do not relapse. These could be from clones with very slow kinetics (in check from immune mechanisms?), mutations occurring in other lineages (lymphoid), among others. The box next to the blue arrow suggests predictive models that add clinical variables, therapy, and other genetic/biological parameters that could be combined with MRD kinetics to improve the predictive capabilities of MRD monitoring. Professional illustration by Patrick Lane, ScEYEnce Studios.
MRD is a measure of the entirety the factors that determine disease response: the evolving cancer ecosystem of the cancer biology, microenvironment, therapy, the immune system. Given this complexity, we should not be discouraged that the association of MRD and outcome is not stronger; maybe it is a minor miracle that it is so strong at all.
Conflict-of-interest disclosure: J.R. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal