Key Points
NSD2 overexpression in MM diverts S-adenosylmethionine to the epigenome and disrupts creatine synthesis, rendering MM cells dependent on AK2.
Targeting AK2 in MM cells disrupts mitochondrial energy distribution, induces DNA replication stress, and enhances sensitivity to bortezomib.
Visual Abstract
Chromosomal translocation (4;14), an adverse prognostic factor in multiple myeloma (MM), drives overexpression of the histone methyltransferase nuclear receptor binding SET domain protein 2 (NSD2). A genome-wide CRISPR screen in MM cells identified adenylate kinase 2 (AK2), an enzyme critical for high-energy phosphate transfer from the mitochondria, as an NSD2-driven vulnerability. AK2 suppression in t(4;14) MM cells decreased nicotinamide adenine dinucleotide phosphate (NADP[H]) critical for conversion of ribonucleotides to deoxyribonucleosides, leading to replication stress, DNA damage, and apoptosis. Driving a large genome-wide increase in chromatin methylation, NSD2 overexpression depletes S-adenosylmethionine, compromising the synthesis of creatine from its precursor, guanidinoacetate. Creatine supplementation restored NADP(H) levels, reduced DNA damage, and rescued AK2-deficient t(4;14) MM cells. As the creatine phosphate shuttle constitutes an alternative means for mitochondrial high-energy phosphate transport, these results indicate that NSD2-driven creatine depletion underlies the hypersensitivity of t(4;14) MM cells to AK2 loss. Furthermore, AK2 depletion in t(4;14) cells impaired protein folding in the endoplasmic reticulum, consistent with impaired use of mitochondrial adenosine triphosphate (ATP). Accordingly, AK2 suppression increased the sensitivity of MM cells to proteasome inhibition. These findings delineate a novel mechanism in which aberrant transfer of carbon to the epigenome creates a metabolic vulnerability, with direct therapeutic implications for t(4;14) MM.
Introduction
Multiple myeloma (MM) is the second-most frequent blood malignancy.1 The prognosis for patients with MM has greatly improved over the years because of therapies targeting the distinctive characteristics of the plasma cell.2 The combination of proteasome inhibitors (PIs), immunomodulating drugs, and glucocorticoids, followed by autologous stem cell transplantation, is an efficacious first-line treatment for patients with MM.3 However, most patients eventually relapse because of acquired drug resistance, necessitating the identification of additional MM-specific targets or strategies to combat therapy resistance.
The histone methyltransferase NSD2 is overexpressed in ∼15% of MM patients because of chromosomal translocation (4;14) and represents a negative prognostic factor.4 NSD2 overexpression is associated with a global increase in histone H3 lysine 36 dimethylation and concomitant loss of the repressive histone H3 lysine 27 trimethyl mark, thereby promoting an oncogenic gene expression program.5 NSD2 overexpression in MM has been linked to increased cell adhesion, migration, clonogenic growth, and in vivo tumorigenicity.5-8 Because of the oncogenic nature of NSD2, it is designated as an appealing therapeutic target. However, most attempts to develop specific and potent NSD2 inhibitors have been largely unsuccessful. Therefore, identification of pathways driven by NSD2 or novel vulnerabilities associated with NSD2 overexpression in MM represents an alternative therapeutic approach.
Aberrant cellular metabolism is common in MM and can contribute to pathogenesis and resistance to therapeutic agents, including PIs.9-11 Subtype-specific recurrent oncogenic lesions in MM have metabolic consequences and introduce novel dependencies. Genes frequently dysregulated in MM, such as MYC, CCND1, FGFR3, and MAF, drive alterations in diverse metabolic processes including glucose, glutamine, lipid, nucleotide, and energy metabolism.12 Although its role in MM cell metabolism had been unexplored, NSD2 overexpression in breast cancer induces the expression of key enzymes for glycolysis and the pentose phosphate pathway.13 Here, using a CRISPR-based, genome-wide loss of function genetic screen in NSD2 high and low isogenic MM cells, we identified several NSD2-associated metabolic dependencies. We identified the mitochondrial adenine nucleotide regulator adenylate kinase 2 (AK2) as a selective dependency in MM cells that becomes more essential when NSD2 is overexpressed. Our findings present AK2 as a novel therapeutic target for MM, especially in patients with t(4;14) chromosomal translocations.
Methods
Genome-wide CRISPR screening
Genome-wide CRISPR-based loss-of-function dropout screens were performed as previously described.14 Briefly, cells were transduced with the human lentiviral CRISPR knockout pooled library Brunello in LentiCRISPRv2 (RRID: Addgene_73179) at a multiplicity of infection of 0.4 and selected with puromycin for 3 days. Cells from each pool were collected immediately after puromycin selection and after a period corresponding to 12 doublings. Single-guide RNAs from initial and final pools were amplified from genomic DNA and analyzed by next-generation sequencing on an Illumina HiSeq 4000. Candidate gene ranking was performed using MAGeCK-MLE.15
In vitro competitive growth assays
Cells were transduced with lentiviral CRISPR (LentiCRISPRv2-GFP) or short hairpin RNA (shRNA) (psi-LVRU6GP) vectors expressing green fluorescent protein (GFP) at a multiplicity of infection <1. The fraction of GFP-positive cells was measured at different time points by flowcytometry using a BD Accuri C6 (BD Biosciences) (RRID: SCR_019591).
RNAseq analysis
Poly A RNAseq Library preparation was performed at the Gene Expression Core of the University of Florida (UF) Interdisciplinary Center for Biotechnology Research using the Illumina Stranded messenger RNA Preparation protocol. Next-generation sequencing was done at the UF Interdisciplinary Center for Biotechnology Research NextGen DNA sequencing core on NovaSeq 6000 (Illumina) (RRID: SCR_016387). Differential expression analysis was conducted using DESeq2 (RRID: SCR_015687). A fold change threshold of 1.5 and a false discovery rate (FDR)-corrected P value threshold of 0.05 were used to define differentially expressed genes. Venn diagrams were created using DeepVenn.16 Pathway and Gene Ontology analyses were performed using Enrichr (RRID: SCR_001575) and STRING (RRID: SCR_005223).
Metabolomics analysis
Soluble metabolites were extracted directly from snap-frozen cell pellets using cold acetonitrile/water (80/20, vol/vol) at a dilution of 10,000 cells/μL of solvent. Samples were vortexed and incubated at −80°C overnight to precipitate proteins and subsequently thawed, vortexed, and centrifuged at 18 000 × g for 30 min at 4°C to pellet debris. The supernatants were transferred to new vials and analyzed by high-performance liquid chromatography, high-resolution mass spectrometry, and tandem mass spectrometry. Raw values were normalized to the total ion count. Differential metabolite abundance analysis, statistics, and pathway analysis were performed using MetaboAnalyst 5.0 (RRID: SCR_015539).
Xenograft mouse models
KMS11 and LP1 cells were transduced with a scrambled or AK2-targeting, doxycycline-inducible shRNA vector and selected with puromycin. A total of 5 × 106 cells were suspended in phosphate-buffered saline (PBS), mixed with lactose dehydrogenase elevating virus (LDEV)-free Geltrex basement membrane matrix (ThermoFisher, #A1413201), and injected subcutaneously into the flanks of 6-week-old non-obese diabetic (NOD)-severe combined immunodeficiency (SCID) female mice (Jackson Laboratory, RRID: IMSR_JAX:001303). When tumors became visible, mice were euthanized, the tumors were isolated, and the dimensions were measured using a caliper. Tumor volumes were calculated using the formula V = ½ L × W2 (where V is the tumor volume, L is the tumor length and W is the tumor width). The experimental protocol (#202100000078) was approved by the University of Florida Institutional Animal Care and Use Committee.
Protein folding capacity measurements
The mammalian endoplasmic reticulum (ER)–localized redox-sensitive green-fluorescent protein (Mero-GFP) lentiviral vector was a gift from the laboratory of Fumihiko Urano (Washington University). Cells were transduced with Mero-GFP and selected with puromycin for 5 days. After recovery, cells were transduced with the shRNA-mCherry vector expressing scrambled or AK2 shRNAs. Fluorescence of unfolded (reduced) or folded (oxidized) GFP was measured in mCherry-positive cells using a Sony SP6800 Spectral Analyzer (RRID: SCR_018067).
dNTP quantification
Cells (2 × 106) were washed in PBS and lysed by suspending in 200 μL of ice-cold 65% methanol, vortexing for 2 minutes, and incubating at 95°C for 3 minutes. Samples were then centrifuged at 18 000g in a Beckman Coulter Microfuge 20R for 3 minutes. Supernatants were transferred to new tubes and dried by SpeedVac (ThermoFisher). 2′-deoxyribonucleoside 5'-triphosphate (dNTP) concentrations were determined using an HIV reverse transcriptase–based dNTP assay as previously described.17
NADP(H) and creatine assays
For NADP(H) assays, 1-4 × 105 cells were plated in a white-walled tissue culture plate. Levels of NADP(H) levels were measured using NADP/NADPH-Glo Assay Kit (Promega, #G9081), following the manufacturer’s protocol. Intracellular creatine levels were measured using the Colorimetric/Fluorometric Creatine Assay Kit (Abcam, #ab65339). A total of 2 × 106 cells were washed with ice-cold PBS, homogenized in assay buffer, and the lysates were deproteinized using the deproteinizing sample kit (Abcam, #ab284939).
Additional methods and data analysis are provided in supplemental Materials and Methods, available on the Blood website.
Results
CRISPR screens reveal NSD2-driven dependencies in MM
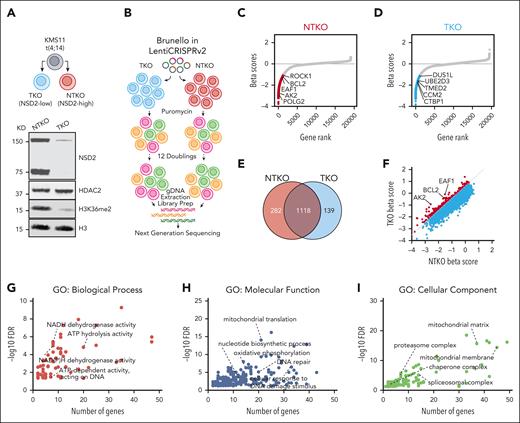
We performed CRISPR-based loss-of-function genetic screens in isogenic cells with high (non-translocated allele knockout; NTKO) and low (translocated allele knockout; TKO) NSD2 expression,6 derived from the t(4;14) MM cell line KMS11 (Figure 1A, B). In addition to common essential genes, the screen identified multiple genes more important for the survival of NSD2-overexpressing MM cells (Figure 1C-F; supplemental Figure 1A). Pathway enrichment analysis revealed that NSD2-overexpressing MM cells are more dependent on metabolic processes, including oxidative phosphorylation and NADPH metabolism, in addition to different adenosine triphosphate (ATP)–dependent processes, such as RNA splicing, DNA repair, protein folding, and proteasomal degradation (Figure 1G-I; supplemental Figure 1B). Other top genes determined to be more essential in NTKO cells (Beta score difference > 0.5) included the transcription elongation component ELL-associated factor 1 (EAF1) and the antiapoptotic protein B-cell lymphoma 2 (BCL2), which were validated by genomic and pharmacological approaches (supplemental Figure 1C-D). The increased dependence on proteasomal degradation driven by NSD2 overexpression was also confirmed by the reduced cytotoxicity of the PI bortezomib in NSD2-low TKO cells and increased sensitivity upon re-expression of NSD2 in these cells (supplemental Figure 1E). Collectively, these results confirmed the efficacy of our screening approach in identifying vulnerabilities caused by NSD2 overexpression in MM.
NSD2 overexpression drives metabolic dependencies in MM. (A) Immunoblot analysis confirming the decrease in NSD2 levels and H3K36me2 levels in TKO compared to NTKO isogenic MM cells derived from KMS11. (B) Workflow of genome wide CRISPR screening in NTKO and TKO MM cells. (C-D) MAGeCK-generated gene essentiality (Beta) scores plotted vs gene ranks. Essential genes for NTKO and TKO cells (MAGeCK FDR < 0.1) are indicated in red and blue, respectively. (E) Venn diagram showing common and differentially essential genes between NTKO and TKO cells. (F) Scatter plot of NTKO and TKO Beta scores. Genes that are more essential for NTKO cells (difference in Beta scores > 0.5; FDR < 0.1) are shown in red. (G-I) ShinyGO gene ontology (GO) analyses of the 282 genes defined to be more essential in NTKO than TKO cells. FDRs of the significant GO terms are plotted against the corresponding number of gene hits in each term.
NSD2 overexpression drives metabolic dependencies in MM. (A) Immunoblot analysis confirming the decrease in NSD2 levels and H3K36me2 levels in TKO compared to NTKO isogenic MM cells derived from KMS11. (B) Workflow of genome wide CRISPR screening in NTKO and TKO MM cells. (C-D) MAGeCK-generated gene essentiality (Beta) scores plotted vs gene ranks. Essential genes for NTKO and TKO cells (MAGeCK FDR < 0.1) are indicated in red and blue, respectively. (E) Venn diagram showing common and differentially essential genes between NTKO and TKO cells. (F) Scatter plot of NTKO and TKO Beta scores. Genes that are more essential for NTKO cells (difference in Beta scores > 0.5; FDR < 0.1) are shown in red. (G-I) ShinyGO gene ontology (GO) analyses of the 282 genes defined to be more essential in NTKO than TKO cells. FDRs of the significant GO terms are plotted against the corresponding number of gene hits in each term.
NSD2 overexpression increases AK2 dependence in MM cells
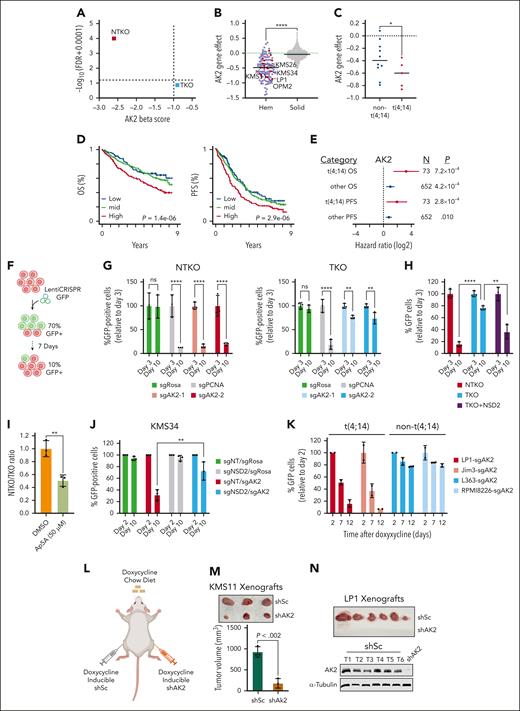
To prioritize screening hits for subsequent investigation, we ranked genes by differences in MAGeCK beta scores. AK2 ranked top with a score of −2.6 (FDR = 0) in NTKO and −0.9 (FDR = 0.133) in TKO cells (Figure 2A). Analysis of Broad Institute dependency map (DepMap) data indicated that AK2 loss has a mild negative effect on the growth of most solid tumor cell lines, whereas hematologic malignancies, including MM,18 showed significantly higher dependence on AK2 activity (Figure 2B). Comparing Gene Effect scores between the t(4;14) and non-t(4;14) MM cell lines available in Depmap showed a significantly lower median score in the t(4;14) group (P <. 05, Mann-Whitney one-tailed test), indicating higher dependence on AK2 (Figure 2C). Investigation of Multiple Myeloma Research Foundation CoMMpass data revealed that MM patients with higher AK2 expression exhibited inferior survival (supplemental Figure 2A; Figure 2D). Higher AK2 levels were significantly associated with worse overall and progression-free (PFS) survival in both non-t(4;14) and t(4;14) subsets (hazard ratio >1). The hazard ratios tended to be higher in patients with t(4;14) with greater significance for PFS, despite only representing 10% (73 of 725) of patients (Figure 2E). To confirm the increased dependence of NSD2 overexpressing MM cells on AK2, we used CRISPR-Cas9 to disrupt AK2 in NTKO and TKO cells (supplemental Figure 2B). In vitro competitive growth assays showed a striking depletion of AK2-disrupted (GFP-positive) NTKO cells and a less pronounced depletion of AK2-disrupted TKO cells (Figure 2F-G). Intriguingly, enforced overexpression of NSD2 in TKO cells restored the hypersensitivity to AK2 loss (supplemental Figure 2B; Figure 2H). To further corroborate the increased sensitivity of NSD2 overexpressing MM cells to the loss of AK2 activity, cells were treated with the AK2 inhibitor P1, P5-Di (adenosine-5') pentaphosphate (Ap5A). We first confirmed binding of AP5A to AK2 in vitro using a cellular thermal shift assay (supplemental Figure 2C). Next, treating a mixed population of GFP-expressing TKO and nonfluorescent NTKO cells with Ap5A, we observed selective depletion of NTKO cells (Figure 2I). Similarly, the growth of NSD2-high NTKO cells was more profoundly inhibited by shRNA depletion or induced knockout of AK2 than TKO cells (supplemental Figure 2D-E). We also confirmed the NSD2-driven dependence on AK2 in another t(4;14) MM cell line, KMS34. In these cells, AK2 disruption led to significant growth suppression but had a much more modest effect when NSD2 was knocked out (supplemental Figure 2F; Figure 2J). Furthermore, induced AK2 loss in other t(4;14) MM cell lines (Jim3, LP1, and OPM2) resulted in striking depletion of cells, whereas the non-t(4;14) cell lines L363 and RPMI8226 exhibited only ∼20% inhibition when AK2 was disrupted (supplemental Figure 2G-H; Figure 2K). To examine the role of AK2 in t(4;14) MM cell growth in vivo, KMS11 and LP1 cells containing doxycycline-inducible shRNA-targeting AK2 or scramble shRNA were injected into NOD-SCID mice that were fed a diet containing doxycycline (Figure 2L). Induced AK2 depletion suppressed tumor growth in the KMS11 xenograft model and completely inhibited tumor formation in LP1 xenografts (Figure 2M-N).
NSD2 overexpression increases MM cell dependence on AK2. (A) Plot of MAGeCK-generated AK2 Beta scores and the corresponding FDRs in NTKO and TKO cells. (B) Depmap AK2 dependency scores across different cancer cell line lineages (red - MM cell lines; blue - other hematologic cell lines; grey - solid cancer cell lines). t(4;14) MM cell lines are labeled. (C) Comparison of AK2 dependency scores between t(4;14) and non-t(4;14) MM cell lines. In B and C, horizontal lines represent median gene effect scores. ∗P < .05 Mann-Whitney, one-sided. (D) Kaplan-Meier curves showing overall survival (OS and PFS of patients with MM stratified by AK2 expression into low (Q1; blue), mid (Q2+Q3; green), and high (Q4; red). P value were calculated by Cox proportional hazards regression. (E) Hazard ratio of AK2 expression associated with OS and PFS in t(4;14) (red) and other (blue) patients with MM as determined using Cox proportional hazards regression. Lines represent 95% confidence intervals RNAseq and WGS. P value and cohort sizes are shown (total N = 725, baseline patients with CoMMpass). (F) Schematic representation of confirmatory in vitro competitive growth assays used for validating essential genes. (G) Time-dependent depletion of GFP-positive NTKO and TKO isogenic cells after transduction with LentiCRISPRv2-GFP expressing the indicated sgRNAs. sgROSA and sgPCNA were used as negative and positive controls, respectively. (H) Comparison of time-dependent depletion of GFP-positive cells harboring sgAK2 in NTKO, TKO, and NSD2-replete TKO cells. (I) Competitive growth of nonlabeled NTKO and GFP-labeled TKO cells treated with DMSO or the AK2 inhibitor AP5A (50 μM) for 72 h. (J) Time-dependent depletion of control and NSD2-depleted KMS34 cells after transduction with LentiCRISPRv2-GFP expressing the indicated sgRNAs. Cells were transduced with LentiCRISPRv2 expressing a nontargeting (sgNT) or NSD2-targeting (sgNSD2) sgRNA and selected with puromycin before transduction with the LentiCRISPRv2-GFP constructs. All experiments were performed in biological triplicate. (K) Time-dependent depletion of t(4;14) and non-t(4;14) MM cell lines transduced with inducible Cas9/GFP/sgAK2 (TLCV2) vectors. Percentage of GFP-positive cells were measured at the indicated times by flow cytometry. Experiments were performed in biological duplicate. (L) Subcutaneous flank injections of cells expressing doxycycline-inducible scrambled (shSc) or AK2-targeting (shAK2) shRNA into NOD/SCID mice. Mice were fed a doxycycline chow diet. (M) KMS11-derived tumors isolated from the left (shSc) and right (shAK2) mouse flanks. Tumor dimensions were measured using a caliper and the volumes were calculated using the following formula: V = ½ L x W2. (N) Tumors derived from shSc-expressing LP1 cells. No tumors from AK2-depleted LP1 cells were detected. Immunoblots confirming continued AK2 expression in isolated LP1-derived tumors are shown. ∗∗P < . 01, ∗∗∗P < . 001, and ∗∗∗∗P < . 0001. DMSO, dimethyl sulfoxide; sgROSA, sgRNA targeting mouse Rosa26 locus; sgPCNA; TLCV2, LentiCRISPRv2 with Tet Response Element Promoter; WGS, whole genome sequencing.
NSD2 overexpression increases MM cell dependence on AK2. (A) Plot of MAGeCK-generated AK2 Beta scores and the corresponding FDRs in NTKO and TKO cells. (B) Depmap AK2 dependency scores across different cancer cell line lineages (red - MM cell lines; blue - other hematologic cell lines; grey - solid cancer cell lines). t(4;14) MM cell lines are labeled. (C) Comparison of AK2 dependency scores between t(4;14) and non-t(4;14) MM cell lines. In B and C, horizontal lines represent median gene effect scores. ∗P < .05 Mann-Whitney, one-sided. (D) Kaplan-Meier curves showing overall survival (OS and PFS of patients with MM stratified by AK2 expression into low (Q1; blue), mid (Q2+Q3; green), and high (Q4; red). P value were calculated by Cox proportional hazards regression. (E) Hazard ratio of AK2 expression associated with OS and PFS in t(4;14) (red) and other (blue) patients with MM as determined using Cox proportional hazards regression. Lines represent 95% confidence intervals RNAseq and WGS. P value and cohort sizes are shown (total N = 725, baseline patients with CoMMpass). (F) Schematic representation of confirmatory in vitro competitive growth assays used for validating essential genes. (G) Time-dependent depletion of GFP-positive NTKO and TKO isogenic cells after transduction with LentiCRISPRv2-GFP expressing the indicated sgRNAs. sgROSA and sgPCNA were used as negative and positive controls, respectively. (H) Comparison of time-dependent depletion of GFP-positive cells harboring sgAK2 in NTKO, TKO, and NSD2-replete TKO cells. (I) Competitive growth of nonlabeled NTKO and GFP-labeled TKO cells treated with DMSO or the AK2 inhibitor AP5A (50 μM) for 72 h. (J) Time-dependent depletion of control and NSD2-depleted KMS34 cells after transduction with LentiCRISPRv2-GFP expressing the indicated sgRNAs. Cells were transduced with LentiCRISPRv2 expressing a nontargeting (sgNT) or NSD2-targeting (sgNSD2) sgRNA and selected with puromycin before transduction with the LentiCRISPRv2-GFP constructs. All experiments were performed in biological triplicate. (K) Time-dependent depletion of t(4;14) and non-t(4;14) MM cell lines transduced with inducible Cas9/GFP/sgAK2 (TLCV2) vectors. Percentage of GFP-positive cells were measured at the indicated times by flow cytometry. Experiments were performed in biological duplicate. (L) Subcutaneous flank injections of cells expressing doxycycline-inducible scrambled (shSc) or AK2-targeting (shAK2) shRNA into NOD/SCID mice. Mice were fed a doxycycline chow diet. (M) KMS11-derived tumors isolated from the left (shSc) and right (shAK2) mouse flanks. Tumor dimensions were measured using a caliper and the volumes were calculated using the following formula: V = ½ L x W2. (N) Tumors derived from shSc-expressing LP1 cells. No tumors from AK2-depleted LP1 cells were detected. Immunoblots confirming continued AK2 expression in isolated LP1-derived tumors are shown. ∗∗P < . 01, ∗∗∗P < . 001, and ∗∗∗∗P < . 0001. DMSO, dimethyl sulfoxide; sgROSA, sgRNA targeting mouse Rosa26 locus; sgPCNA; TLCV2, LentiCRISPRv2 with Tet Response Element Promoter; WGS, whole genome sequencing.
AK2 suppression impairs protein folding and increases MM cell sensitivity to PIs
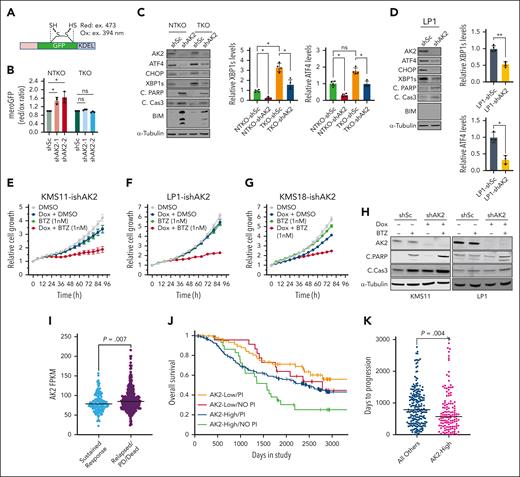
The established function of AK2 in energy metabolism led to the prediction that AK2 deficiency in MM would disrupt ATP-dependent processes, including protein folding in the ER. We used the mammalian ER-localized redox-sensitive green fluorescent protein (mero-GFP) system19 (Figure 3A) to investigate the role of AK2 in protein folding. AK2 suppression led to an increase in unfolded protein load in NSD2-high (NTKO) but not in NSD2-low (TKO) KMS11 cells (Figure 3B). Because the accumulation of unfolded proteins activates the unfolded protein response (UPR), we investigated the effect of AK2 inhibition on UPR signaling. Unexpectedly, AK2 depletion dramatically decreased the levels of UPR signaling components, including spliced X-box binding protein 1 (XBP1s) as well as the proapoptotic UPR arm components activating transcription factor 4 (ATF4) and C/EBP homologous protein (CHOP), but still induced apoptosis in MM cells (Figure 3C-D), suggesting that the AK2 effect on cell viability occurs through a different mechanism. Nevertheless, AK2 depletion increased MM cell sensitivity to the PI bortezomib (Figure 3E-H). In addition, analysis of Multiple Myeloma Research Foundation CoMMpass data from patients with MM revealed that these patients with a sustained response to PI therapy had lower AK2 expression than patients who are relapsed/deceased (Figure 3I). Patients with lower AK2 expression (first quartile) treated with PIs appear to have a survival advantage over patients who are AK2-high (fourth quartile)/PI-treated or those with low AK2 levels who did not receive PI therapy (Figure 3J), although more patients are needed in each of these groups for firm statistical inference. Furthermore, PFS in patients treated with PIs was significantly shorter in patients in the highest quartile of AK2 expression (577 days) compared with the rest of the group (788 days) (Figure 3K).
AK2 depletion increases MM cell sensitivity to proteasome inhibitors. (A) Schematic representation of the MeroGFP system. (B) Evaluation of unfolded protein load indicated by the ratio of reduced/oxidized GFP disulfide bonds measured at the indicated excitation wavelengths by flow cytometry 72 hours after transduction with the indicated shRNAs. (C-D) Immunoblot analyses of proteins involved in unfolded protein response and apoptosis signaling. Four independent experiments were performed 72 hours after transduction with the indicated IPTG induced shRNAs. (E-G) Incucyte cell proliferation assays in KMS11, LP1, and KMS18 cells expressing doxycycline-inducible shRNA-targeting AK2. Cells were treated with doxycycline (0.2 μg/mL), bortezomib (1nM), or a combination of both. (H) Immunoblot analysis showing levels of PARP and caspase 3 cleavage in KMS11 and LP1 cells with doxycycline-inducible shAK2 in the presence or absence of bortezomib. Immunoblots were performed 72 hours after doxycycline induction of AK2 knockdown. (I) Analysis of data from the MMRF CoMMpass study of patients treated with proteasome inhibitor comparing levels of AK2 expression between patients showing sustained response and those that are relapsed or deceased. (J) Kaplan-Meier curves showing overall survival of patients with MMwith high (fourthquartile) or low (first quartile) AK2 expression with or without PI therapy. (K) Comparison of relapse times between AK2-high (fourth quartile) and all other patients with MM treated with PIs. Data were analyzed using the MMRF Researcher Gateway. ∗P < .05, ∗∗P < .01. IPTG, isopropyl ß-D-1-thiogalactopyranoside; MMRF, Multiple Myeloma Research Foundation; PARP, poly(ADP-ribose) polymerase.
AK2 depletion increases MM cell sensitivity to proteasome inhibitors. (A) Schematic representation of the MeroGFP system. (B) Evaluation of unfolded protein load indicated by the ratio of reduced/oxidized GFP disulfide bonds measured at the indicated excitation wavelengths by flow cytometry 72 hours after transduction with the indicated shRNAs. (C-D) Immunoblot analyses of proteins involved in unfolded protein response and apoptosis signaling. Four independent experiments were performed 72 hours after transduction with the indicated IPTG induced shRNAs. (E-G) Incucyte cell proliferation assays in KMS11, LP1, and KMS18 cells expressing doxycycline-inducible shRNA-targeting AK2. Cells were treated with doxycycline (0.2 μg/mL), bortezomib (1nM), or a combination of both. (H) Immunoblot analysis showing levels of PARP and caspase 3 cleavage in KMS11 and LP1 cells with doxycycline-inducible shAK2 in the presence or absence of bortezomib. Immunoblots were performed 72 hours after doxycycline induction of AK2 knockdown. (I) Analysis of data from the MMRF CoMMpass study of patients treated with proteasome inhibitor comparing levels of AK2 expression between patients showing sustained response and those that are relapsed or deceased. (J) Kaplan-Meier curves showing overall survival of patients with MMwith high (fourthquartile) or low (first quartile) AK2 expression with or without PI therapy. (K) Comparison of relapse times between AK2-high (fourth quartile) and all other patients with MM treated with PIs. Data were analyzed using the MMRF Researcher Gateway. ∗P < .05, ∗∗P < .01. IPTG, isopropyl ß-D-1-thiogalactopyranoside; MMRF, Multiple Myeloma Research Foundation; PARP, poly(ADP-ribose) polymerase.
AK2 depletion in NSD2-high MM cells induces DNA damage response
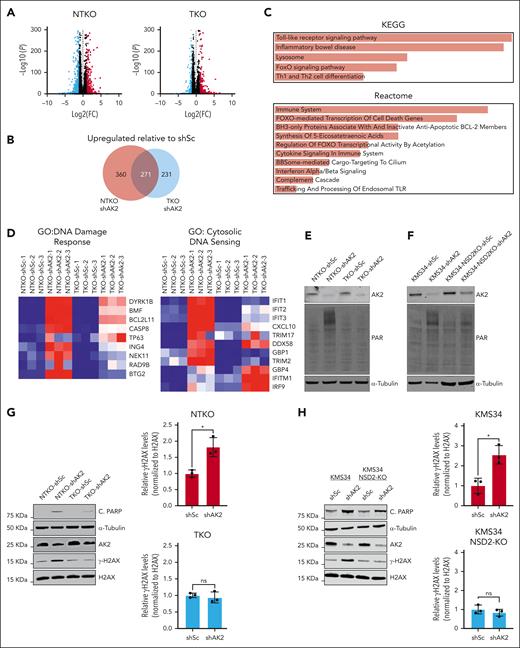
RNAseq analysis of KMS11-derived MM cell lines revealed that AK2 suppression induces more pronounced gene expression changes in NSD2-overexpressing cells (Figure 4A-B). Among the genes upregulated by AK2 suppression are those implicated in apoptotic signaling, DNA damage response, cytosolic DNA sensing (Figure 4C-D), and oxidative stress response (supplemental Figure 2I). Induction of apoptosis in AK2-deficient cells was confirmed by elevated Bcl-2 interacting mediator of cell death (BIM) levels and increased poly(ADP-ribose) polymerase (PARP) cleavage and rescue by treatment with the caspase inhibitor carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone (Z-VAD-FMK) (supplemental Figure 2J-K). However, there was no difference in the formation of reactive oxygen species when AK2 was suppressed in MM cells, suggesting that apoptosis was induced through a reactive oxygen species-independent mechanism (supplemental Figure 2L). To further evaluate the effect of AK2 depletion on DNA damage, we measured protein PARylation in MM cells after shRNA-mediated depletion of AK2. AK2 suppression induced PARylation in the NSD2-overexpressing t(4;14) MM cells KMS11-NTKO and KMS34 but not in the NSD2-low KMS11-TKO cells or NSD2-deficient KMS34 cells (Figure 4E-F). We further confirmed induction of DNA damage response in AK2-depleted cells by measuring H2AX phosphorylation (γH2AX) in NSD2-high and -low MM cell pairs after AK2 suppression. Elevated γH2AX levels were detected in AK2-depleted NTKO, KMS34, and LP1 cells in association with increased PARP cleavage (Figure 4G-H; supplemental Figure 3A). In contrast, γH2AX levels were unaffected by AK2 suppression in NSD2-low TKO cells or NSD2-deficient KMS34 cells (Figure 4G-H). However, the induction of γH2AX in response to AK2 depletion was restored upon repleting NSD2 in TKO cells (supplemental Figure 3B). These findings imply that AK2 depletion induces MM cell apoptosis in part through increasing DNA damage and/or inhibiting DNA damage repair.
AK2 suppression induces DNA damage response in NSD2-overexpressing MM cells. (A) Volcano plots showing genes differentially expressed between AK2-depleted (shAK2) and control (shSc) NTKO and TKO MM cells plotted from RNA-seq data. FC: fold change shAK2/shSc. FDR: false discovery rate. (B) Area proportional Venn diagram of genes upregulated by AK2 suppression in NTKO and TKO MM cells plotted from RNA-seq data. (C) EnrichR pathway analysis of genes upregulated by AK2 suppression in NTKO cells. (D) Heatmaps of genes involved in DNA damage response and cytosolic DNA sensing plotted from RNAseq data of AK2-suppressed NTKO and TKO cells. (E-F) Immunoblot analysis showing protein PARylation in control and AK2-depleted KMS11 NTKO and TKO (E) or parental and NSD2-deficient KMS34 (F) isogenic cells. (G-H) Immunoblot analysis of basal levels of H2AX phosphorylation and cleaved PARP in control and AK2-depleted KMS11 NTKO and TKO (G) or parental and NSD2-deficient KMS34 (H) isogenic cells. Three independent experiments were performed 72 hours after transduction with the indicated shRNA constructs. ∗P <. 05.
AK2 suppression induces DNA damage response in NSD2-overexpressing MM cells. (A) Volcano plots showing genes differentially expressed between AK2-depleted (shAK2) and control (shSc) NTKO and TKO MM cells plotted from RNA-seq data. FC: fold change shAK2/shSc. FDR: false discovery rate. (B) Area proportional Venn diagram of genes upregulated by AK2 suppression in NTKO and TKO MM cells plotted from RNA-seq data. (C) EnrichR pathway analysis of genes upregulated by AK2 suppression in NTKO cells. (D) Heatmaps of genes involved in DNA damage response and cytosolic DNA sensing plotted from RNAseq data of AK2-suppressed NTKO and TKO cells. (E-F) Immunoblot analysis showing protein PARylation in control and AK2-depleted KMS11 NTKO and TKO (E) or parental and NSD2-deficient KMS34 (F) isogenic cells. (G-H) Immunoblot analysis of basal levels of H2AX phosphorylation and cleaved PARP in control and AK2-depleted KMS11 NTKO and TKO (G) or parental and NSD2-deficient KMS34 (H) isogenic cells. Three independent experiments were performed 72 hours after transduction with the indicated shRNA constructs. ∗P <. 05.
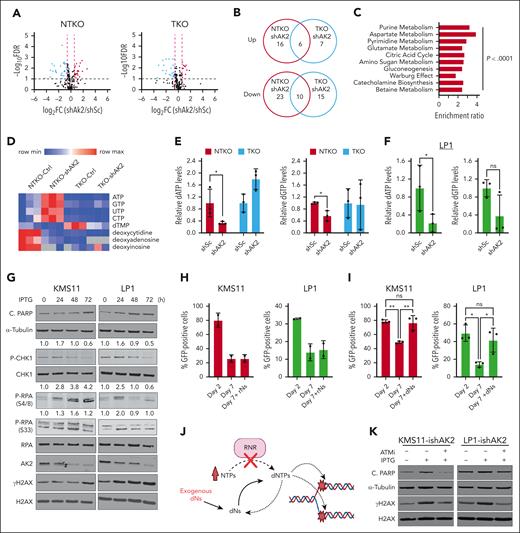
AK2 suppression in NSD2-high MM cells depletes dNTP pools, resulting in increased replication stress
To investigate the role of AK2 in MM cell metabolism and DNA damage and repair, untargeted metabolomic profiling was performed on AK2-depleted KMS11 NTKO and TKO cells. Correlating with the growth suppressive phenotype of AK2 depletion in NSD2-high cells, AK2 depletion altered a greater number of metabolites in NSD2-overexpressing NTKO cells than in isogenic TKO cells (Figure 5A-B; supplemental Figure 3C). Nucleotide metabolism and energy metabolism pathways were among the primary metabolic pathways altered by AK2 suppression (Figure 5C). Analysis of individual metabolites altered by AK2 knockdown exclusively in NSD2-high NTKO cells revealed accumulation of nucleoside triphosphates (NTPs) and depletion of deoxynucleoside triphosphate (dNTP) metabolites (Figure 5D), suggesting a defect in ribonucleotide reductase (RNR) activity. Depletion of dATP and dGTP levels upon AK2 suppression was further confirmed by reverse transcriptase–based dNTP quantification assays (Figure 5E-F). Prolonged dNTP deprivation can result in replication fork stalling and the generation of DNA double-strand breaks.20 Consequently, we demonstrated that AK2 suppression induces transient phosphorylation of replication protein A (RPA) and checkpoint kinase 1 (CHK1), sensors of damaged replication forks, followed by γH2AX phosphorylation and induction of apoptosis as indicated by increased PARP cleavage (Figure 5G). AK2-deficient cells displayed reduced S phase entry (supplemental Figure 3D), consistent with the cell cycle arrest induced by replication stress. Importantly, exogenous deoxyribonucleosides, but not ribonucleosides, rescued the viability of AK2-deficient MM cells, supporting the role of AK2 in regulating RNR activity (Figure 5H-J). ATM kinase plays a key role in the induction of apoptosis in response to DNA damage.21 Treatment of AK2-suppressed cells with the selective ATM inhibitor KU55933 alleviated γH2AX phosphorylation and prevented the induction of apoptosis (Figure 5K). Collectively, these data indicate that AK2 depletion induces cell death through the induction of DNA damage.
AK2 suppression in NSD2-overexpressing MM cells results in DNA replication stress by depleting dNTPs. (A) Volcano plots from metabolomic profiling showing metabolites altered by AK2 suppression in NTKO and TKO cells. (B) Venn diagrams showing metabolites upregulated and downregulated by AK2 depletion NTKO and TKO cells. (C) Pathway analysis of metabolites significantly altered by AK2 suppression in NTKO but not TKO cells. (D) Heatmap of the relative abundance of ribonucleotides and deoxyribonucleotides in AK2-depleted NTKO and TKO cells plotted from the metabolomics data. (E-F) Quantification of dATP and dGTP levels in control and AK2-depleted NSD2-high and -low MM cells by RT-based dNTP assays. All metabolite measurements were performed 72 hours after transduction with the indicated shRNAs in 3 biological replicates. (G) Time-course analysis of replication stress and apoptosis markers after IPTG induction of shAK2 in KMS11 and LP1 MM cell lines. Immunoblots for phosphorylated CHK1 and RPA were quantified by Image J and the signal was normalized to total CHK1 or RPA proteins, respectively. (H-I) In vitro competitive growth assays showing time-dependent depletion of GFP-positive cells after transduction with LentiCRISPRv2-GFP-sgAK2 in the presence and absence of 10× Embryomax nucleoside mix (rNs; 300 μM) or deoxynucleoside pools (dNs; 250 μM). Experiments were performed in 2 or 3 biological replicates as indicated by the individual replicate symbols. (J) Schematic representation of DNA replication fork stalling due to depletion of dNTP pools resulting from impaired RNR activity. (K) Immunoblot analysis of H2AX phosphorylation and PARP cleavage 72 hours after IPTG-induced AK2 depletion in KMS11 and LP1 cells in the presence and absence of 10 μM of the ATM kinase inhibitor (ATMi) KU55933. ∗P< .05, ∗∗P<.01.
AK2 suppression in NSD2-overexpressing MM cells results in DNA replication stress by depleting dNTPs. (A) Volcano plots from metabolomic profiling showing metabolites altered by AK2 suppression in NTKO and TKO cells. (B) Venn diagrams showing metabolites upregulated and downregulated by AK2 depletion NTKO and TKO cells. (C) Pathway analysis of metabolites significantly altered by AK2 suppression in NTKO but not TKO cells. (D) Heatmap of the relative abundance of ribonucleotides and deoxyribonucleotides in AK2-depleted NTKO and TKO cells plotted from the metabolomics data. (E-F) Quantification of dATP and dGTP levels in control and AK2-depleted NSD2-high and -low MM cells by RT-based dNTP assays. All metabolite measurements were performed 72 hours after transduction with the indicated shRNAs in 3 biological replicates. (G) Time-course analysis of replication stress and apoptosis markers after IPTG induction of shAK2 in KMS11 and LP1 MM cell lines. Immunoblots for phosphorylated CHK1 and RPA were quantified by Image J and the signal was normalized to total CHK1 or RPA proteins, respectively. (H-I) In vitro competitive growth assays showing time-dependent depletion of GFP-positive cells after transduction with LentiCRISPRv2-GFP-sgAK2 in the presence and absence of 10× Embryomax nucleoside mix (rNs; 300 μM) or deoxynucleoside pools (dNs; 250 μM). Experiments were performed in 2 or 3 biological replicates as indicated by the individual replicate symbols. (J) Schematic representation of DNA replication fork stalling due to depletion of dNTP pools resulting from impaired RNR activity. (K) Immunoblot analysis of H2AX phosphorylation and PARP cleavage 72 hours after IPTG-induced AK2 depletion in KMS11 and LP1 cells in the presence and absence of 10 μM of the ATM kinase inhibitor (ATMi) KU55933. ∗P< .05, ∗∗P<.01.
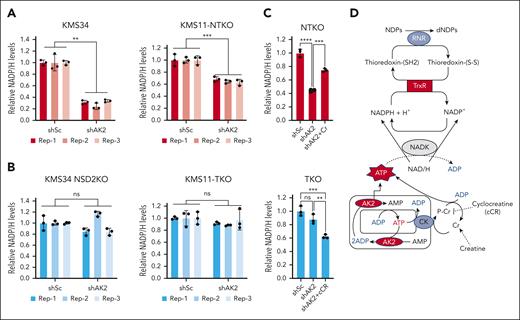
AK2 depletion in NSD2-high MM cells decreases cellular NADP/H levels
Electrons required for the reduction of ribonucleotides by RNR are ultimately provided by NADPH.22 Cytosolic NADP/H is created by phosphorylation of NAD/H by NAD kinase using ATP as the phosphate donor.23 Accordingly, NADPH depletion due to a relative deficiency of cytoplasmic ATP could underlie the compromised conversion of ribonucleotides to deoxyribonucleotides in AK2-deficient t(4;14) MM cells. Indeed, AK2 suppression diminished NADP/H levels in all the t(4;14) MM cell lines examined (Figure 6A; supplemental Figure 4A). However, when NSD2 is depleted, AK2 suppression has no effect on NADP/H levels (Figure 6B), correlating with the diminished effect of AK2 loss on dNTP levels. By catalyzing the transfer of high-energy phosphate, AK2 facilitates the shuttling of mitochondrial ATP to different subcellular compartments, including the cytosol. Defective mitochondrial ATP transport can result in the accumulation of ATP in the mitochondrial matrix, which would compromise mitochondrial respiration. Accordingly, Seahorse flux analysis showed that depletion of AK2 in t(4;14) MM cell lines impaired basal mitochondrial respiration (supplemental Fig. 4B-C), suggesting accumulation of ATP within the mitochondria. To investigate whether NADPH depletion in AK2-deficient cells is due to poor ATP efflux to the cytosol, we treated cells with creatine, which can transfer high-energy phosphate to the cytosol via the creatine/phosphocreatine shuttle, to restore cytosolic ATP levels. Remarkably, creatine supplementation of AK2-deficient NTKO cells rescued NADPH levels (Figure 6C), whereas treatment of AK2-depleted TKO cells with cycloreatine, which blocks the creatine/phosphocreatine shuttle, exacerbated NADPH depletion (Figure 6D). Taken together, these results indicate that energy transfer from the mitochondria, facilitated by AK2 and/or the creatine/phosphocreatine shuttle, is required for NADP/H homeostasis. A proposed model for the role of AK2 in modulating NADP/H is shown in Figure 6D.
AK2 suppression depletes NADP/H levels in NSD2-overexpressing MM cells. (A-B) Relative quantification of NADP/H in control (shSc) and AK2-depleted (shAK2) KMS11 and KMS34-derived NSD2-high (A) and NSD2-low (B) isogenic cells. The different colors indicate different biological replicates, and the dots represent technical replicates. (C) Relative quantification of NADP/H in control and AK2-depleted NTKO cells with and without creatine (Cr; 50mM) supplementation. (D) Relative quantification of NADP/H in control and AK2-depleted TKO cells with and without cyclocreatine (cCR; 10mM) treatment. NADP/H levels were measured by enzyme-based bioluminescent assays 72 hours after IPTG-induced AK2 knockdown. Experiments were performed in biological triplicate. (E) Schematic diagram of the proposed role of AK2 in dNTP homeostasis. RNR: ribonucleotide reductase. TrxR, thioredoxin reductase; NADK: NAD+ kinase; CK, creatine kinase. ∗∗P, 0.01, ∗∗∗P, 0.001.
AK2 suppression depletes NADP/H levels in NSD2-overexpressing MM cells. (A-B) Relative quantification of NADP/H in control (shSc) and AK2-depleted (shAK2) KMS11 and KMS34-derived NSD2-high (A) and NSD2-low (B) isogenic cells. The different colors indicate different biological replicates, and the dots represent technical replicates. (C) Relative quantification of NADP/H in control and AK2-depleted NTKO cells with and without creatine (Cr; 50mM) supplementation. (D) Relative quantification of NADP/H in control and AK2-depleted TKO cells with and without cyclocreatine (cCR; 10mM) treatment. NADP/H levels were measured by enzyme-based bioluminescent assays 72 hours after IPTG-induced AK2 knockdown. Experiments were performed in biological triplicate. (E) Schematic diagram of the proposed role of AK2 in dNTP homeostasis. RNR: ribonucleotide reductase. TrxR, thioredoxin reductase; NADK: NAD+ kinase; CK, creatine kinase. ∗∗P, 0.01, ∗∗∗P, 0.001.
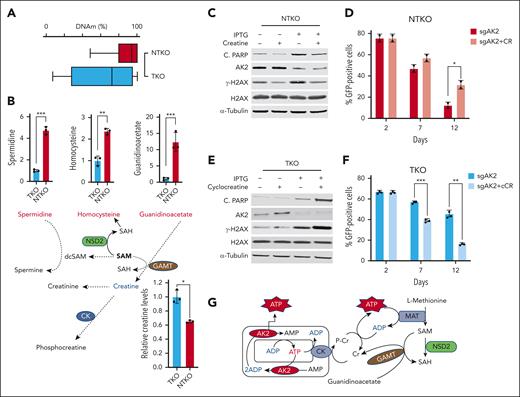
Depletion of creatine levels by NSD2 underlies increased dependence on AK2
Previous studies demonstrated that NSD2-high MM cells exhibit a sixfold to eightfold increase in H3K36 dimethylation compared with NSD2-low cells.5 In addition, we showed that DNA methylation levels were 23% higher in NTKO than TKO cells (Figure 7A). These findings suggest an NSD2-driven increased flux of methyl groups into the genome. All histone and DNA methyltransferases use S-adenosylmethionine (SAM) as the methyl donor. Metabolomic profiling of isogenic NSD2-high (NTKO) and NSD2-low (TKO) MM cells revealed significant changes in metabolites related to one carbon metabolism, including increased levels of homocysteine in NTKO cells, which indicates depletion of SAM (Figure 7B). There was also the accumulation of spermidine (Figure 7B), which requires decarboxylated SAM to be metabolized to spermine, and 2-ketoisovaleric acid (supplemental Figure 5A), which accumulates when there is a lack of carbon available for its hydroxymethylation to ketopantoic acid.24 In addition, there was a striking accumulation of the creatine precursor guanidinoacetate in NSD2-high cells (Figure 7B). Guanidinoacetate conversion to creatine is catalyzed by guanidinoacetate N-methyltransferase, which uses SAM as a methyl donor. Importantly, guanidinoacetate accumulation associated with NSD2 overexpression correlated with a drop in creatine levels (Figure 7B). Collectively, these results suggest that increased SAM consumption in NSD2-overexpressing MM cells impedes creatine synthesis.
NSD2-derived creatine depletion underlies increased dependence on AK2. (A) DNA methylation in KMS11 NTKO and TKO cells measured by whole-genome bisulfite sequencing (coverage at 19,190,528). (B) Differential abundance of metabolites related to SAM/creatine metabolism in NTKO and TKO MM cells. Relative guanidinoacetate, homocysteine, and spermidine levels were calculated from the metabolomics data. Creatine levels were determined using an enzyme-based fluorometric assay. (C) Immunoblot analysis of H2AX phosphorylation and PARP cleavage in KMS11 NTKO cells 72 hours after IPTG induction of AK2 suppression with and without creatine (50 mM) supplementation. (D) Time-dependent depletion of GFP-positive NTKO cells after transduction with LentiCRISPRv2-GFP-sgAK2 in the presence and absence of creatine (50 mM). (E) Immunoblot analysis of H2AX phosphorylation and PARP cleavage in KMS11 TKO cells 72 hours after IPTG induction of AK2 suppression with or without cyclocreatine (10 mM) treatment. (F) Time-dependent depletion of GFP-positive TKO cells after transduction with LentiCRISPRv2-GFP-sgAK2 in the presence and absence of cyclocreatine (10 mM). Quantitative experiments were performed in 2 or 3 biological replicates as indicated by the individual replicate symbols. (G) Schematic diagram showing the compensatory role of creatine in replenishing cytosolic ATP levels. dcSAM, decarboxylated SAM; SAH, S-adenosyl homocysteine; GAMT, guanidinoacetate N-methyltransferase; CK, creatine kinase. MAT, methionine adenosyltransferase. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
NSD2-derived creatine depletion underlies increased dependence on AK2. (A) DNA methylation in KMS11 NTKO and TKO cells measured by whole-genome bisulfite sequencing (coverage at 19,190,528). (B) Differential abundance of metabolites related to SAM/creatine metabolism in NTKO and TKO MM cells. Relative guanidinoacetate, homocysteine, and spermidine levels were calculated from the metabolomics data. Creatine levels were determined using an enzyme-based fluorometric assay. (C) Immunoblot analysis of H2AX phosphorylation and PARP cleavage in KMS11 NTKO cells 72 hours after IPTG induction of AK2 suppression with and without creatine (50 mM) supplementation. (D) Time-dependent depletion of GFP-positive NTKO cells after transduction with LentiCRISPRv2-GFP-sgAK2 in the presence and absence of creatine (50 mM). (E) Immunoblot analysis of H2AX phosphorylation and PARP cleavage in KMS11 TKO cells 72 hours after IPTG induction of AK2 suppression with or without cyclocreatine (10 mM) treatment. (F) Time-dependent depletion of GFP-positive TKO cells after transduction with LentiCRISPRv2-GFP-sgAK2 in the presence and absence of cyclocreatine (10 mM). Quantitative experiments were performed in 2 or 3 biological replicates as indicated by the individual replicate symbols. (G) Schematic diagram showing the compensatory role of creatine in replenishing cytosolic ATP levels. dcSAM, decarboxylated SAM; SAH, S-adenosyl homocysteine; GAMT, guanidinoacetate N-methyltransferase; CK, creatine kinase. MAT, methionine adenosyltransferase. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
As creatine phosphorylation by mitochondrial creatine kinase (CK) provides a means of replenishing cytosolic ATP, depletion of cellular creatine due to the shift in one-carbon metabolism resulting from NSD2 overexpression appears to account for the increased dependence on AK2 in t(4;14) MM cells. Exogenous creatine restores NADP/H levels in NSD2-high, AK2-deficient MM cells (Figure 6C). Accordingly, creatine supplementation prevented the ability of AK2 depletion to induce DNA damage and apoptosis in NSD2-high cells (Figure 7C; supplemental Figure 5B-C) and partially reversed the impact of AK2 loss on cell survival (Figure 7D). The effect of creatine supplementation was specific for AK2-depleted NSD2-high MM cells, as exogenous creatine did not rescue NTKO MM cells from dexamethasone-induced apoptosis (supplemental Figure 5D). By contrast, treatment of NSD2-low TKO MM cells with cyclocreatine, which inhibits CK activity, exacerbated the effect of AK2 depletion, increasing DNA damage as measured by H2AX phosphorylation and cell death (Figure 7E-F).
Collectively, these findings indicate that NSD2 overexpression creates a new synthetic lethal vulnerability. Loss of AK2 is more lethal to NSD2-high cells because of the shift of one-carbon metabolism to the epigenome, leading to creatine deficiency and a subsequent failure to efficiently transfer high-energy phosphates from mitochondria to the cytosol (Figure 7G).
Discussion
The genetic heterogeneity of MM has rendered strategies targeting the cancer biology of the disease less successful25 than other tumor types. Personalized therapies targeting vulnerabilities emerging from specific genetic lesions represent prospective MM treatments. However, for most molecular subtypes of MM, such liabilities have yet to be identified. The t(4;14) chromosomal translocation, a driver of NSD2 overexpression, remains a predictor of poor prognosis in MM, even in the era of novel therapies. Thus, selective targeting of NSD2-overexpressing t(4;14) MM cells remains of great interest.
We interrogated the genome for genes more essential for MM cell survival when NSD2 is overexpressed. Our approach revealed several metabolic perturbations whose impact on MM cell viability is exacerbated by NSD2 overexpression. In addition to metabolic dependencies, NSD2-overexpressing myeloma cells appeared to be more reliant on processes related to DNA repair. This result is consistent with the previously demonstrated increased DNA damage and improved DNA repair in MM cells overexpressing NSD2.26 Enrichment analysis of the identified gene candidates also indicated that NSD2-overexpressing MM cells are more dependent on proteasomal degradation, which was further confirmed by increased sensitivity to bortezomib. This result is in line with the finding that PIs improved the prognosis of patients with t(4;14) MM.27
The mitochondrial adenine nucleotide regulator AK2 was identified by our screening approach as one of the chief NSD2-driven dependencies in MM. The critical role of AK2 in the hematological system was first manifested in reticular dysgenesis (RD), a form of SCID associated with inactivating AK2 mutations.28 Extreme lymphocytopenia in patients with RD suggests that AK2 is critical for both B and T lymphocytes. A recent report on patients with hypomorphic AK2 variants, however, indicated that B cells were more dependent on AK2 function.29 Furthermore, T-cell acute lymphoblastic leukemia cells were found to be highly dependent on AK2, as the depletion of AK2 in these cells disrupted mitochondrial bioenergetics, a finding we also noted in MM cells, and induced apoptosis.30 However, until this point, the function of AK2 in B-cell malignancies, including MM, was unclear.
Localized in the mitochondrial intermembrane space, AK2 catalyzes the transfer of high-energy phosphate to sites of energy use within the cell.31 As a major location for protein folding and trafficking, the ER is a key site for ATP use. We found that AK2 depletion in NSD2-high MM cells impedes protein folding, consistent with a role for AK2 in shuttling ATP from the mitochondria to the ER. Nevertheless, a direct link of AK2 to protein folding requires further investigation. Moreover, despite increasing the burden of unfolded proteins, AK2 suppression in MM cells dampens the UPR, indicating that the induction of apoptosis by AK2 loss in MM cells is not mediated by ER stress signaling. Consistent with this finding, AK2 depletion in differentiated B cells was shown to inhibit activation of the UPR, which was attributed to a defect in ATP-dependent XBP1 splicing.32 We, therefore, reasoned that ER stress triggered by the increased load of unfolded proteins in AK2-deficient MM cells is alleviated by high proteasomal degradation rates in MM cells. In line with this notion, bortezomib collaborated with AK2 depletion to induce apoptosis of MM cells, which represents a potential novel therapeutic strategy.
Additional investigation identified DNA replication stress as the mechanism underlying the adverse impact of AK2 depletion on t(4;14) MM cells. AK2 suppression impaired the conversion of ribonucleotides to deoxyribonucleotides by RNR, which was inferred by the depletion of intracellular deoxyribonucleotides and concomitant accumulation of ribonucleotides. Insufficient cellular levels of dNTPs can cause premature termination of DNA synthesis, replication stress, and subsequent DNA damage.33 Accordingly, we found that AK2 depletion in NSD2-high MM cells induces replication stress, DNA damage, and apoptosis. Moreover, we showed that supplementation with exogenous deoxyribonucleosides but not ribonucleosides rescues MM cells from apoptosis induced by AK2 depletion. This finding is in accordance with the established role of deoxyribonucleosides in RNR-independent dNTP salvage.34
Electrons for the reductive conversion of nucleotides to deoxynucleotides are provided by thioredoxin, which is in turn reduced by thioredoxin reductase using NADPH as an electron donor.35,36 As NADPH is key for RNR activity, we postulated that AK2 suppression disrupts the conversion of nucleotides to deoxyribonucleotides by decreasing NADPH levels. Our results showed that AK2 suppression was not associated with elevated oxidative stress, indicating that the observed defect in RNR activity is not due to an increased NADP+/NADPH ratio. Nonetheless, quantification of total NADP/H revealed a substantial decrease in NADP/H levels, exclusively in NSD2-overexpressing MM cells, when AK2 levels are suppressed. These findings suggested that NADP/H depletion underlies disrupted nucleotide homeostasis and replication stress in AK2-deficient t(4;14) MM cells.
An alternative mechanism to AK2-mediated mitochondrial high-energy phosphate transport is the creatine/phosphocreatine shuttle, by which the mitochondrial CK uses ATP in the intermembrane space to phosphorylate creatine in the cytosol.37 Accordingly, creatine supplementation of AK2-deficient t(4;14) MM cell lines restored NADP/H levels, confirming the critical role of mitochondrial ATP export in NADP/H homeostasis. The mechanism by which the cytosolic ATP supply maintains NADP/H levels likely involves NAD/H phosphorylation by the cytosolic NAD kinase, in which ATP serves as the phosphate donor.23
Epigenomic and metabolic studies in NSD2-high and -low MM cells demonstrated that NSD2 overexpression blocks SAM-dependent creatine synthesis by diverting SAM to the epigenome. The resulting creatine deficiency underlies the increased dependence of t(4;14) MM cells on AK2, as creatine supplementation rescued these cells from replication stress and apoptosis induced by AK2 suppression. Consistent with the observed compensatory interplay between AK2 and the creatine/phosphocreatine system, redistribution of phosphotransfer through adenylate kinase networks was reported to complement the loss of creatine kinase function in the muscles of CK-deficient mice.38 Similarly, cyclocreatine-mediated disruption of the creatine/phosphocreatine system in NSD2-low MM cells, where creatine levels are sufficient, exacerbates the effect of AK2 depletion, further supporting the functional redundancy of the 2 energy distribution systems.
Our study elucidates the mechanism by which NSD2 drives increased dependence of MM cells on AK2 and provides insight on potential tissue-specific determinants of AK2 dependence in other cancers. AK2 deficiency underlies RD, and our findings suggest that replication stress, DNA damage, and apoptosis may underlie the loss of lymphocytes noted in this disease. Why hematopoietic and particularly lymphoid malignancies as well as normal lymphocytes show an increased sensitivity to AK2 loss remains uncertain but may be related to both a high dependence on oxidative phosphorylation for energy production and the relatively low levels of CK found in hematopoietic cells versus other tissues (GTex Portal).
Acknowledgments
The authors thank S. Wohlgemuth for help with Seahorse XF experiments and data analysis and members of the Interdisciplinary Center for Biotechnology Research (ICBR) at UF especially the Bioinformatics (RRID:SCR_019120) Cytometry (RRID:SCR_019119), Gene Expression (RRID:SCR_019145) and Next Generation Sequencing (RRID:SCR_019152) Core Facilities.
This work was supported by Florida Department of Health Grant 22B10, Leukemia and Lymphoma Society (LLS) Specialized Center of Research Grant 7021-20, Translational Research Program grant 6657-23 (J.D.L.), Myeloma Solutions Fund IMMPACT award (J.D.L., L.H.B., B.G.B., C.S.M.), American Association for Cancer Research (AACR)-Takeda Oncology Myeloma Research Fellow grant 19-40-38-SOBH, LLS Special Fellow grant 3417-22 (A.S.), the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases AI162633 and AI136581 (B.K.), NIH, National Cancer Institute (NCI) R21CA280584 and K22CA266739 (B.G.B.), The Paula and Roger Riney Foundation (L.H.B.), NIH/NCI R01CA19836 and R01241191 (D.Z.), NIH/NCI 5F30CA250236 (K.V.), and NIH/NCI 1R35CA197532 (N.S.C.). The University of Florida Health Cancer Center, supported in part by state appropriations provided in Fla. Stat. § 381.915 and NIH/NCI (P30CA247796). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authorship
Contribution: A.S., and J.D.L. conceived the study and designed experiments; A.S., E.E., A.P., G.S., E.B., C.K., J.P., M.C., K.V., T.F., D.D-.R., and D.L. performed experiments; A.S., K.V., T.F., A.R., and B.G.B. performed data analysis; D.Z., L.H.B., C.S.M., B.K., R.L.B. N.S.C., and J.D.L. supervised the work; and A.S., and J.D.L., wrote the manuscript with input from all authors.
Conflict-of-intreset disclosure: C.S.M. serves on the scientific advisory board of Adicet Bio and discloses consultant/honoraria from Genentech, Nerviano, Secura Bio and Oncopeptides, and research funding from EMD Serono, Karyopharm, Sanofi, Nurix, Bristol Myers Squibb, H3 Biomedicine/Eisai, Springworks, Abcuro, Novartis, and Opna Bio. The remaning authors declare no competing financial intresets.
Correspondence: Jonathan D. Licht, University of Florida Health Cancer Center, Cancer/Genetics Research Complex, 2033 Mowry Rd, Suite 145, Gainesville, FL 32610; email: jdlicht@ufl.edu.
References
Author notes
Data Availability Next generation sequencing data corresponding to the CRISPR screens and RNAseq experiments were deposited to the Gene Expression Omnibus (accession number GSE245150).
Untargeted metabolomics data have been uploaded to the National Institutes of Health Common Fund’s National Metabolomics Data Repository (ST002876).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal