Novel complementary functions between β-catenin and Hoxa9 in regulating Prmt1 to protect stem cell quiescence and DNA replication dynamics.
Prmt1 KO phenocopies and its re-expression partially rescues cellular and molecular defects of β-catenin/Hoxa9 KO.
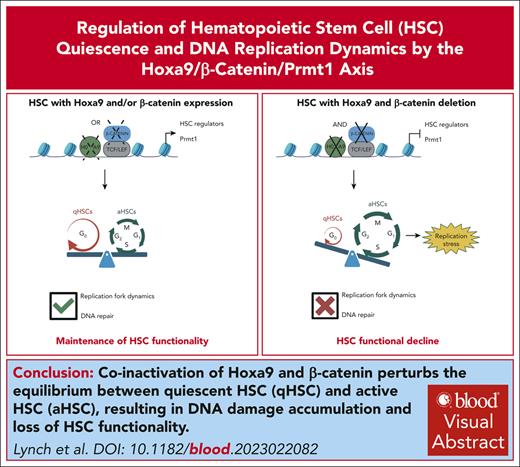
Visual Abstract
Maintenance of quiescence and DNA replication dynamics are 2 paradoxical requirements for the distinct states of dormant and active hematopoietic stem cells (HSCs), which are required to preserve the stem cell reservoir and replenish the blood cell system in response to hematopoietic stress, respectively. Here, we show that key self-renewal factors, β-catenin or Hoxa9, largely dispensable for HSC integrity, in fact, have dual functions in maintaining quiescence and enabling efficient DNA replication fork dynamics to preserve the functionality of hematopoietic stem and progenitor cells (HSPCs). Although β-catenin or Hoxa9 single knockout (KO) exhibited mostly normal hematopoiesis, their coinactivation led to severe hematopoietic defects stemmed from aberrant cell cycle, DNA replication, and damage in HSPCs. Mechanistically, β-catenin and Hoxa9 function in a compensatory manner to sustain key transcriptional programs that converge on the pivotal downstream target and epigenetic modifying enzyme, Prmt1, which protects the quiescent state and ensures an adequate supply of DNA replication and repair factors to maintain robust replication fork dynamics. Inactivation of Prmt1 phenocopied both cellular and molecular phenotypes of β-catenin/Hoxa9 combined KO, which at the same time could also be partially rescued by Prmt1 expression. The discovery of the highly resilient β-catenin/Hoxa9/Prmt1 axis in protecting both quiescence and DNA replication dynamics essential for HSCs at different key states provides not only novel mechanistic insights into their intricate regulation but also a potential tractable target for therapeutic intervention.
Introduction
Life-long regeneration of the blood system relies on a rare population of self-renewing hematopoietic stem cells (HSCs) that are capable of upkeeping faithful genome replication and integrity.1-6 Maintenance of quiescence/dormancy not only allows cells to persist in nondividing state over extended time to preserve self-renewal potential, but also protects against DNA damage and functional exhaustion.1,2,7 However, HSCs must also respond efficiently to hematological stress such as infection, blood loss, or exposure to cytotoxic agents by re-entering active cell cycle to rapidly generate functional progenitors and progenies of different lineages. In contrast to quiescent HSCs (qHSCs), active HSCs (aHSCs) must robustly replicate their genome to replenish the hematopoietic system without depleting the stem population.8-10 So far, factors have been identified to enable either but not both features (eg, cyclin-dependent kinase inhibitors for quiescence and minichromosome maintenance [MCM] proteins for genome replication),3,11,12 suggesting the maintenance of each cell status by distinct molecular pathways. Although this stepwise regulation of different pathways in governing status switching is in line with linear discrete hematopoietic differentiation model, emerging evidence indicate that HSC differentiation and acquisition of lineage-specific fates is a continuous process, characterized by a highly coordinated transcriptional program.4,13,14 Given the interfluent requirement of qHSC and aHSC states that enable swift adaptation to changing requirements,13,15 it remains to be determined if there are indeed common pathways operating to safeguard both quiescence and efficient DNA replication.4,7,12
To gain novel insights into the molecular regulation of HSC functions, signaling pathways such as Homeobox, Wnt, Notch, and Hedgehog, which are frequently dysregulated in cancer, have been shown to govern self-renewal in hematopoietic stem and progenitor cells (HSPCs).16 Ectopic expression of Hoxa917-20 or β-catenin,21-26 the key component of canonical Wnt pathway,27 promotes HSC expansion and eventually leukemic transformation.16 Importantly, overexpression of either factor is highly predictive of poor patient prognosis, whereas their inhibitions suppress leukemia development, leading to intense therapeutic interests in targeting these factors.28,29 However, inactivation of β-catenin or Hoxa9 has only modest impacts on adult HSCs, suggesting (1) the existence of alternative pathways that support hematopoietic self-renewal in the absence of either factor or (2) their otherwise distinct physiological functions in normal HSCs that are different from those upon their overexpression. Extensive searches for such alternative pathways over the past decade (eg, γ-catenin in β-catenin knockout (KO) and Hoxb3/4 in Hoxa9 KO) have so far yielded little success, casting doubts about the physiological roles of these key self-renewal molecules in HSC functions.30,31 Clearly, resolving this dilemma will have profound implications on stem cell biology and also strategic therapeutic design. How HSCs sustain their functions in the absence of either factor or what the underlying mechanisms are remains enigmatic.
Here we establish, for the first time, how endogenous β-catenin and Hoxa9 function in a novel complementary manner with dual roles in protecting HSCs from exit of quiescence and accumulation of replication stress, in which activation of key downstream epigenetic enzyme Prmt1 is responsible, at least in part, for circumventing functional decline of HSCs.
Methods
Mouse models and transplantation studies
β-cateninfl/fl mice32 were crossed with Hoxa9−/− mice19,33 to generate Hoxa9−/−β-cateninfl/fl Rosa-CreER mice. Conditional Prmt1 KO mouse models were generated using targeted enrichment score cell clones of C57BL/6, as described previously.34 Further details are provided in supplemental Methods, available on the Blood website. A total of 1 × 106 CD45.2+ donor bone marrow cells were transplanted IV into lethally irradiated (13 Gy total body γ-irradiation) C57BL/6 congenic strain expressing pan-leukocyte marker CD45.1 (Jackson Laboratory; JAX:002014) recipient mice along with 2 × 105 CD45.1+ helper bone marrow cells. Deletion of floxed alleles were achieved via daily intraperitoneal injection of tamoxifen (60 μL at 20 mg/mL) for 5 consecutive days.
Immunophenotype analysis and sorting
Femora and tibiae were isolated from mice that received transplant and gently crushed in staining medium (Phosphate buffered saline + 2% fetal bovine serum) before red blood cell lysis. Detailed staining procedure for stem and progenitor populations can be found in supplemental Methods. Stained cells were analyzed on a BD LSRII flow cytometer using the gating strategy illustrated in supplemental Figure 1F and described previously.35
Ki67 and BrdU immunostaining
For in vivo bromodeoxyuridine (BrdU) incorporation analysis, mice that received transplant were intraperitoneally injected with 150 μL (10 mg/mL solution) BrdU (BD Biosciences) 2 weeks after induction of target gene deletion. After 48 hours, mice were euthanized and femora, tibiae, and pelvis were isolated, and LSKs (Lin–c-Kit+Sca1+) were sorted by fluorescence-activated cell sorting. For Ki67 staining, LSK cells were isolated from transplanted mice 2 weeks after targeted gene deletion. Isolated LSKs were fixed, permeabilized, and stained as outlined in supplemental Methods.
Immunofluorescent staining
Bone marrow isolated from KO mouse models were enriched for cKit+ cells using CD117 Micro-Beads (Miltenyi Biotec) by positive selection according to manufacturer’s instructions. ckit+ HSPCs were treated with either 4-hydroxytamoxifen (25 ng/mL; Sigma) or ethanol (vehicle) for 48 hours before fixation and staining as outlined in supplemental Methods.
DNA fiber assay
CD45.2+ ckit+ HSPCs were isolated from mice that received transplant, and 5000 cells per sample were incubated at 37°C for 30 minutes in StemSpan medium (StemCell Technologies). Subsequently, 19 mM 5-Chloro-20-deoxyuridine (CldU; Sigma) was added to the medium for 30 minutes. Medium was exchanged, and cells were incubated with 28 mM 5-iodo-20-deoxyuridine (IdU; Sigma) for 30 minutes. DNA fibers were then spread on glass slides and stained as described previously.36,37
RNA sequencing
Total RNA from LSK cells was isolated using the RNeasy Plus Micro Kit (Qiagen), and Library preparation was performed using Ultra II Directional RNA Library Prep Kit for Illumina (New England BioLabs, E7760L), according to manufacturer’s instructions. Detailed information in supplemental Methods.
Assay for Transposase Accessible Chromatin sequencing
A total of 50 000 LSK cells from wild-type (WT) or KO mouse models were used for assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq), which was performed as described previously with minor modifications.38 Peaks were detected using function callpeak from the MACS2 tool with ‘--format BAMPE’ option.39 Profile plots were generated with computeMatrix and plotProfile from deepTools.40
CUT&RUN sequencing
Cleavage under targets and release using nuclease (CUT&RUN) sequencing was performed using 0.5 million HSPCs from WT BL6 mice or Ctnnb1-Biotin-3xFLAG knockin mice (JAX:029511) according to Epicypher CUT&RUN Protocol V2.0. Detailed information is provided in supplemental Methods.
Results
Coinactivation of Hoxa9 and β-catenin induces severe defects in hematopoietic stem populations
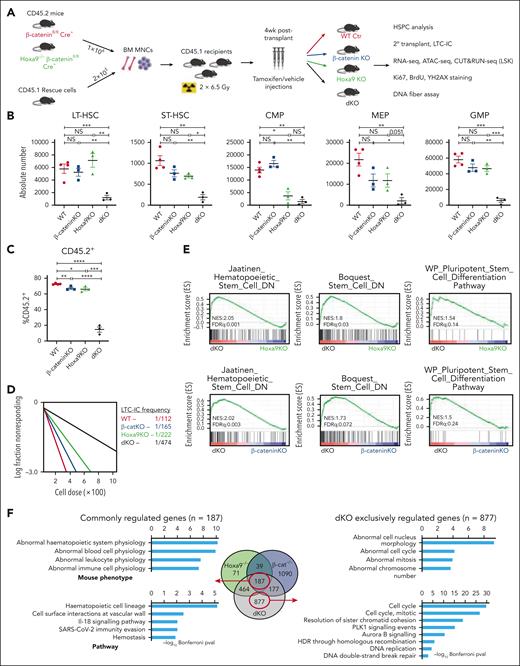
To identify potential molecular pathways that sustain stem cell integrity in the absence of Hoxa9 or β-catenin, RNA-sequencing was performed on LSKs harboring inactivation of either factor. To our surprise, among the top candidate pathways involved in maintaining HSC functions are canonical Wnt signaling and Hoxa9 pathways upon the reciprocal inactivation of Hoxa9 and β-catenin respectively (supplemental Figure 1A-B). Although no prior molecular link has been reported in HSCs between these 2 pathways, the hypothesis of their novel cross talk in preserving HSC functionality is consistent with our recent discovery of the coregulation of posterior Hoxa loci and Ctnnb by long noncoding RNA, HOTTIP.17,26 To further investigate this novel cross talk, we generated a Hoxa9-/-Ctnnb1fl/fl Cre ER mouse model by crossing our previously described Hoxa9 KO mice33 with Ctnnb1 floxed mice carrying Rosa26-CreER allele23 for serial bone marrow transplant assays (Figure 1A; supplemental Figure 1C-E). In contrast to single Hoxa9 or β-catenin inactivation, in which we observed only mild hematopoietic phenotypes with a noticeable reduction of common myeloid progenitor as previously reported,33,41 combined inactivation of Hoxa9/β-catenin (dKO) resulted in severe hematopoietic defects at all measured time points (Figure 1B; supplemental Figure 1F-P). Defects originated as early as the long-term HSC stage, with a substantial sixfold reduction and a fivefold reduction in short-term HSCs at 12-week time point compared with WT counterparts. Consistent with a stem cell defect, we also observed significant reductions in downstream myeloid progenitor populations including common myeloid progenitor (eightfold), granulocyte-monocyte progenitor (10-fold), and megakaryocyte and erythrocyte progenitors (sevenfold). In line with these findings, the absolute number and percentage of total CD45.2+ donor cells were drastically reduced in the dKO bone marrow (17%) compared with single KOs (67%) (Figure 1C; supplemental Figure 1Q-R). At a functional level, in vitro clonogenic assays revealed that dKO bone marrow cells generated markedly reduced myeloid colony numbers, of which mature CFU-G and CFU-E were the predominant composition (supplemental Figure 1S). LSKs isolated from dKO mice generated significantly fewer colonies and were devoid of multipotential progenitors (CFU-GEMM), in contrast to the diverse colony composition of control and single KO cells (supplemental Figure 1T). The frequency of long-term culture initiating cells (LTC-ICs) was also significantly diminished in dKO HSPCs (Figure 1D), consistent with functional decline of HSCs. Finally, the ability of dKO cells to engraft and reconstitute the bone marrow of secondary recipient mice was severely impaired compared with WT and single KO cells (supplemental Figure 1U-V). Together, these data suggest that coinactivation of β-catenin/Hoxa9 results in significant functional defects in the stem population.
Coinactivation of Hoxa9/β-catenin induces severe HSC defects. (A) Schematic representation of experimental procedures. Absolute number of HSPC populations (B) and percentage CD45.2+ donor cells (C) from indicated mice at 12 week time point (at least n = 3 mice/group). Floxed allele deletion was achieved by intraperitoneal injection of tamoxifen for 5 consecutive days. β-cateninfl/fl mice treated with corn oil (vehicle) served as WT control as indicated. (D) LTC-IC frequency from HSPCs as indicated. Enrichment in stem related gene sets (E) and gene ontology analysis (F) of LSKs isolated from mice that received transplant. All data represent mean ± standard error of the mean (SEM). P values calculated by t test. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001; NS, P > .05.
Coinactivation of Hoxa9/β-catenin induces severe HSC defects. (A) Schematic representation of experimental procedures. Absolute number of HSPC populations (B) and percentage CD45.2+ donor cells (C) from indicated mice at 12 week time point (at least n = 3 mice/group). Floxed allele deletion was achieved by intraperitoneal injection of tamoxifen for 5 consecutive days. β-cateninfl/fl mice treated with corn oil (vehicle) served as WT control as indicated. (D) LTC-IC frequency from HSPCs as indicated. Enrichment in stem related gene sets (E) and gene ontology analysis (F) of LSKs isolated from mice that received transplant. All data represent mean ± standard error of the mean (SEM). P values calculated by t test. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001; NS, P > .05.
To gain further insight into the mechanisms underlying the stem cell defects in dKO HSCs, RNA-sequencing analysis was performed and revealed a significant reduction of stem-related gene sets in dKO LSKs compared with either β-catenin KO or Hoxa9 KO alone. Indeed, genes downregulated in HSCs (Jaatinen_Hematopoeietic_Stem_Cell_DN and Boquest_Stem_Cell_DN) were positively enriched in dKO LSKs, with additional enrichment in stem cell differentiation pathways (WP_Stem_Cell_Differentiation) (Figure 1E; supplemental Table 1), suggesting their roles in maintaining stemness. Comparison of differentially expressed genes (DEGs ≥ 1.5-fold; P adjusted ≤ .05) in Hoxa9 KO and β-catenin KO LSKs revealed that a small subset (n = 226) is coordinately regulated by the single KOs, consistent with an independent and compensatory relationship between these 2 pathways in governing HSC functions (Figure 1F). Strikingly, Gene Ontology analysis revealed that the DEGs exclusively regulated by dKO LSKs (n = 877) were overwhelmingly correlated with cellular processes involved in regulating cell cycle, DNA replication, double-strand break repair, and genomic stability (Figure 1F), suggesting their key functions in regulating cell cycle, DNA replication, and repair. Further mechanistic insight into the Hoxa9/β-catenin cross talk was revealed by CUT&RUN sequencing on endogenous HOXA9 and endogenously FLAG-tagged β-CATENIN in HSPCs. Genome-wide chromatin mapping of HOXA9 and β-CATENIN binding demonstrated a striking correlation in binding site occupancy (supplemental Figure 1W). Consistent with RNA sequencing analysis, the regions of HOXA9/β-CATENIN co-occupancy were enriched in pathways regulating cell cycle, DNA repair, and replication in addition to stem cell pluripotency pathways, with co-occupancy detected at the loci of HSC regulators including Myc, Fos, Jarid2, and Stat3 (supplemental Figure 1X-Y).
Concurrent Hoxa9/β-catenin deletion disrupts HSC quiescence and induces replication stress and DNA damage accumulation
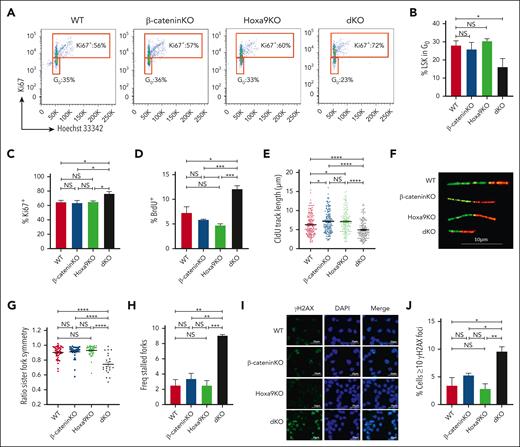
We next sought to functionally investigate the effect of β-catenin/Hoxa9 coinactivation on quiescent state of HSCs, which greatly aids their ability to preserve their genomic integrity and that of all future downstream lineages. LSKs were isolated from single and dKO transplanted mice 2 weeks after deletion and stained with the proliferation marker Ki67 and the DNA content dye Hoechst 33342 to examine their quiescent vs proliferation status. As a control, WT mice were treated with the thymidylate synthase inhibitor, fluorouracil (5-FU), known to induce replication stress resulting in the exit of HSCs from quiescence and entry into cell activation programs.42 Similarly, dKO LSKs displayed a marked reduction in the frequency of cells within the G0 subset or Ki67– fraction and a concomitant increase in Ki67+ fraction compared with WT or single Hoxa9/β-catenin KO (Figure 2A-C; supplemental Figure 2A). To further confirm the increase in the fraction of actively cycling LSKs, in vivo BrdU incorporation analysis was performed. Consistent with Ki67 staining, dKO LSKs exhibited a significantly greater frequency of BrdU+ cells than WT or single β-catenin/Hoxa9 KO (Figure 2D; supplemental Figure 2B). Together, these findings indicate that although the complementary functions between β-catenin and Hoxa9 are capable of resiliently preserving HSC functionality upon its single KO, their combined inactivation drive HSCs out from their quiescent state into active cycling.
Disruption of stem cell quiescence and DNA replication dynamics by concurrent Hoxa9/β-catenin deletion. (A) Representative fluorescence-activated cell sorting plots of Ki67 and Hoechst 33342 staining of LSKs isolated from indicated mice. (B) Percentage of LSKs in G0 cell cycle phase. Percentage Ki67+ (C) and BrdU+ (D) LSKs (n = 3 mice per group). (E) CldU track length of DNA fibers from HSPCs isolated from indicated mice. (F) Representative images of replication fork lengths. Scale bar, 10 μm. (G) Ratio of IdU track lengths of bidirectional forks. (H) Percentage of stalled forks (green only) present in single/combined KO cells. Fiber data from 1 representative experiment is shown (n = 3; data is shown in supplemental Figure 2). Immunofluorescent images (I) and focus counts (J) of ƳH2AX staining on HSPCs isolated from indicated mice. Several fields of cells were scored from 2 independent experiments. Scale bars, 25 μm. WT BL6 mice treated with corn oil (vehicle) are indicated as WT control. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001; NS, P > .05.
Disruption of stem cell quiescence and DNA replication dynamics by concurrent Hoxa9/β-catenin deletion. (A) Representative fluorescence-activated cell sorting plots of Ki67 and Hoechst 33342 staining of LSKs isolated from indicated mice. (B) Percentage of LSKs in G0 cell cycle phase. Percentage Ki67+ (C) and BrdU+ (D) LSKs (n = 3 mice per group). (E) CldU track length of DNA fibers from HSPCs isolated from indicated mice. (F) Representative images of replication fork lengths. Scale bar, 10 μm. (G) Ratio of IdU track lengths of bidirectional forks. (H) Percentage of stalled forks (green only) present in single/combined KO cells. Fiber data from 1 representative experiment is shown (n = 3; data is shown in supplemental Figure 2). Immunofluorescent images (I) and focus counts (J) of ƳH2AX staining on HSPCs isolated from indicated mice. Several fields of cells were scored from 2 independent experiments. Scale bars, 25 μm. WT BL6 mice treated with corn oil (vehicle) are indicated as WT control. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001; NS, P > .05.
Effective and faithful DNA replication is fundamental for genomic integrity and cell cycle progression, which is particularly important after HSCs exit from quiescence. In order to functionally investigate whether DNA replication dynamics were indeed altered upon inactivation of β-catenin/Hoxa9 as suggested by the RNA-sequencing analyses, HSPCs isolated from primary mice that received transplant were pulse-labeled with nucleotide analogues (CldU and IdU), and DNA fibers were spread to monitor replication fork progression at the single-molecule level (supplemental Figure 2C-D). Combined inactivation of β-catenin and Hoxa9 significantly compromised the speed of replication forks evident by reduced fork track length in dKO cells compared with WT or single KO cells (Figure 2E-F; supplemental Figure 2E). Consistent with their heightened levels of replication stress, dKO HSPCs displayed asymmetrical progression of sister forks from recently fired origins and an overall higher frequency of stalled forks in the population of events (Figure 2G-H; supplemental Figure 2F-H). Moreover, dKO HSPCs displayed an accumulation of γH2AX foci indicative of double-strand breaks and fork collapse (Figure 2I-J; supplemental Figure 2I). Together, these results are consistent with a novel function of β-catenin/Hoxa9 in protecting HSPCs from replication stress resulting in replication fork stalling, accumulating DNA damage, and functional decline of HSCs.
Prmt1 KO phenocopies the severe hematopoietic, quiescent, DNA replication and damage defects associated with β-catenin/Hoxa9 KO
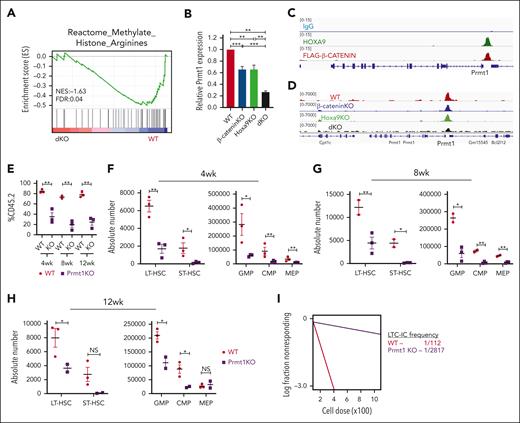
To identify key downstream players that mediate the dKO phenotype, further investigation of gene set enrichment analysis of RNA-sequencing data generated from dKO LSKs revealed a significant negative enrichment in arginine histone methylation (Figure 3A). Protein arginine methyltransferase 1 (PRMT1) is the predominant epigenetic enzyme that catalyzes the asymmetric dimethylation of arginine, and its overexpression has been implicated in numerous solid and hematological cancers leading to significant clinical interest in developing PRMT-targeted therapeutics.34,43,44 Further evaluation of Prmt1 in dKO LSKs revealed a substantial reduction in Prmt1 expression compared with single KO LSKs (Figure 3B; supplemental Figure 3A). CUT&RUN sequencing revealed co-occupancy of HOXA9 and β-CATENIN at Prmt1 transcription start site (Figure 3C). At the same time, ATAC-sequencing analysis revealed reduced chromatin accessibility at the Prmt1 promoter region in dKO LSKs (Figure 3D), consistent with epigenetic downregulation of Prmt1 expression upon coinactivation of β-catenin/Hoxa9. To further investigate the role of Prmt1 in hematopoietic development, we generated a novel Prmt1 KO model in which Prmt1 can be conditionally inactivated in HSCs upon tamoxifen treatment. Deletion of Prmt1 alone phenocopied the β-catenin/Hoxa9 dKO, resulting in severe reductions in long-term HSC, short-term HSC, and downstream progenitor populations at all analyzed time points (Figure 3E-H; supplemental Figure 3B-F). Both in vitro clonogenic assays and LTC-IC assays demonstrated functional decline of HSPCs with impaired colony forming ability and significantly reduced frequency of LTC-IC (supplemental Figure 3G; Figure 3I). KO of Prmt1 resulted in severe stem cell defects as evidenced by the inability of Prmt1 KO cells to engraft and reconstitute in secondary recipient mice (supplemental Figure 3H).
Prmt1 KO phenocopies the hematopoietic defects associated with dKO. (A) Negative enrichment in arginine methyltransferase activity in isolated dKO LSKs. (B) Independent qPCR validation of Prmt1 expression in indicated LSKs. Data represent 2 independent experiments. (C) Co-occupancy of HOXA9 and β-CATENIN at the Prmt1 transcription start site detected by CUT&RUN-sequencing in HSPCs. (D) Chromatin accessibility at the Prmt1 locus visualized by ATAC-sequencing of LSKs. (E) Percentage of donor cells in bone marrow of transplanted mice at indicated time points. Absolute number of HSPCs at 4-week (F), 8-week (G), and 12-week (H) harvests (n = 3 mice per group per time point). (I) LTC-IC frequency of HSPCs isolated from mice that received transplant. Prmt1fl/fl mice treated with corn oil (vehicle) are indicated as WT control. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; NS, P > .05. qPCR, quantitative polymerase chain reaction.
Prmt1 KO phenocopies the hematopoietic defects associated with dKO. (A) Negative enrichment in arginine methyltransferase activity in isolated dKO LSKs. (B) Independent qPCR validation of Prmt1 expression in indicated LSKs. Data represent 2 independent experiments. (C) Co-occupancy of HOXA9 and β-CATENIN at the Prmt1 transcription start site detected by CUT&RUN-sequencing in HSPCs. (D) Chromatin accessibility at the Prmt1 locus visualized by ATAC-sequencing of LSKs. (E) Percentage of donor cells in bone marrow of transplanted mice at indicated time points. Absolute number of HSPCs at 4-week (F), 8-week (G), and 12-week (H) harvests (n = 3 mice per group per time point). (I) LTC-IC frequency of HSPCs isolated from mice that received transplant. Prmt1fl/fl mice treated with corn oil (vehicle) are indicated as WT control. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; NS, P > .05. qPCR, quantitative polymerase chain reaction.
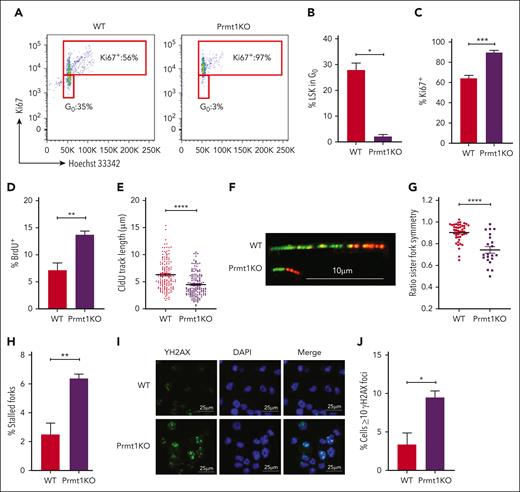
Importantly, Prmt1-deficient LSKs displayed features of hematopoietic and replication stress, consistent with the observed β-catenin/Hoxa9 (dKO) phenotypes. These included a diminishing G0 (quiescent) fraction (Figure 4A-B) and corresponding increases in Ki67+ (Figure 4C) and BrdU+ (Figure 4D; supplemental Figure 4A) fractions. Perturbations in replication dynamics were evidenced by reduced replication fork speed (Figure 4E-F; supplemental Figure 4B) and stability (Figure 4G-H; supplemental Figure 4C-D), leading to the accumulation of DNA breaks (increased γH2AX foci; Figure 4I,J; supplemental Figure 4E). These findings consistently suggest that Prmt1 may serve as a functional downstream mediator of the β-catenin/Hoxa9 complementary signaling axis that preserves DNA replication dynamics and long-term self-renewal potential that underpin stem cell functionality.
Impaired quiescence and DNA replication dynamics induced by Prmt1 KO. (A) Representative fluorescence-activated cell sorting plots of Ki67 and Hoechst 33342 staining of LSKs isolated from indicated mice 2 weeks after tamoxifen treatment. (B) Percentage of LSKs in G0 cell cycle. Percentage Ki67+ (C) and BrdU+ (D) LSKs from indicated mice (n = 3 per group). (E) CldU track length of DNA fibers from HSPCs isolated from indicated mice. (F) Representative images of elongating fork lengths. Scale bar, 10 μm. (G) Ratio of IdU (red) track lengths of bidirectional forks. (H) Percentage of stalled forks (green only). Fiber data from 1 representative experiment are shown (n = 3, data are shown in supplemental Figure 4). Representative immunofluorescent images (I) and ƳH2AX focus counts (J) of ckit+ Prmt1 WT and KO cells. Several fields of cells were scored from 2 independent experiments. Scale bars, 25 μm. WT BL6 mice treated with corn oil (vehicle) are indicated as WT control. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001.
Impaired quiescence and DNA replication dynamics induced by Prmt1 KO. (A) Representative fluorescence-activated cell sorting plots of Ki67 and Hoechst 33342 staining of LSKs isolated from indicated mice 2 weeks after tamoxifen treatment. (B) Percentage of LSKs in G0 cell cycle. Percentage Ki67+ (C) and BrdU+ (D) LSKs from indicated mice (n = 3 per group). (E) CldU track length of DNA fibers from HSPCs isolated from indicated mice. (F) Representative images of elongating fork lengths. Scale bar, 10 μm. (G) Ratio of IdU (red) track lengths of bidirectional forks. (H) Percentage of stalled forks (green only). Fiber data from 1 representative experiment are shown (n = 3, data are shown in supplemental Figure 4). Representative immunofluorescent images (I) and ƳH2AX focus counts (J) of ckit+ Prmt1 WT and KO cells. Several fields of cells were scored from 2 independent experiments. Scale bars, 25 μm. WT BL6 mice treated with corn oil (vehicle) are indicated as WT control. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001.
Prmt1 KO replicates the transcriptomic and epigenetic alterations induced by dKO
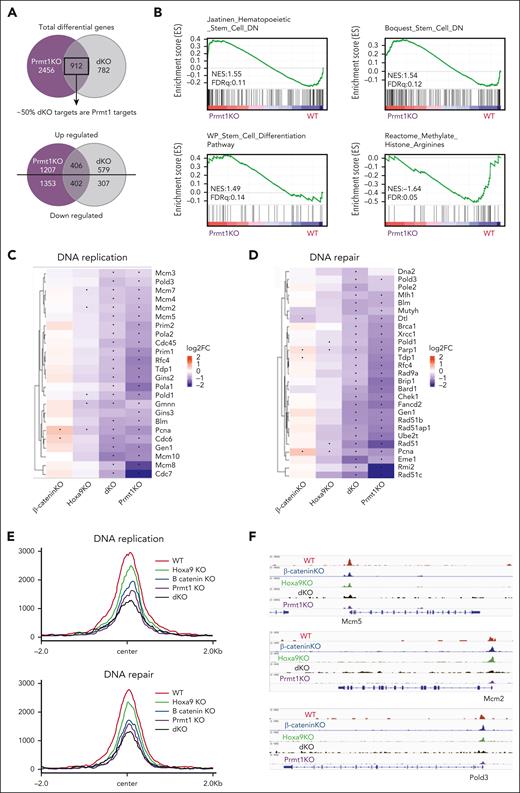
Further comparison of the DEGs revealed a striking correlation between β-catenin/Hoxa9 and Prmt1-regulated targets in LSKs, in which >50% of β-catenin/Hoxa9 targets were also identified as Prmt1 targets, with ∼90% of their common targets regulated in the same direction, consistently suggesting Prmt1 as a key transcriptional mediator for β-catenin/Hoxa9 in HSCs (Figure 5A). In addition to the expected negative enrichment in histone arginine methylation, Prmt1 KO replicated the pathways deregulated in dKO LSKs including abnormal cell cycle, DNA replication, DNA repair (GO: pathway), and chromosomal instability (GO: mouse phenotype) (supplemental Figure 5A). Gene set enrichment analysis of Prmt1 KO LSKs also revealed enrichment in stem cell differentiation pathways and positive enrichment in genes normally downregulated in HSCs (Figure 5B), although it can be possible that this reduction in stemness occurs as a consequence of dysfunctional DNA replication and repair rather than direct regulation of stemness-related genes.
Prmt1 KO replicates the transcriptomic alterations induced by dKO. (A) Overlap of DEGs from Prmt1 KO and dKO LSKs. (B) Gene set enrichment analysis of Prmt1 KO LSKs. Heatmap illustrating the log2fold suppression of DNA replication genes (C) and DNA repair genes (D) in single and dKO LSKs compared with WT LSKs. Prmt1fl/fl and β-cateninfl/fl mice treated with corn oil (vehicle) served as WT control for Prmt1 KO and Hoxa9/β-catenin KO, respectively. Asterisks (∗) indicate significant change in gene expression (P adjusted ≤ .05). (E) Profile plots of ATAC-seq data (RPKM-normalized) around peak center of DNA replication and repair genes. (F) Chromatin accessibility at Mcm5, Mcm2, and Pold3 loci. WT control represents BL6 mice treated with corn oil (vehicle).
Prmt1 KO replicates the transcriptomic alterations induced by dKO. (A) Overlap of DEGs from Prmt1 KO and dKO LSKs. (B) Gene set enrichment analysis of Prmt1 KO LSKs. Heatmap illustrating the log2fold suppression of DNA replication genes (C) and DNA repair genes (D) in single and dKO LSKs compared with WT LSKs. Prmt1fl/fl and β-cateninfl/fl mice treated with corn oil (vehicle) served as WT control for Prmt1 KO and Hoxa9/β-catenin KO, respectively. Asterisks (∗) indicate significant change in gene expression (P adjusted ≤ .05). (E) Profile plots of ATAC-seq data (RPKM-normalized) around peak center of DNA replication and repair genes. (F) Chromatin accessibility at Mcm5, Mcm2, and Pold3 loci. WT control represents BL6 mice treated with corn oil (vehicle).
We then focused our analysis to identify possible transcriptomic alterations that may underpin the observed perturbations to replication dynamics and genomic stability. ATAC- and RNA-sequencing analyzes revealed that both dKO and Prmt1 KO LSKs were characterized by significant deficiencies in both chromatin accessibility and expression of molecules involved in DNA replication and repair compared with WT and single KOs (Figure 5C-F; supplemental Figure 5B-C). Of note, numerous members of the minichromosome maintenance complex (MCM2-7) which binds and licenses origins of replication essential for HSC functionality,11 were among the targets coregulated by β-catenin/Hoxa9 and Prmt1 including Mcm2, Mcm3, Mcm4, Mcm5, and Mcm7. Additionally, targets such as Fancd2, Brca1, and Pcna play a dual role in canonical HR-mediated repair of stalled forks and replication stress tolerance reflecting the extensive crosstalk between the processes of DNA replication and repair. These findings indicate that Prmt1 functions as a pivotal target for the convergence of the β-catenin and Hoxa9 compensatory pathways to maintain an adequate repertoire of replication and repair factors required for protection against the detrimental effects of replication stress after HSCs exit from quiescence.
Prmt1 rescues both cellular and molecular defects of β-catenin/Hoxa9 KO HSPCs
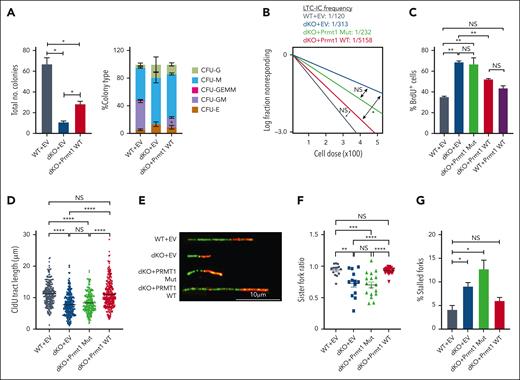
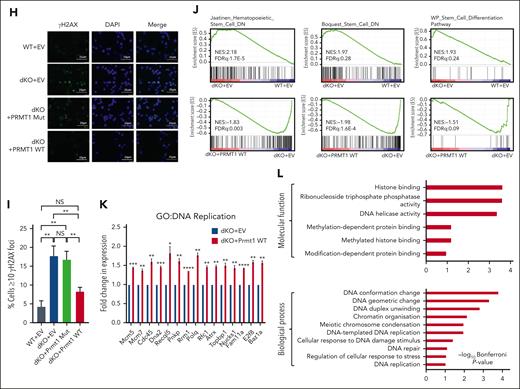
To further interrogate the integral downstream role of Prmt1 in the β-catenin/Hoxa9 signaling axis, we performed ectopic expression of PRMT1 in dKO HSPCs (supplemental Figure 6A). Functional assays demonstrated that PRMT1 could significantly rescue the colony forming ability of dKO LSK cells (Figure 6A) and markedly restore the frequency of LTC-IC (Figure 6B). In addition, dKO HSPCs transduced with WT but not catalytically dead mutant of PRMT1 (Mut) displayed a significantly reduced shift in BrdU incorporation and maintained a profile highly reflective of WT HSPCs, whereas the expression of WT PRMT1 in WT HSPCs did not significantly alter the fraction of BrdU+ cells (Figure 6C; supplemental Figure 6B). Importantly, ectopic expression of WT PRMT1 also protected against perturbations to replication dynamics seen in dKO HSPCs transduced with empty vector or PRMT1 Mut. Replication track length in dKO HSPCs was fully restored by expression of WT but not PRMT1 Mut (Figure 6D-E; supplemental Figure 6C). Similarly, sister fork progression and the frequency of stalled forks in dKO HSPCs were rescued to levels observed in control HSPCs when WT, but not PRMT1 Mut, was ectopically expressed (Figure 6F-G; supplemental Figure 6D-E). Intriguingly, similar to the results from BrdU studies above, overexpression of WT PRMT1 in WT HSPCs did not confer a replicative advantage because no increase in fork speed and no alteration in bidirectional fork symmetry was observed, suggesting that PRMT1 functions in a protective manner to safeguard against replication stress rather than conferring a generalized proliferative or replicative advantage (supplemental Figure 6F-G). Correspondingly, PRMT1 protected against the accumulation of ƳH2AX foci induced by β-catenin/Hoxa9 coinactivation in HSPCs (Figure 6H-I; supplemental Figure 6H).
At the gene expression level, we also demonstrated that enforced expression of WT PRMT1 could restore all the major transcriptomic alterations in the dKO. In addition to stem related gene sets (Figure 6J), PRMT1 also rescued gene expression programs for DNA replication and repair processes including those associated with DNA helicase activity, methylation-dependent protein binding (GO: Molecular Function), and DNA replication and repair (GO: Biological Process) (Figure 6K-L; supplemental Figure 6I-J). In contrast, genes upregulated upon expression of PRMT1 Mut were not associated with any of these functions or processes (supplemental Figure 6K). Together, these functional and transcriptomic data indicate that Prmt1 regulates DNA replication dynamics, genomic integrity, and gene expression programs of HSPCs, thereby preserving stem cell functionality.
Discussion
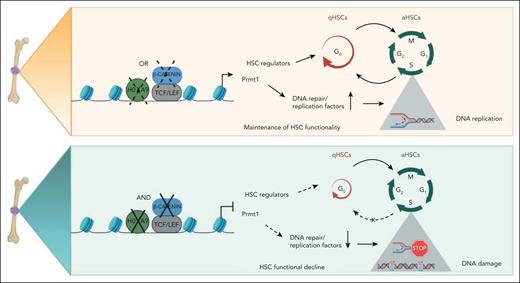
The maintenance of quiescence and replication potential are 2 paradoxical requirements of HSCs that underpin their functionally distinctive dormant and active states, imperative to the lifetime supply of hematopoietic cells. In this study, we depict novel functions of the β-catenin/Hoxa9/Prmt1 axis that robustly preserves HSC functionality by protecting both stem cell quiescence and DNA replication dynamics. We propose that the complementary regulation17,26 and functions between β-catenin and Hoxa9 can provide a resilient safeguard mechanism to protect HSCs against suppression of either single factor (Figure 7, top panel). Only coinactivation of β-catenin and Hoxa9 drives HSCs out of their quiescent state into active cycling (Figure 7, bottom panel). Upon their exit from quiescence, accurate genome duplication that requires the orchestrated activation and maintenance of replication forks emanating from thousands of origins of replication during S-phase of the cell cycle then immediately becomes one of the most important challenges faced by aHSCs.6,45 Perturbations to normal replication fork progression due to exogenous and endogenous stress such as DNA lesions, insufficient nucleotides, chemical compounds, or a shortage of replication factors lead to fork stalling.46,47 Sustained replication stress or the inability to resolve replication obstacles can result in fork collapse, via fork inactivation or endonucleolytic cleavage leading to double-strand breaks.48 Here, we show that the shortage of replication factors such as Mcm2, Mcm3, Mcm4, Mcm5, Mcm7, Cdc7, Fancd2, and Brca1 as a result of Prmt1 downregulation in the β-catenin/Hoxa9 dKO directly impedes replication fork speed and stability, ultimately resulting in an accumulation of double-strand breaks and functional decline of the stem cell population (Figure 7, bottom panel). Indeed, old cycling HSCs in mice were shown to have heightened levels of replication stress associated with cell cycle defects, chromosome breaks, and altered DNA replication fork dynamics, which have been attributed to decreased expression of key replication factors such as MCM helicase components.3,11 The spontaneous recombination events and genomic instability induced by perturbations to replication dynamics are associated with many pathological conditions. For example, homozygous inactivation of any member of the Fanconi anemia replication factors leads to the pediatric syndrome, Fanconi anemia, characterized by progressive bone marrow failure, chromosomal instability, and high cancer predisposition.49,50
Schematic summary of Hoxa9/β-catenin/Prmt1-mediated regulation of HSC quiescence and DNA replication dynamics. (Top panel) The maintenance of HSC functionality in the absence of either Hoxa9 or β-catenin. (Bottom panel) The perturbance to qHSC and aHSC equilibrium and DNA replication dynamics induced by coinactivation of Hoxa9 and β-catenin.
Schematic summary of Hoxa9/β-catenin/Prmt1-mediated regulation of HSC quiescence and DNA replication dynamics. (Top panel) The maintenance of HSC functionality in the absence of either Hoxa9 or β-catenin. (Bottom panel) The perturbance to qHSC and aHSC equilibrium and DNA replication dynamics induced by coinactivation of Hoxa9 and β-catenin.
Increasing evidence has revealed that protein arginine methylation plays critical roles in regulating DNA damage response including methylation of canonical DDR molecules such as MRE11, USP11, BRCA1, and 53BP1.44,51-54 At the same time, Prmt1 mediated H4R3 methylation has been correlated with active transcription,55-57 and our preliminary unpublished CUT&RUN sequencing data also showing potential direct binding of PRMT1 to key DNA replication/repair loci raise a possibility of transcriptional regulation of DNA replication/repair pathways by PRMT1. Here, we reveal a novel role of Prmt1 as a key, albeit likely not the only, mediator of β-catenin and Hoxa9 functions to preserve stem cell quiescence and DNA replication dynamics, in part, by maintaining adequate expression of DNA replication and repair factors at a transcriptional level, thereby, alleviating DNA damage accumulation and preserving HSC integrity (Figure 7). Given the importance of quiescence and replication dynamics in maintaining stem cell functionality, it is not surprising that HSCs have developed intricate cross talk and compensatory signaling pathways to protect such critical cellular processes in the events of genetic and/or epigenetic inactivation of either of the key players. Our finding of novel functions of the β-catenin/Hoxa9/Prmt1 axis in safeguarding both stem cell quiescence and DNA replication fork processivity also provide support for a continuum model to enable dynamic switching between qHSC and aHSC states.4,58,59 Given that this study was conducted solely using mouse models, further future studies in human HSCs will be required to confirm our findings, although previous studies have shown highly conserved hematopoietic functions of Hoxa9 and β-catenin in both mouse and human.20,23,60-62 While the proto-oncogenic and generally undruggable natures of transcription factors make β-catenin and Hoxa9 challenging targets, identification of Prmt1 with tractable enzymatic activity as a key mediator in safeguarding HSC functionality provides a novel avenue for therapeutic intervention.63 Since many of these molecules are also frequently hijacked during malignant transformation and responsible for treatment resistance,19,23,64 a deeper understanding of their roles in maintaining stem cell functions will also aid in the development of antileukemia targeted therapies.
Acknowledgments
The authors thank Wojciech Niedzwiedz, Jadwiga Nieminuszczy, and Cyril Doerdelmann for advice on single-molecule DNA fiber assay; Quan Zho for Prmt1 retroviral constructs; Paul Lavender for pAG-MNase; and Katie Holmes for the initiation of Prmt1 knockout work.
C.W.E.S. is a Royal Society Wolfson Research Merit Award holder. This study received support from the Kay Kendall Leukaemia Fund Junior Fellowship Programme (J.L.). This work is financially supported by Cancer Research UK programme grant C17197/A29213, National Institutes of Health, National Cancer Institute R01 grant R01 CA264932, SNF Sinergia grant CRSII5_213586/1, Research Grant Council Theme-based Research Scheme T12-702/20-N, and Blood Cancer UK project grant 20002.
Authorship
Contribution: J.L. performed majority of the experiments and prepared the manuscript draft; E.T., T.K.F., C.V., P.N.I.L., and N.C. carried out some of the experiments; K.G., B.Z., and J.W.H.W. conducted bioinformatics analyses; M.L. and S.H. advised on experimental design and data interpretation; and C.W.E.S. designed and oversaw the execution of project and preparation of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Chi Wai Eric So, Leukaemia and Stem Cell Biology Group, School of Cancer and Pharmaceutical Sciences, King's College London, London SE5 9NU, United Kingdom; email: eric.so@kcl.ac.uk.
References
Author notes
E.T. and T.K.F. contributed equally to this work.
All RNA-seq, ATAC-seq, and CUT&RUN-seq data are available in Gene Expression Omnibus database (accession number GSE235821).
Data are available upon reasonable request from the corresponding author, Chi Wai Eric So (eric.so@kcl.ac.uk).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.