Abstract
Despite recent progress in identifying the genetic drivers of acute lymphoblastic leukemia (ALL), prognosis remains poor for those individuals who experience disease recurrence. Moreover, acute leukemias of ambiguous lineage lack a biologically informed framework to guide classification and therapy. These needs have driven the adoption of multiple complementary single-cell sequencing approaches to explore key issues in the biology of these leukemias, including cell of origin, developmental hierarchy and ontogeny, and the molecular heterogeneity driving pathogenesis, progression, and therapeutic responsiveness. There are multiple single-cell techniques for profiling a specific modality, including RNA, DNA, chromatin accessibility and methylation; and an expanding range of approaches for simultaneous analysis of multiple modalities. Single-cell sequencing approaches have also enabled characterization of cell-intrinsic and -extrinsic features of ALL biology. In this review we describe these approaches and highlight the extensive heterogeneity that underpins ALL gene expression, cellular differentiation, and clonal architecture throughout disease pathogenesis and treatment resistance. In addition, we discuss the importance of the dynamic interactions that occur between leukemia cells and the nonleukemia microenvironment. We discuss potential opportunities and limitations of single-cell sequencing for the study of ALL biology and treatment responsiveness.
Introduction
ALL represents one of the best examples of how advances in genomic analysis can transform clinical management and improve outcome. Next-generation sequencing studies performed over the past decade have revolutionized the molecular taxonomy of acute lymphoblastic leukemia (ALL) with >30 different subtypes of B- (B-ALL) or T-lymphoid (T-ALL) cell lineage with prognostic and therapeutic significance.1,2 Despite improvements in treatment outcome, relapse poses a major challenge.3,4 Thus, much work remains to be done to elucidate the mechanisms that influence the development, progression, and treatment response. For this purpose, single-cell sequencing (sc-seq) holds promise by enabling interrogation of gene expression, epigenetic heterogeneity, and DNA subclonal architecture with a resolution that cannot be obtained by bulk sequencing (Figure 1).5-10 There are multiple techniques (Table 1; Figure 1) that enable investigation of a specific modality in single cells, including analysis of the transcriptome (scRNA-seq), genome, chromatin accessibility, DNA methylation, proteome, and metabolome. Targeted DNA sequencing of genes and mutational hotspots has become popular to identify copy number variants, single-nucleotide variants, small deletions, and insertions within single cells. This approach is customizable, and multiple validated panels are commercially available for different subtypes of hematological malignancies.11-14 However, single cell targeted DNA sequencing is limited by the inability to identify gene fusions and structural variations, high-resolution karyotypic changes, or novel DNA mutations (unless in the regions targeted). Whole transcriptome sequencing enables comprehensive characterization of fusion transcript chimeras and mutant allele expression and gene expression profiling to identify ALL subgroups and phenocopies (eg, BCR::ABL1-like ALL and ETV6::RUNX1-like ALL). However, current scRNA-seq approaches lack the ability to sensitively identify expressed gene fusions and/or mutations, unless combined with targeted amplification of specific regions of interest. Thus, current single-cell approaches complement existing subtyping methods, but may not identify novel subtypes on their own.
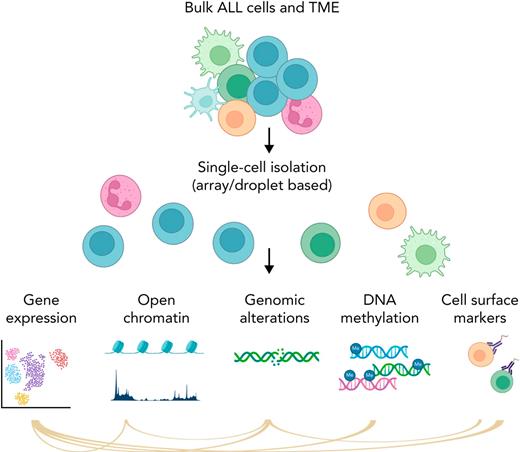
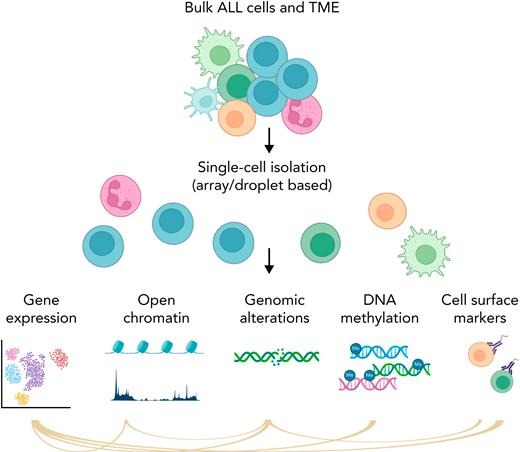
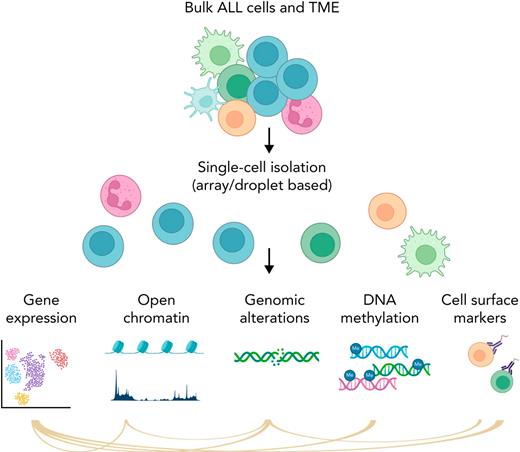
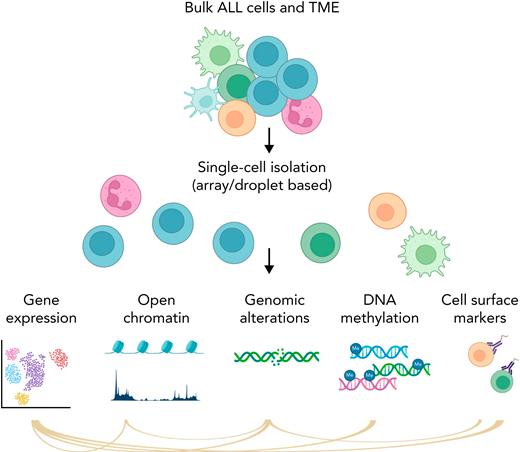
Simplified schema of main single-cell applications. Part of the figure was created in BioRender.com.
Simplified schema of main single-cell applications. Part of the figure was created in BioRender.com.
Multiomic single-cell approaches offer the opportunity to interrogate multiple modalities simultaneously15 and analyze intrasample heterogeneity (Table 1; Figure 1). These include analysis of the DNA methylome and transcriptome,16,17 chromatin accessibility using ATAC-seq (the assay for transposase-accessible chromatin using sequencing) and gene expression18; cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq), which simultaneously measures gene expression and extracellular protein markers by using DNA-barcoded antibodies19; and multiome cell surface protein and DNA sequencing.20-23 These combinatorial approaches provide unprecedented resolution to investigate cellular heterogeneity, and they can improve current leukemia classification by providing insights into cell state and putative cell of origin that bulk sequencing cannot and into interactions with the tumor microenvironment.
Despite the existence of multiple sc-seq methods, these approaches often share a common workflow: separation of single cells, barcoding, lysis, nucleic acid amplification, high-throughput sequencing, and data processing and analysis. Cells can be separated by 2 main methods: droplet-based, in which microfluidics encapsulate and uniquely barcode cells in oil microdroplets; or array based (Table 1), in which cells are dispensed into plate wells or nanowells where they are individually processed.24 Single-cell applications are continuously evolving, and recent advances in spatial transcriptomics not only investigate gene expression but map where it occurs in a tissue sample.25,26 The application of spatial transcriptomics in ALL may further aid characterization of the tumor microenvironment and its role in leukemia development and progression. Multiple recent review articles have described in great detail the up-to-date technologies and applications of sc-seq in cancer research.5,27-30 In this review, we describe new findings from the application of sc-seq approaches to the study of ALL, lineage-ambiguous leukemia, and normal cells (eg, immune microenvironment; Table 2).
Transcriptional heterogeneity in ALL
Several studies have analyzed normal hematopoiesis to gain insights into developmental states of ALL and putative cell of origin. Lee et al31 used CITE-seq to determine differentially expressed gene networks in normal mouse B-cell development and applied them to bulk RNA-seq from different B-ALL subtypes.31 High-risk subtypes (eg, BCL2/MYC, IKZF1 N159Y, and KMT2A-rearranged) were enriched in transcriptional signatures of cycling pro-B–dependent, pre-BCR–dependent, and pre-BCR–independent stages, but these were absent in low-risk subtypes (eg, ETV6::RUNX1 and ZNF384-rearranged). Several ALL subtypes, notably KMT2A-rearranged infant ALL, are thought to arise in utero,32 and Khabirova et al33 analyzed scRNA-seq from ∼60 000 normal fetal bone marrow cells34 and compared the data to those obtained from bulk RNA-seq of 1665 childhood cases of ALL or acute myeloid leukemia (AML). Interestingly, infant KMT2A-rearranged ALL exhibited a gene expression profile similar to that of early lymphocyte precursors, which are fetal-specific oligopotent early lymphoid progenitors that differentiate along different lymphocyte lineages and retain minimal myeloid differentiation capacity in vitro.35 In contrast, NUTM1-rearranged infant ALL, which has a favorable outcome,36 was characterized by transcriptional programs of late-developing B cells.
Caron et al37 used scRNA-seq to examine sources of intraindividual transcriptional heterogeneity in leukemia samples obtained from 4 individuals with ETV6::RUNX1 ALL, 2 with high hyperdiploid B-ALL and 2 with T-ALL. ETV6::RUNX1 samples showed enrichment of expression of genes for B-cell activation/differentiation, cell proliferation/cell death, and regulation of multiple metabolic processes. In contrast, high hyperdiploid and T-ALL samples exhibited enrichment of pathways involved in translation initiation and protein synthesis. The differentiation states of individual leukemic cells were predicted using developmental state classifiers from healthy B and T cells. Interestingly, there was an inverse correlation between the developmental state of the leukemic cell and the expression of ribosomal protein genes.37
Overall, these studies have provided insights into intraindividual transcriptional heterogeneity of pediatric ALL; however, they have analyzed few samples and subtypes. Thus, the mechanisms and patterns of intraindividual transcriptional heterogeneity in ALL and their clinical significance should be determined in larger studies. Recently, Zeng and coauthors38 used single-cell data generated from leukemia stem, progenitor, and mature cell types and determined the leukemia cell hierarchy framework from bulk transcriptomes of >1000 cases of AML through deconvolution. This cell hierarchy composition was then associated with functional, genomic, and clinical features and was linked to therapy response. A similar analysis in ALL is needed to understand the link between cell type composition, genetic heterogeneity, and clinical outcome. The studies so far performed in ALL and normal hematopoietic ontogeny have determined various degrees of differentiation and inferred the putative ALL cell origin, although it should be noted that a comparative analysis of putative stage does not directly implicate a cell of origin but rather the stage at which developmental arrest is seen in leukemia.
Dissection of leukemia lineage plasticity
Single-cell sequencing studies have provided important insights into cell lineage plasticity in the context of acute leukemia of ambiguous lineage (ALAL).39-41 ALAL includes acute undifferentiated leukemia, which lacks specific lineage differentiation, and mixed-phenotype acute leukemia (MPAL), which expresses markers of >1 lineage18,42,43 and for which immunophenotypic classification43,44 and appropriate therapeutic approach remain controversial. Bulk DNA sequencing of MPAL has shown that the immunophenotypic heterogeneity observed in individual cases of MPAL is not determined by subclonal genetic variegation,42,45 but rather that founding lesions arise in primitive hematopoietic progenitors that retain multilineage potential.42 More recently, this finding has been confirmed by integrated single-cell immunophenotypic, transcriptomic, and epigenetic analyses (CITE-seq19 and scATAC-seq) of normal bone marrow and MPAL samples,46 with the identification of transcriptional programs that were shared by immunophenotypically distinct subpopulations within individual patients. Integrative analysis of transcriptomic and chromatin-accessibility maps revealed enrichment of RUNX1 motifs among MPAL cells. The expression of genes identified as putatively regulated by RUNX1 was further investigated in bulk AML RNA-seq data from The Cancer Genome Atlas.47 Patients with high levels of RUNX1 target gene signature had inferior survival compared with those with low levels of RUNX1 target gene signature.
Single-cell studies have also helped resolve longstanding uncertainties regarding the relationship of lineage-ambiguous leukemias that are classified as separate entities but are commonly immunophenotypically similar. These include T/myeloid MPAL and early T-cell precursor ALL (ETP ALL), both of which express T and myeloid antigens, but often differ by the presence (T/myeloid MPAL) or absence (ETP ALL) of myeloperoxidase expression.48-51 Two studies18,52 showed that diverse genomic rearrangements deregulating BCL11B through juxtaposition to hematopoietic stem cell superenhancers define a subtype of ALAL that comprises one-third of cases of T/myeloid MPAL and ETP ALL, and a smaller proportion of cases of AML and acute undifferentiated leukemia. Single-cell multiomic ATAC-seq/RNA-seq demonstrated that BCL11B gene expression correlates with enrichment for a signature of open chromatin in normal human hematopoietic stem/progenitor cells (HSPCs). These findings and the lack of evidence of T-cell antigen receptor rearrangements in BCL11B-rearranged leukemias support the notion that a subset of HSPCs is the cell of origin (and final stage of maturation) of this subtype of leukemia.18
Rearrangement of KMT2A (MLL) is also associated with lineage plasticity in acute leukemia, including lineage switch during disease progression.53,54 Single-cell multiomic ATAC-seq/RNA-seq of KMT2A-r leukemias and normal hematopoietic cells from patients of different ages55 identified subsets of lymphomyeloid-primed progenitor like blasts, which may explain the lineage switch under the pressure of B-cell–directed immunotherapy. Moreover, this study identified HSPC-like cells, enriched for upregulated genes involved in interferon response and promoting an immunosuppressive signaling circuit with cytotoxic lymphocytes, which may favor the immune escape of leukemic cells. Collectively, these data show the utility of sc-seq to nominate cellular origin and mechanisms of lineage plasticity during disease progression in lineage-ambiguous leukemias.
Dissection of ALL clonal architecture by single-cell genomics
Before sc-seq, next-generation sequencing used variant allele frequency to predict the order and clonality of mutations, supporting a stepwise accumulation of mutations during leukemogenesis and clonal evolution.56,57 Mutations in the Ras pathways, CREBBP, TP53, and NT5C2 are commonly enriched at relapse in ALL.56,58 By deep sequencing and droplet digital polymerase chain reaction, some of these mutations (eg, CREBBP) have been found to be preserved from or acquired after diagnosis, and others (eg, NT5C2 and USH2A) have been observed only after initial therapy in minor subclones.59-61 However, the identification of clonal heterogeneity with rare mutations at diagnosis or at earlier time points before relapse remains challenging. For example, mutational co-occurrence can be inferred only by bulk studies, but it is possible to detect it directly by sc-seq.62
Early sc-seq studies in ALL physically separated single cells by microfluidics or cell sorting, allowing for analysis of a limited number of cells and patients. Bulk and targeted sc-seq from pediatric ALL cases were used to identify structural variations, mutations, and immunoglobulin heavy-chain sequences and to reconstruct clonal evolution in each sample.63 From these analyses, most structural variants were inferred to be acquired before single-nucleotide variations, and among the latter, KRAS mutations were late events. De Bie et al64 used microfluidic-based, single-cell–targeted DNA sequencing, and droplet-based scRNA-seq of total bone marrow cells and CD34+CD38− multipotent progenitor cells from 4 patients with T-ALL to investigate clonal heterogeneity and determine the order in which mutations are acquired. scRNA-seq revealed limited heterogeneity across the T-ALL cells, but targeted scDNA sequencing elucidated the order of mutation acquisition with mutations in known oncogenes (MED12, STAT5B) among various preleukemia events, followed by T-cell receptor gene rearrangements, CDKN2A/B deletions, and gene fusions, whereas NOTCH1 mutations were typically subclonal and late events. Analysis of CD34+CD38− multipotent progenitor cells and bulk myeloid cells isolated at diagnosis and during remission revealed that, in half of the cases, somatic mutations started to accumulate in multipotent progenitor cells and were detectable in myeloid cells.
Integrated genomic analyses (fluorescence in situ hybridization, single-cell multiplex quantitative-polymerase chain reaction, and xenotransplant experiments) were used to determine the phylogenetic architecture of STIL::TAL1 in T-ALL.65 Acquisition of STIL::TAL1 and loss of CDKN2A were early events, with STIL::TAL1 occurring first, whereas both NOTCH1 and PTEN mutations were secondary and subclonal.
Simultaneous targeted scDNA sequencing and cell-surface protein expression analysis in B-ALL66 suggest that lineage-related mutations (ETV6, IKZF1, and PAX5) occur early, as they were present in all leukemia cells, whereas kinase-related mutations (FLT3, PTPN11, NRAS, and KRAS) appear later and were most frequently mutually exclusive. In contrast to B-ALL, certain kinase alterations (JAK3 and NRAS) in T-ALL co-occurred in the same clone in 1 patient with HOXA T-ALL, suggesting a convergent evolution or a shared founder cell. A similar pattern has been reported in T-ALL,14 with multiple JAK-STAT pathway mutations that coexist in the same leukemia clone (eg, JAK3 and JAK1 or STA5B and JAK1), confirming an additive effect of such mutations.60 In half of cases, more than 2 NOTCH1 mutations were identified in the same patient but in different clones. In contrast to samples from cases of ALL with ETV6::RUNX1, BCR::ABL1, BCR::ABL1-like, and IKZF1 N159Y mutations, which almost never harbor additional subclonal mutations, high hyperdiploid ALL is more commonly clonally heterogeneous with up to 7 subclones at diagnosis, with frequently mutually exclusive JAK/STAT, Ras, or FLT3 signaling mutations.13 This finding raises the prospect that serial monitoring for selection of such clones may guide tailored therapy in the context of suboptimal treatment response.
Predicting ALL relapse
Multiple sc-seq approaches have attempted to improve prediction of the risk of relapse, which bulk sequencing studies have shown to be propagated from ancestral, major, or minor clones at initial diagnosis.59,60,67 By mass cytometry analysis of 60 B-ALL samples at diagnosis and a machine learning approach,68 relapse was found to be associated with pathway activation (mTOR signaling) rather than mutations.68 Thus, the single-cell identification of developmental states may improve patient risk stratification and identify cell populations at higher risk of relapse. Analysis of longitudinal T-ALL samples has enabled early detection of relapse with identification of residual mutated cells and has revealed a variable clonal response to corticosteroid treatment, with the clone carrying the most mutations showing the greatest reduction.14
To identify the cell subpopulations associated with response to prednisone, Candelli et al69 performed scRNA-seq in 15 diagnostic samples from infants with KMT2A-r B-ALL, by using plate-based (SORT-seq)70 and droplet-based (10X Genomics) approaches. Based on a previously identified prednisone-dependent gene expression signature,71 individual cells were classified as sensitive (metabolically more active) or resistant (with reduced metabolic and cell-cycle activity and more quiescent) to prednisone. Therapy-resistant cells exhibited a partially activated glucocorticoid response before treatment (eg, deregulation of the glucocorticoid receptor gene NR3C1 and several of its downstream targets) and expression of general mediators of drug resistance (CTNBB1 and MCL1), efflux transporters (ABCA1), and stemness markers (eg, CD44, EPC1, SET2D, and SOCS2), suggesting a more general mechanism of resistance to chemotherapy rather than one specific to prednisone.69
The notion that different blast developmental states may affect pathogenesis and treatment responsiveness has also been demonstrated by the subclonal analysis of organ-specific ALL clones before and after therapy in xenografts by integrated synthetic DNA barcode tracking72,73 and scRNA-seq. With this approach, Contreras-Trujillo et al74 observed that engraftment in extramedullary sites was accompanied by expansion of clones with expression of different genes compared with those in the hematopoietic tissues (eg, bone marrow and spleen). Furthermore, although most of the clones were concordant between bone marrow and spleen, few of them were discrepant, with clones expressing BTK, DNAJC, and LRIF1 that locally expanded in the bone marrow at single anatomical sites. The analysis of clonal architecture before and after various treatments revealed expansion of new clones after chemotherapy.
Another application of how sc-seq can improve treatment by deciphering intratumoral heterogeneity and identification of resistant cells was described by Mehtonen et al,75 who used sc-seq to elucidate cell states and transcription factor activities during B-lineage differentiation of normal and leukemic cells from 6 patients with ETV6::RUNX1 B-ALL at diagnosis and during standard induction chemotherapy (day 15).75 At diagnosis, ETV6::RUNX1 leukemic blasts resembled the pro-B differentiation state, displayed heterogeneity in cell cycle activity, and expressed several genes (cytokine, chemokine, and growth factor genes) that are normally downregulated during differentiation but remained expressed in leukemic cells.75 Upon induction chemotherapy, mild changes in transcription factor activity and expression and partial differentiation toward the pre–B-cell state were features of chemoresistance.
Single-cell analysis of the ALL microenvironment
During lymphoid progenitor transformation and expansion, ALL blasts actively remodel both nonhematopoietic (eg, in the mesenchymal, endothelial, and osteoblast niche) and surrounding immune cells.76 The dynamic interactions occurring between tumor cells and infiltrating immune cells represents a precarious balance between immune evasion and immune rejection of malignant cells. For example, tumor neoepitope expression may elicit an antigen-specific adaptive immune response capable of clearing tumor cells.77,78 Tumor progression, however, may result from immune evasion through multiple mechanisms including, for example, establishment of an immunosuppressive tumor microenvironment and reduction tumor neoantigen expression. Numerous single-cell studies of the tumor microenvironment have described tumor-specific T-cell activity within spatially demarcated, immunogenic solid tumors that harbor a high somatic mutational burden.79-84 ALL blasts, in contrast, typically harbor relatively few genetic alterations and have often been considered immunogenically inert. This notion is now being challenged in light of recent studies that have identified leukemia-specific effector T cells and T-cell activation and dysfunction in infiltrating leukemia T cells. Multiplexed immunohistochemistry analysis, bulk RNA-seq, and scRNA-seq analysis of primary B-ALL samples have shown highly variable cytotoxicity and exhaustion profiles of the infiltrating B-ALL T cells; however, these approaches did not decipher antigen-driven expansion of T-cell clones.85-88 Using combined scRNA/T-cell receptor sequencing (TCR-seq) of peripheral blood T cells from 3 patients with primary B-ALL, Wang and colleagues89 identified patient-specific and clonally expanded effector-like T-cell subpopulations, suggesting the presence of a leukemia-reactive T-cell population, but without formal identification of leukemia-specific antigen recognition.
To elucidate the importance of expression of ALL neoantigens in driving leukemia-specific T-cell expansion, Zamora and colleagues88 used RNA-seq data from DUX4-rearranged and ETV6::RUNX1-diagnostic B-ALL samples to predict patient-specific, tumor-derived neoepitopes resulting from somatic nonsynonymous and gene fusions (eg, the ETV6::RUNX1 fusion junction). Based on this, isolated B-ALL infiltrating CD8+ T cells were cocultured with patient-specific, neoepitope-expressing, artificial antigen-presenting cells (aAPCs). All patient samples harbored tumor-infiltrating T cells responsive to stimulation by predicted neoantigens. Fluidigm-based single-cell analysis of tumor-reactive CD8+CD45RO+CCR7− effector T cells, as identified by neoepitope-specific tetramer binding, revealed significant intrapatient and interpatient gene expression heterogeneity in the proportion of TBX21-expressing functional T cells and STAT1/3/4-expressing dysfunctional effector T-cell states. In addition to B-ALL, a study of primary ETP-ALL showed that BCL11BhighLCKhighIL7RAhigh nontransformed T-cell populations were found to coexist with ETP-ALL “stemlike” cells. Nontransformed, patient-derived CD8+ T cells displayed an oligoclonal TCR repertoire and a prominent exhaustion signature (HAVCR2highPDCD1high) suggestive of clonal T-cell expansion preceding effector dysfunction. Immunohistochemistry and predictive receptor-ligand analysis confirmed that both ETP-ALL and T-ALL express high levels of galectin-9, the ligand of TIM-3 (encoded by HAVCR2). Furthermore, leukemia-derived, secreted galectin-9 (LGALS9) was sufficient to upregulate HAVCR2 levels on nontransformed primary CD8+ T cells ex vivo, suggesting that ETP-ALL promotes leukemia-specific T-cell dysfunction through secretion of galectin-9.
Single-cell analyses of response to ALL-directed immunotherapy
B-ALL–directed immunotherapy, such as chimeric antigen receptor (CAR) T-cell and Bispecific T-cell Engager (BiTE) therapy, co-opt patient T cells to drive tumor antigen–specific, T-cell–mediated killing of ALL blasts (eg, CD19 and/or CD22 CAR T cells and BiTE, blinatumomab). The functional heterogeneity of leukemia-associated T cells may profoundly impact ex vivo CAR T-cell manufacturing and the persistence of antileukemia activity. Recent studies have contributed to unraveling the clinical importance of patient-specific T-cell heterogeneity before blinatumomab infusion and throughout CAR T-cell manufacturing and infusion for treatment of primary B-cell malignancies. Zhao and colleagues90 used scRNA-seq and TCR-seq to analyze the T-cell composition of B-ALL bone marrow before blinatumomab infusion. Strikingly, patients responsive to subsequent blinatumomab therapy harbored a higher proportion of TCF7-expressing stemlike CD4+ T cells, central memory CD8+ T cells, and TCR diversity when compared with nonresponders, who were enriched for exhausted CD8+ T cells.90 In addition, Chen and colleagues91 performed CITE-seq and scATAC-seq analysis of premanufactured T cells isolated from 6 patients with B-cell leukemia or lymphoma treated with anti-CD19 CAR T-cell therapy. T-cell persistence ranged from 2 to 22 months in the analyzed cohort, with CITE-seq protein data highlighting a higher proportion of naive and memory T cells in persistent CAR T-cell cases when compared with short-lived counterparts, a finding consistent with single-cell analysis of anti-CD19 CAR T-cell infusion products targeting large B-cell lymphomas.92 Combination bulk RNA-seq data analysis of an expanded patient cohort with scATAC-seq data showed that gene expression and chromatin accessibility patterns regulated by TCF1 (encoded by TCF7), a transcription factor involved in thymocyte maturation93 and driving stemlike properties in effector T-cell subsets,94 were associated with naive, memory, and, to a lesser extent, effector T-cell subsets, and long-term persistent anti-CD19 CAR T-cell products.
The association between the composition of T-cell subsets in the infusion product and CAR T-cell persistence stimulated research that addressed how CAR T-cells functionally respond in vivo after infusion. Sheih et al95 performed serial analyses of preinfusion CD8+ CAR T cells with matched peripheral blood CD8+ CAR T cells during both early (days 7-14 after infusion) and late (days 26-30 after infusion) post expansion time points after infusion of adult patients with B-ALL, chronic lymphocytic leukemia, or non-Hodgkin lymphoma. Bulk TCR sequencing identified progressive reduction in TCR diversity after CAR T-cell infusion; however, the magnitude of change in clonal diversity was highly variable among patients. Additional single-cell analysis of CD8+ CAR T cells showed a progressive loss of gene expression heterogeneity after infusion, which was associated with increased proliferation and cytotoxicity signatures, as well as progressive increases in expression of T-cell inhibitory marker (eg, PD-1, LAG-3, and TIGIT) genes. Consistent with these findings, a genome-wide methylation analysis96 of preinfusion and postinfusion CD8+ CD19-CAR T cells in pediatric B-ALL showed rapid demethylation of CpGs associated with T-cell cytotoxicity and progressive methylation of stem-associated loci (eg, LEF1).
DNMT3A-deficient CD8+ T cells97 exhibit reduced TCF7 methylation, increased TCF1 expression, and stemlike gene expression programs that lead to prolonged persistence of antitumor DNMT3A-knockout CAR T-cells in vivo when compared with DNMT3A wild-type CAR T-cells.
Several groups have used scRNA-seq analysis of primary B-ALL samples throughout the course of conventional chemotherapy86 and blinatumomab treatment90 and observed a dramatic remodeling of leukemia-associated myeloid cells. Both studies identified a significant skewing of monocyte subset representation; specifically, there was an overrepresentation of CD14−CD16+ nonclassic monocytes within the B-ALL immune microenvironment. The importance of leukemia-associated myeloid cells in promoting ALL survival remains unclear; however, incorporation of immune microenvironment characterization may extend standard biospecimen flow cytometry beyond ALL immunophenotyping.
Future directions
Spatial dissection of the ALL niche
ALL dissemination from the bone marrow and establishment of numerous specialized niches throughout various organs, including the central nervous system98; spleen, stromal, and endothelial niches90,99; adipose tissue100; and testis.101,102 Isolation and analysis of these rare niche populations from primary patient biospecimens remains a significant challenge. With the promise of emerging spatial transcriptomic platforms that enable analysis of frozen and paraffin-embedded specimens, spatially resolved maps of intact hematopoietic and nonhematopoietic interactions may present an exciting avenue for exploring organ-specific ALL-niche interactions without tissue disaggregation and cell sorting.103
Impact of genetic mutations on ALL microenvironment development and function
The contribution of the host genotype to development of both microenvironment and leukemia remains unclear. Numerous studies have identified germline variants associated with B-cell development and transformation (eg, PAX5,104IKZF1105), inherited aneuploidies that predispose to leukemic transformation106 (eg, trisomy 21), and racial disparities in immune cell development107 and ALL incidence.108 For example, loss-of-function mutations affecting the lymphoid tumor suppressor109,110 IKAROS are associated with poor prognosis and increased ALL dependence on cellular adhesion for survival. Notably, IKAROS also regulates mature T-cell differentiation and function,111 playing a prominent repressive role in Th1 cell differentiation through the repression of T-bet expression.112 Using emerging tools, such as single-cell genotyping113 and mitochondrial variant tracking,113,114 we can begin to investigate how different genetic backgrounds shape microenvironment development and, potentially, extrinsic interactions regulating treatment responsiveness.
Predicting ALL relapse
Relapse still represents a major obstacle to increasing cure rates in ALL. Preliminary single-cell studies in ALL enabled analysis of clonal architecture and discrimination of founder alterations (present in all leukemic cells) and secondary and subclonal events, which may not be suitable candidates for robust monitoring,65 but may be informative in predicting relapse and/or in suggesting novel therapeutic strategies (eg, ABL1 mutations in BCR-ABL1 ALL). Although current costs for sc-seq are still prohibitive for routine analyses in the clinic, we envision that they will decline in the near future, and thus scDNA-seq applications will be more accessible to screening large cohorts of patients and tracking measurable residual disease. Such results may reveal relapse-seeding clones and potentially guide clinical management of disease to mitigate relapse.
Conclusions
Single-cell studies have built on existing bulk approaches to add resolution to our understanding of ALL biology. These studies have refined clonal heterogeneity, elucidated cell-type composition and developmental states in relation to pathogenesis and therapeutic response, and explored the role of microenvironment and cell immunity (Table 2). The exciting prospect of expanded multiomic approaches, such as single-cell cellular barcoding coupled with transcriptomic/epigenetic profiling,115 will allow for deeper examination of the ALL cell of origin and cellular hierarchy composition across the different molecular ALL subtypes. In addition, emerging single-cell approaches that enable interrogation of genome-wide changes in copy number, structural variations, sequence mutations, and changes in gene expression may further expand clinical approaches designed to monitor measurable residual disease and detect the early clones responsible for relapse.
Acknowledgments
This work was supported by the American and Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital and the National Institutes of Health, National Cancer Institute, grants R35 CA197695 (C.G.M.) and K22 CA258520 (M.T.W.).
Authorship
Contribution: I.I., M.T.W., and C.G.M. wrote the manuscript; and I.I., M.T.W., and C.G.M. edited and provided final approval of the manuscript.
Conflict-of-interest disclosure: I.I. has received honoraria from Amgen and Mission Bio; M.T.W. has received royalties from the Walter and Eliza Hall Institute of Medical Research (Melbourne, Australia); C.G.M. has received research funding from Loxo Oncology, Pfizer, AbbVie and honoraria from Amgen and Illumina and holds stock in Amgen. There are no financial conflicts of interest in the work presented in this article.
Correspondence: Charles G. Mullighan, Department of Pathology, Hematological Malignancies Program, St Jude Children’s Research Hospital, 262 Danny Thomas Place, Mail Stop 342, Memphis, TN 38105; e-mail: charles.mullighan@stjude.org.