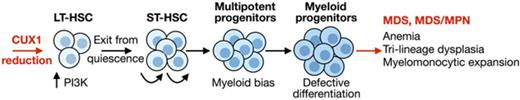
Key Points
CUX1 deficiency leads to transient clonal expansion followed by HSC depletion, anemia, and trilineage dysplasia.
CUX1 transcriptionally regulates HSC quiescence, proliferation, and lineage specification.
Abstract
Monosomy 7 (−7) and del(7q) are high-risk cytogenetic abnormalities common in myeloid malignancies. We previously reported that CUX1, a homeodomain-containing transcription factor encoded on 7q22, is frequently inactivated in myeloid neoplasms, and CUX1 myeloid tumor suppressor activity is conserved from humans to Drosophila. CUX1-inactivating mutations are recurrent in clonal hematopoiesis of indeterminate potential as well as myeloid malignancies, in which they independently carry a poor prognosis. To determine the role for CUX1 in hematopoiesis, we generated 2 short hairpin RNA-based mouse models with ∼54% (Cux1mid) or ∼12% (Cux1low) residual CUX1 protein. Cux1mid mice develop myelodysplastic syndrome (MDS) with anemia and trilineage dysplasia, whereas CUX1low mice developed MDS/myeloproliferative neoplasms and anemia. In diseased mice, restoration of CUX1 expression was sufficient to reverse the disease. CUX1 knockdown bone marrow transplant recipients exhibited a transient hematopoietic expansion, followed by a reduction of hematopoietic stem cells (HSCs), and fatal bone marrow failure, in a dose-dependent manner. RNA-sequencing after CUX1 knockdown in human CD34+ cells identified a −7/del(7q) MDS gene signature and altered differentiation, proliferative, and phosphatidylinositol 3-kinase (PI3K) signaling pathways. In functional assays, CUX1 maintained HSC quiescence and repressed proliferation. These homeostatic changes occurred in parallel with decreased expression of the PI3K inhibitor, Pik3ip1, and elevated PI3K/AKT signaling upon CUX1 knockdown. Our data support a model wherein CUX1 knockdown promotes PI3K signaling, drives HSC exit from quiescence and proliferation, and results in HSC exhaustion. Our results also demonstrate that reduction of a single 7q gene, Cux1, is sufficient to cause MDS in mice.
Introduction
Monosomy 7 (−7) and del(7q) are frequent, adverse-risk cytogenetic abnormalities in diseases of hematopoietic stem and progenitor cells (HSPCs). −7/del(7q) occurs across the spectrum of myeloid disorders, from bone marrow (BM) failure to overt leukemia, and typically carries an adverse prognosis. Fourteen percent of adult and 30% of pediatric myelodysplastic syndrome (MDS) patients have −7/del(7q).1,2 Among inherited and acquired BM failure syndromes, −7/del(7q) develops in 40% to 80% of cases that evolve to MDS or acute myeloid leukemia (AML).3 −7/del(7q) occurs in 10% to 15% of de novo AML4 and 49% of therapy-related myeloid neoplasms.5 In addition, −7/del(7q) is identified in 33% of juvenile myelomonocytic leukemia (JMML) and 14% of chronic myelomonocytic leukemia (CMML), which are myelodysplastic/myeloproliferative neoplasms (MDS/MPN).2,6 Regardless of the disease subtype, −7/del(7q) typically portends chemoresistance and poor survival. Despite this clear clinical importance, the key tumor suppressor genes (TSGs) encoded on 7q have remained unclear.
We previously identified a haploinsufficient, myeloid TSG on chromosome band 7q22, CUX1.7 CUX1 is a nonclustered homeobox transcription factor.8 The full-length p200 protein has 1 homeodomain and 3 cut repeat DNA-binding domains. CUX1 is highly conserved, ubiquitously expressed, and essential for survival in mice and Drosophila.9-12 We demonstrated that haploinsufficiency of the ortholog, cut, led to hemocyte overgrowth and tumor formation in Drosophila melanogaster. Similarly, CUX1 haploinsufficiency gave human HSPC an engraftment advantage in mouse xenotransplants.7
The genetic evidence for CUX1 as a critical TSG is overwhelming. CUX1-inactivating mutations have been reported in numerous analyses of myeloid neoplasms.7,13-17 CUX1 mutations that occur in the context of normal chromosome 7 alleles independently carry a poor prognosis.14,17 CUX1 is also recurrently mutated in clonal hematopoiesis of indeterminate potential (CHIP),18-21 indicating that CUX1 mutations provide HSPC with a competitive advantage and predisposition for transformation.22 Further, CUX1 is mutated in a variety of solid tumors; thus, it may regulate tumorigenesis more generally.7,17
We reported CUX1 global genomic targets by chromatin immunoprecipitation-sequencing in 3 human cell types, including the K562 myeloid leukemia cell line.23 CUX1 preferentially bound distal, active enhancers and controlled expression of cell-cycle regulators.23 CUX1 fit an analog model of dose-sensitive gene regulation,23 which may provide a nuanced means for CUX1 control of a disparate array of cellular programs (including proliferation, differentiation, and apoptosis) across a broad array of cell types, and even with a single cell type.24-26
To identify the in vivo functions for CUX1 in mice, it was essential for us to take a nontraditional genetic approach because of the complexity of the gene. Cux1 is large, ranked in the top 2.1% of mammalian genes by size. Cux1 has 33 exons, more than ∼97% of other murine genes. Cux1 shares exons with Casp, a gene encoding a Golgi-associated protein.8,27 In addition, Cux1 has 3 alternative start sites and 7 RNA splice isoforms.27 Traditional knockout mice had unanticipated alternative splicing that removed the inserted STOP cassettes, causing hypomorphic protein expression and incomplete Cux1 knockouts.12,28-30 Previous models also had perinatal lethality and extrahematopoietic effects.12,28-30 To circumvent these issues, we engineered inducible Cux1 short hairpin RNA (shRNA) knock-in mice.31 We report that CUX1 knockdown led to MDS and MDS/MPN with features of human disease. Our results also demonstrate that reduction of a single 7q gene, Cux1, is sufficient to cause myeloid disorders, and Cux1 is a critical regulator of hematopoietic stem cell (HSC) homoeostasis.
Methods
Complete methods are provided in the supplemental Methods, available on the Blood Web site.
Results
CUX1 is haploinsufficient in human myeloid malignancies
We previously reported that CUX1 is expressed at ∼50% levels in de novo AML and therapy-related myeloid neoplasms with −7/del(7q).7 We extended this finding by first analyzing The Cancer Genome Atlas AML data.32 CUX1 has 7 RefSeq isoforms; 3 encode the CUX1 DNA-binding transcription factor and the others encode the Golgi-associated CASP protein.8,27 The Cancer Genome Atlas AML RNA-sequencing data were provided at the gene level, wherein CUX1 and CASP isoforms were aggregated to a single gene. This analysis showed 60% residual CUX1/CASP in samples with a deletion spanning CUX1 (supplemental Figure 1A). To look at this finding in more detail, we parsed the CUX1 and CASP encoding isoforms from our published RNA-sequencing data.7 The CUX1 isoform, NM_181552, is expressed at 66% residual levels in −7/del(7q) patients (supplemental Figure 1B). We interrogated MDS and JMML microarray datasets.33,34 We divided patients into those with or without −7/del(7q) and removed CASP-specific probes. Across datasets, CUX1 probes showed a range of −0.3 to −0.9 log2 fold-change reduction (supplemental Figure 1C-E). In other words, for the majority of patients with −7/del(7q), CUX1 levels do not fall below 50% of the levels expressed in patients with diploid chromosome 7 alleles. These results suggest that for patients with −7/del(7q), the remaining CUX1 allele is expressed and CUX1 is haploinsufficient.
Generation of CUX1 knockdown mouse models
We sought to determine the level of Cux1 in murine hematopoietic lineages, and if the expression pattern is conserved between humans and mice. By quantitative reverse transcription polymerase chain reaction (qRT-PCR), Cux1 is high in long-term HSC (LT-HSC) and lower in short-term HSC (ST-HSC) and myeloid progenitors (MPs). Cux1 increases in differentiated granulocytes and monocytes, compared with granulocyte-monocyte progenitors (GMPs) (Figure 1A). This pattern is similar to what we observed in human counterparts.7
Description of CUX1 knockdown mouse models and characterization of MDS in Cux1midmice. (A) Cux1 has cell type–specific expression levels. Cells were sorted as follows from the BM of wild-type C57BL/6 mice: LT-HSC (LSK+/CD34−/Flt3−), ST-HSC (LSK+/CD34+/Flt3−), MPP (LSK+/CD34+/Flt3+), CMP (Lin−/Sca1−/c-Kit+/CD34+/CD16/32low), MEP (Lin−/c-Kit+/Sca1−/CD34−/CD16/32−), and GMP (Lin−/c-Kit+/Sca1−/CD34+/CD16/32high). The following cells were sorted from the spleen: granulocytes (CD11b+/Gr1+), monocytes (CD11b+/Gr1−), B cells (B220+), and T cells (CD3+). qRT-PCR for Cux1 mRNA was performed. The data represent the mean ± SEM from 4 biological replicates. *Student t test, P < .05. (B) The Cux1 genomic locus is shown with all 7 RefSeq mRNA isoforms. shRNA (Cux1low and Cux1mid) targeting the indicated exons (red arrows) were used to generate transgenic mice. (C) A representative immunoblot for CUX1 protein in thymocytes isolated from Cux1low, Cux1mid, and Ren mice after 7 days of dox. The mean ± SD level of residual CUX1 protein quantified from 4 biological replicates for Cux1low was 12% ± 9% and 54% ± 17% for Cux1mid. (D) Cux1 mRNA level in sorted populations from Cux1mid and Cux1low BM compared with Ren littermate control. One representative replicate of 3 biological replicates is shown. LSK are Lin−/c-Kit+/Sca1+), and myeloid progenitors are Lin−/Sca1−/c-Kit+. (E) At 6 to 10 weeks of age, Cux1mid and Ren littermate control mice were started on continuous dox treatment and monitored for up to 18 months. Complete blood counts of mice at 8 and 18 months of dox are shown. (F) Flow cytometric analysis of peripheral blood (PB) samples for monocyte (CD11b+/Gr1−) and granulocyte (CD11b+/Gr1+) populations at 8 and 18 months of dox treatment. (G) Spleen weights after 18 months of dox. The mean ± SD is shown. *P ≤ .05, **P ≤ .01, Wilcoxon rank test. (H-P) Representative morphology images are shown for mice after 18 months of dox. (H) Wright-Giemsa staining of the peripheral blood in (Hi) Ren and (Hii-iii) Cux1mid mice. (Hii) Cux1mid mice have an increase in circulating mature granulocytes (arrowheads). (Hiii) A granulocyte with pseudo Pelger-Huet anomaly is shown (yellow arrowhead). Red blood cells in Cux1mid mice have increased Howell-Jolly bodies (arrow) and increased reticulocytes with basophilic stippling (black arrowheads). (Ii-ii) H&E staining of the bone marrow of aged Cux1mid mice shows a marked increase in megakaryocytes. (Iiii) At high power, clusters of megakaryocytes (arrowheads) with dysplastic features can be appreciated, including micromegakaryocytes, nuclear hypolobation, and condensed, hyperchromatic nuclei. (Ji-ii) H&E staining shows an expansion of the red pulp at the expense of the white pulp. The white pulp is outlined by a yellow dashed line for clarity. (Jiii) High-power analysis of Cux1mid spleens illustrates increased megakaryocytes (arrowheads) with some micromegakaryocytes with condensed, hyperchromatic nuclei. (K-L) Additional examples of peripheral blood granulocytes with pseudo Pelger-Huet anomaly. (M) Neutrophil with hypersegmentation. (N) A representative giant platelet is shown (arrow). (O) Touch preparation of Cux1mid spleen reveals erythroid dysplasia as evidenced by binucleated erythroblasts (arrowheads) and abnormal nuclear contours (arrow). (P) H&E staining shows a myelomonocytic cell infiltrate (dashed yellow line) in the liver of Cux1mid mice. Images were taken with a Zeiss Axioskop microscope. Chr, chromosome; H&E, hematoxylin and eosin; mRNA, messenger RNA; PB, peripheral blood; SD, standard deviation; SEM, standard error of the mean.
Description of CUX1 knockdown mouse models and characterization of MDS in Cux1midmice. (A) Cux1 has cell type–specific expression levels. Cells were sorted as follows from the BM of wild-type C57BL/6 mice: LT-HSC (LSK+/CD34−/Flt3−), ST-HSC (LSK+/CD34+/Flt3−), MPP (LSK+/CD34+/Flt3+), CMP (Lin−/Sca1−/c-Kit+/CD34+/CD16/32low), MEP (Lin−/c-Kit+/Sca1−/CD34−/CD16/32−), and GMP (Lin−/c-Kit+/Sca1−/CD34+/CD16/32high). The following cells were sorted from the spleen: granulocytes (CD11b+/Gr1+), monocytes (CD11b+/Gr1−), B cells (B220+), and T cells (CD3+). qRT-PCR for Cux1 mRNA was performed. The data represent the mean ± SEM from 4 biological replicates. *Student t test, P < .05. (B) The Cux1 genomic locus is shown with all 7 RefSeq mRNA isoforms. shRNA (Cux1low and Cux1mid) targeting the indicated exons (red arrows) were used to generate transgenic mice. (C) A representative immunoblot for CUX1 protein in thymocytes isolated from Cux1low, Cux1mid, and Ren mice after 7 days of dox. The mean ± SD level of residual CUX1 protein quantified from 4 biological replicates for Cux1low was 12% ± 9% and 54% ± 17% for Cux1mid. (D) Cux1 mRNA level in sorted populations from Cux1mid and Cux1low BM compared with Ren littermate control. One representative replicate of 3 biological replicates is shown. LSK are Lin−/c-Kit+/Sca1+), and myeloid progenitors are Lin−/Sca1−/c-Kit+. (E) At 6 to 10 weeks of age, Cux1mid and Ren littermate control mice were started on continuous dox treatment and monitored for up to 18 months. Complete blood counts of mice at 8 and 18 months of dox are shown. (F) Flow cytometric analysis of peripheral blood (PB) samples for monocyte (CD11b+/Gr1−) and granulocyte (CD11b+/Gr1+) populations at 8 and 18 months of dox treatment. (G) Spleen weights after 18 months of dox. The mean ± SD is shown. *P ≤ .05, **P ≤ .01, Wilcoxon rank test. (H-P) Representative morphology images are shown for mice after 18 months of dox. (H) Wright-Giemsa staining of the peripheral blood in (Hi) Ren and (Hii-iii) Cux1mid mice. (Hii) Cux1mid mice have an increase in circulating mature granulocytes (arrowheads). (Hiii) A granulocyte with pseudo Pelger-Huet anomaly is shown (yellow arrowhead). Red blood cells in Cux1mid mice have increased Howell-Jolly bodies (arrow) and increased reticulocytes with basophilic stippling (black arrowheads). (Ii-ii) H&E staining of the bone marrow of aged Cux1mid mice shows a marked increase in megakaryocytes. (Iiii) At high power, clusters of megakaryocytes (arrowheads) with dysplastic features can be appreciated, including micromegakaryocytes, nuclear hypolobation, and condensed, hyperchromatic nuclei. (Ji-ii) H&E staining shows an expansion of the red pulp at the expense of the white pulp. The white pulp is outlined by a yellow dashed line for clarity. (Jiii) High-power analysis of Cux1mid spleens illustrates increased megakaryocytes (arrowheads) with some micromegakaryocytes with condensed, hyperchromatic nuclei. (K-L) Additional examples of peripheral blood granulocytes with pseudo Pelger-Huet anomaly. (M) Neutrophil with hypersegmentation. (N) A representative giant platelet is shown (arrow). (O) Touch preparation of Cux1mid spleen reveals erythroid dysplasia as evidenced by binucleated erythroblasts (arrowheads) and abnormal nuclear contours (arrow). (P) H&E staining shows a myelomonocytic cell infiltrate (dashed yellow line) in the liver of Cux1mid mice. Images were taken with a Zeiss Axioskop microscope. Chr, chromosome; H&E, hematoxylin and eosin; mRNA, messenger RNA; PB, peripheral blood; SD, standard deviation; SEM, standard error of the mean.
To determine the in vivo role for CUX1 in hematopoiesis, we generated CUX1 knockdown mice using an shRNA approach.31 We engineered 2 knock-in lines, expressing shRNAs of different specificity, to control for potential off-target effects. Cux1mid targets exon 5, an exon shared by all Cux1 and Casp isoforms (Figure 1B). Cux1low targets the 3′ untranslated region of exon 24, shared only by CUX1-encoding transcripts and not CASP-encoding isoforms (Figure 1B). The transgenes were knocked in to the endogenous Col1a1 locus and under the control of a tetracycline response element (supplemental Figure 2). A second-generation reverse tet-transactivator, m2-rtTA, is expressed under the ubiquitous ROSA26 promoter. In the presence of doxycycline (dox), GFP is turned on, along with the shRNA embedded within the GFP 3′ untranslated region. As a control, we used littermates expressing an shRNA targeting renilla luciferase (Ren.713+/−;M2rtTA+/+ [Ren]).35 Cux1low (Cux1low+/−;M2rtTA+/+) had more potent knockdown, with 12% ± 9% residual CUX1 protein in thymocytes (Figure 1C). Cux1mid (Cux1mid+/−;M2rtTA+/+) had 54% ± 17% residual CUX1 protein. CUX1 protein was too low to reliably detect in the BM and spleen by western blot. qRT-PCR of Lin−/c-Kit+/Sca1+ (LSK) showed 58% ± 2% and 42% ± 2% residual Cux1 transcripts in Cux1mid and Cux1low mice, respectively. MPs (Lin−/c-Kit+/Sca1−) from Cux1mid and Cux1low mice showed 31% ± 8% and 25% ± 18% residual Cux1 transcripts, respectively (Figure 1D).
Cux1mid mice develop MDS with anemia and trilineage dysplasia
We first characterized Cux1mid mice, which approximate CUX1 haploinsufficiency. Because Cux1 is essential for development,12 we induced the transgene in 6- to 10-week-old mice. Cux1mid mice had normal appearance and lifespan. Aged Cux1mid mice developed a normocytic anemia and thrombocytosis (Figure 1E; supplemental Figure 3A). Although the white blood cell count (WBC) was unchanged, the monocyte and granulocyte frequencies were increased (Figure 1F). By 18 months of dox, Cux1mid mice had mild splenomegaly (Figure 1G). Monocytes were expanded in the BM, whereas B lymphocytes were reduced (supplemental Figure 3D). CUX1 knockdown also led to an expansion of LSK and ST-HSC (supplemental Figure 3E).
Aged Cux1mid mice had red blood cell polychromasia, with increased reticulocytes, basophilic stippling, and increased Howell-Jolly bodies (Figure 1H). Evidence of dysgranulopoiesis included pseudo Pelger-Huet anomalies and hypersegmentation (Figure 1Hiii, K-M). Cux1mid BM had normal cellularity with a marginal increase in erythroid precursors (P = .10; Figure 1I; supplemental Figure 3D,F). Cux1mid BM had a marked increase in megakaryocytes, many with dysplasia, including micromegakaryocytes, nuclear hypolobation, and condensed hyperchromatic nuclei (Figure 1I). The myelomonocytic cells were maturing with no morphologic increase in blasts.
Cux1mid splenic architecture was partially disrupted because of red pulp expansion (Figure 1J). Although Ren mice had occasional subcapsular megakaryocytes, Cux1mid mice had increased megakaryocytes throughout the spleen and some micromegakaryocytes with condensed, hyperchromatic nuclei (Figure 1J). Giant platelets, which can accompany megakaryocyte dysplasia, were present (Figure 1N). The splenic touch preparation revealed dyserythropoiesis, including erythroblasts with binucleation or irregular nuclear contours (Figure 1O). There was myeloid cell infiltration of the livers of some Cux1mid mice (Figure 1P). In summary, the findings of anemia with trilineage dysplasia are consistent with a diagnosis of MDS, most similar to refractory cytopenia with multilineage dysplasia (RCMD).36-38
Spontaneous and fatal MDS/MPN in Cux1low mice
Compared with Cux1mid, the Cux1low line had more rapid and aggressive hematopoietic abnormalities, with a median survival of 275 days of dox (Figure 2A). Within 1 month, Cux1low mice had splenomegaly and an elevated red cell distribution width (supplemental Figure 4A-C). Over time, hemoglobin and red cell counts decreased, and the mice developed a severe, normocytic anemia (Figures 2B; supplemental Figure 4C). The platelet count was normal or elevated. The WBC count became elevated within 3 months because of an expansion of monocytes (CD11b+/Gr1−) and granulocytes (CD11b+/Gr1+; Figure 2B-C). B cells were decreased and T cells were unchanged.
Cux1lowmice develop a fatal MDS/MPN that is reversed by CUX1 reexpression. Adult Cux1low and littermate Ren mice were continuously treated with dox starting at 6 to 10 weeks of age. (A) Kaplan-Meier survival plot of Cux1low (n = 20), Cux1mid (n = 11), and Ren mice (n = 19) shows that Cux1low mice have significantly decreased survival (log-rank test). (B) Complete blood cell counts of peripheral blood samples collected over time demonstrate a progressive anemia and WBC expansion. (C) Flow cytometric analysis of PB granulocytes (CD11b+/Gr1+), monocytes (CD11b+/Gr1−), B cells (B220+), and T cells (CD3+) over time. (D) Representative images of spleens collected at the time that Cux1low mice were euthanized because of disease. (E) Spleens and (F) livers were weighed and normalized to body weight. (G) Total leukocyte counts from BM (femurs, tibias, and hipbones) and spleens were enumerated at the time of euthanization. (H) Absolute monocyte and granulocyte cell counts are shown from the BM and spleen at the time of euthanization. (I) Absolute lymphoid cell counts from the BM are provided. (J) HSPC populations were quantified at the time of euthanization. (J, left) LSK+ (Lin−/Sca1+/c-Kit+) cells and MP (Lin−/Sca1−/c-Kit+) are shown. (Center) LSK were further gated for LT-HSC (LSK+/CD34−/Flt3−), ST-HSC (LSK+/CD34+/Flt3−), and MPP (LSK+/CD34+/Flt3+). (Right) Within the MP population, cells were separated into CMP (CD34+/CD16/32low), GMP (Lin−/c-Kit+/Sca1−/CD34+/CD16/32high), and MEP (Lin−/c-Kit+/Sca1−/CD34−/CD16/32−). (K) After 7 months of continuous dox exposure, dox treatment was ceased for a cohort of mice, and hematopoietic populations were assessed over time. PB indices show a normalization of RBC and WBC counts. (L) Flow cytometry shows a normalization of PB granulocyte frequency. Additional data are provided in supplemental Figure 5. The mean ± SD is shown. Student t test *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Cux1lowmice develop a fatal MDS/MPN that is reversed by CUX1 reexpression. Adult Cux1low and littermate Ren mice were continuously treated with dox starting at 6 to 10 weeks of age. (A) Kaplan-Meier survival plot of Cux1low (n = 20), Cux1mid (n = 11), and Ren mice (n = 19) shows that Cux1low mice have significantly decreased survival (log-rank test). (B) Complete blood cell counts of peripheral blood samples collected over time demonstrate a progressive anemia and WBC expansion. (C) Flow cytometric analysis of PB granulocytes (CD11b+/Gr1+), monocytes (CD11b+/Gr1−), B cells (B220+), and T cells (CD3+) over time. (D) Representative images of spleens collected at the time that Cux1low mice were euthanized because of disease. (E) Spleens and (F) livers were weighed and normalized to body weight. (G) Total leukocyte counts from BM (femurs, tibias, and hipbones) and spleens were enumerated at the time of euthanization. (H) Absolute monocyte and granulocyte cell counts are shown from the BM and spleen at the time of euthanization. (I) Absolute lymphoid cell counts from the BM are provided. (J) HSPC populations were quantified at the time of euthanization. (J, left) LSK+ (Lin−/Sca1+/c-Kit+) cells and MP (Lin−/Sca1−/c-Kit+) are shown. (Center) LSK were further gated for LT-HSC (LSK+/CD34−/Flt3−), ST-HSC (LSK+/CD34+/Flt3−), and MPP (LSK+/CD34+/Flt3+). (Right) Within the MP population, cells were separated into CMP (CD34+/CD16/32low), GMP (Lin−/c-Kit+/Sca1−/CD34+/CD16/32high), and MEP (Lin−/c-Kit+/Sca1−/CD34−/CD16/32−). (K) After 7 months of continuous dox exposure, dox treatment was ceased for a cohort of mice, and hematopoietic populations were assessed over time. PB indices show a normalization of RBC and WBC counts. (L) Flow cytometry shows a normalization of PB granulocyte frequency. Additional data are provided in supplemental Figure 5. The mean ± SD is shown. Student t test *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Unlike Cux1mid, Cux1low mice fail to gain weight and have fur loss because of the role of CUX1 in hair follicle morphogenesis (supplemental Figure 5A-B).29,39 Almost all Cux1low mice were euthanized because of weakness and anemia after 9 months of dox. At euthanization, Cux1low mice had hepatosplenomegaly (Figure 2D-F). Cux1low mice had splenic and BM hypercellularity (Figure 2G), driven largely by an expansion of monocytes and granulocytes (Figure 2H; supplemental Figure 5C). The T-cell count was also increased in the BM and spleen, comprising an expansion of both CD4+ and CD8+ T cells (Figures 2I; supplemental Figure 5D). In comparison, the B-cell count was decreased in the BM (Figure 2I), consistent with the decreased B-cell frequency in the blood (Figure 2C). Thus, CUX1 knockdown leads to a spontaneous MPN, anemia, and premature mortality.
We next quantified HSPCs. As with Cux1mid, Cux1low mice had an expansion of the LSK compartment (Figure 2J; supplemental Figure 5F). Within this population, ST-HSC and multipotent progenitors (MPPs) had increased numbers, whereas LT-HSC did not. Among the MP, common myeloid progenitors (CMPs) and GMPs exhibited increases numbers, whereas megakaryocyte-erythroid progenitors (MEPs) were unchanged (Figure 2J). The GMP expansion occurred as early as 1 month after CUX1 knockdown (supplemental Figure 4D). The LSK expansion was not from decreased apoptosis (supplemental Figure 4E). These results are consistent with a regulatory function for CUX1 in HSPC homeostasis. Overall, this second mouse line confirms the role of CUX1 in diverse hematopoietic lineages and that the level of CUX1 deficiency affects the severity of hematopoietic and nonhematopoietic phenotypes.
To determine if restoration of CUX1 can reverse the disease process, we reexpressed CUX1 in mice with established disease. By 7 months of dox, Cux1low mice have anemia and leukocytosis, becoming moribund by 8 to 9 months; thus, we withdrew dox after 7 months for a cohort of mice. Within 3 weeks, the peripheral blood indices began to normalize (Figure 2K). By 7 weeks after dox withdrawal, hemoglobin levels rise to control levels, the WBC decreases to normal levels, and the mice gain weight (Figures 2K-L; supplemental Figure 6). All hematopoietic populations assessed, including HSPC, also normalize (supplemental Figure 6). Thus, the disease process in Cux1low mice is dependent on continued CUX1-deficiency and is reversible upon CUX1 restoration.
Trilineage dysplasia and myelomonocytic organ infiltration in Cux1low mice
Hematopathologic assessment of Cux1low mice demonstrated increased circulating myelomonocytic cells, occasional myeloblasts (1% to 2% of WBC), and granulocytic dysplasia (Figure 3Ai-iv). Red cells exhibited polychromasia and anisopoikilocytosis, and giant platelets were observed (Figure 3Av). Erythroblast dysplasia was evident in splenic touch preparations, similar to Cux1mid (Figure 3B). Cux1low BM demonstrated decreased erythropoiesis (supplemental Figure 5E) and maturing myelomonocytic cells with no morphologic increase in blasts (Figure 3C). Megakaryocytes were increased, many with dysplasia (Figure 3C). The splenic architecture was completely effaced in Cux1low mice (Figure 3D). The expanded red pulp comprised maturing erythroid and myeloid cells and a remarkable expansion of megakaryocytes to a greater extent than Cux1mid. Myelomonocytic cells were observed infiltrating lymph nodes and nonhematopoietic organs, including liver periportal and sinusoidal areas, lung, and kidney (Figure 3E-H).
Myelomonocytic expansion and myelodysplasia in Cux1lowmice. Representative morphology images are shown from mice at the time of euthanization because of disease. (A) Wright-Giemsa staining of the peripheral blood in (Ai) Ren and (Aii-Av) Cux1low mice. (Aii) The increased white blood cell count is primarily composed of mature granulocytes (arrowheads), monocytes (arrow), and (Aiii) occasional circulating myeloblasts (arrows). (Aiv) Dysplasia in the granulocyte lineage as evidenced by pseudo Pelger-Huet anomaly (arrows). (Av) Red blood cells in Cux1low mice have anisopoikilocytosis: (1) macrocyte; (2) microcyte; (3) target cell; (4) spherocyte; (5) tear drop cell; (6) and red cell fragment. (7) Polychromatophilic reticulocyte. (8) Giant platelet. (B) Touch preparation of Cux1low spleen shows erythroid dysplasia, including (Bi) binucleated erythroblast (arrow) and nuclear budding (arrowheads), (Bii) nuclear budding, and (Biii) abnormal nuclear contours. (Biv) Pseudo Chediak-Higashi granule in a maturing granulocyte. (C) H&E of Cux1low BM demonstrates numerous megakaryocytes. Nine megakaryocytes (yellow arrowheads) are shown in a single high-power field, several of which are atypically small, with hypolobation and condensed, hyperchromatic nuclei. (D) Compared with Ren, Cux1low spleen H&E staining demonstrates complete effacement of normal splenic architecture with expansion of red pulp and increased megakaryocytes. White pulp is outlined with a yellow dashed line for clarity. There is a small portion of residual white pulp in the bottom left corner of the Cux1low spleen (D, middle panel). Micromegakaryocytes are present, as well as megakaryocytes with hypolobation, abnormally widely spaced nuclear lobes, and hyperchromatic nuclei. H&E staining shows a myelomonocytic cell infiltrate in the following organs of Cux1low mice: (E) lymph nodes; (F) liver periportal and sinusoidal areas; (G) kidney; and (H) lung. Images were taken with a Zeiss Axioskop microscope.
Myelomonocytic expansion and myelodysplasia in Cux1lowmice. Representative morphology images are shown from mice at the time of euthanization because of disease. (A) Wright-Giemsa staining of the peripheral blood in (Ai) Ren and (Aii-Av) Cux1low mice. (Aii) The increased white blood cell count is primarily composed of mature granulocytes (arrowheads), monocytes (arrow), and (Aiii) occasional circulating myeloblasts (arrows). (Aiv) Dysplasia in the granulocyte lineage as evidenced by pseudo Pelger-Huet anomaly (arrows). (Av) Red blood cells in Cux1low mice have anisopoikilocytosis: (1) macrocyte; (2) microcyte; (3) target cell; (4) spherocyte; (5) tear drop cell; (6) and red cell fragment. (7) Polychromatophilic reticulocyte. (8) Giant platelet. (B) Touch preparation of Cux1low spleen shows erythroid dysplasia, including (Bi) binucleated erythroblast (arrow) and nuclear budding (arrowheads), (Bii) nuclear budding, and (Biii) abnormal nuclear contours. (Biv) Pseudo Chediak-Higashi granule in a maturing granulocyte. (C) H&E of Cux1low BM demonstrates numerous megakaryocytes. Nine megakaryocytes (yellow arrowheads) are shown in a single high-power field, several of which are atypically small, with hypolobation and condensed, hyperchromatic nuclei. (D) Compared with Ren, Cux1low spleen H&E staining demonstrates complete effacement of normal splenic architecture with expansion of red pulp and increased megakaryocytes. White pulp is outlined with a yellow dashed line for clarity. There is a small portion of residual white pulp in the bottom left corner of the Cux1low spleen (D, middle panel). Micromegakaryocytes are present, as well as megakaryocytes with hypolobation, abnormally widely spaced nuclear lobes, and hyperchromatic nuclei. H&E staining shows a myelomonocytic cell infiltrate in the following organs of Cux1low mice: (E) lymph nodes; (F) liver periportal and sinusoidal areas; (G) kidney; and (H) lung. Images were taken with a Zeiss Axioskop microscope.
In summary, low levels of CUX1 led to a striking expansion of mature granulocytes and monocytes in hematopoietic and nonhematopoietic tissues, accompanied by anemia and trilineage dysplasia. In human disease, CMML often has hepatosplenomegaly, normocytic anemia, giant platelets, micromegakaryocytes with abnormal lobation, and myeloid infiltrate of lymph nodes.38 JMML patients also often have hepatosplenomegaly, normocytic anemia, and myelomonocytic infiltrates of the liver, lung, and lymph nodes38 ; thus, the disease in Cux1low mice is classified as a MDS/MPN, akin to CMML-1 and JMML.36,38
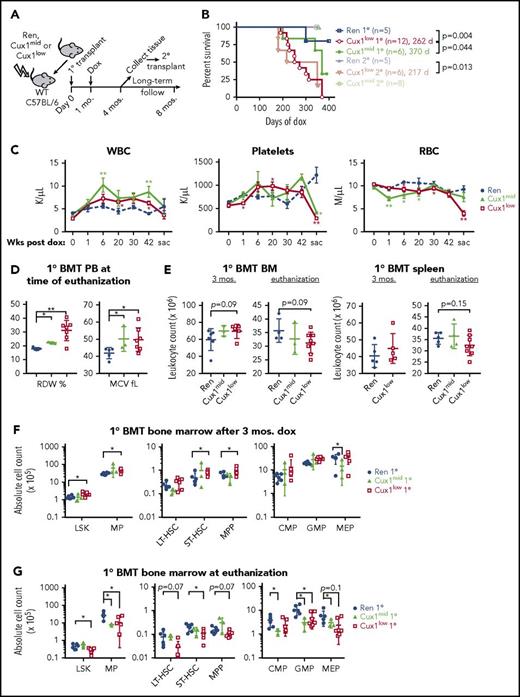
BM transplants develop transient hematopoietic expansion followed by CUX1 dose-dependent BM failure
To determine the hematopoietic-intrinsic role for CUX1, we performed BM transplants (BMTs) of Cux1mid, Cux1low, or Ren BM into lethally irradiated, wild-type recipients. We waited 4 weeks for the transplants to engraft before starting dox (Figure 4A). One cohort was euthanized after 3 months for analysis and to harvest BM for secondary transplantation. Compared with Ren BMTs, Cux1mid BMTs had shorter survival (median, 370 days of dox; Figure 4B). Cux1low primary BMT recipients had more rapid disease onset (median, 262 days of dox), which was not significantly different from nontransplanted Cux1low mice (median, 275 days of dox). Secondary Cux1low BMT recipients had the most rapid onset of disease (217 days), although this did not achieve statistical significance compared with primary Cux1low BMTs. Secondary Cux1mid BMTs survived at least 350 days, at which time they were euthanized for analysis.
Transient hematopoietic expansion is followed by HSC exhaustion and BM failure in CUX1-knockdown BM transplant recipients. (A) Schematic of the BM transplant (BMT) time course. BM from Cux1mid, Cux1low, or Ren mice was transplanted into lethally irradiated C57BL/6 wild-type recipients. Four weeks after transplant, recipient mice were treated continuously with dox. Most mice were monitored long-term, up to 350 to 370 days of dox or time of euthanization because of disease. A cohort of mice was euthanization after 3 months of dox for analysis and to harvest BM for secondary transplantation. Secondary transplant recipients were given dox starting on the day of transplantation. (B) Kaplan-Meier plot shows decreased survival of Cux1mid and Cux1low BM transplant recipients (log-rank test). (C) Complete blood count analysis shows a transient WBC and platelet count expansion in Cux1 knockdown BM transplant recipients, whereas WBC, platelets, and RBCs were decreased by the time of euthanization. The mean ± SEM is shown. (D) Blood cell indices at the time of euthanization show a macrocytic anemia. The mean ± SD is shown. (E) BM from hind legs and hips, and spleen leukocyte cellularity at the indicated time points. Cell counts for Cux1mid spleens were not determined for the 3-month time point. (F) Cux1low BM transplant recipients have greater numbers of HSPC after 3 months of dox, as determined by flow cytometry. The mean ± SD is shown. (G) At the time that diseased mice were euthanized because of anemia and weakness, HSPC were decreased in Cux1 knockdown BM transplant recipients compared with Ren BM transplant recipients. The mean ± SD is shown. (H) Absolute LT-HSC from the BM of secondary BMT recipients is shown at the time of euthanization for disease in Cux1low recipients and after 350 days of dox for Cux1mid recipients. (I) Competitive BM transplantation was performed with Cux1low, Cux1mid, or Ren BM (CD45.2) at a 1:10 ratio with competitor BM (CD45.1). Dox was given on the day of transplant. One replicate of 3 biological replicates is shown, n = 6 mice per experimental group. Flow cytometry of the peripheral blood shows that Cux1mid and Cux1low HSPCs outcompete controls in the total WBC population and myeloid lineages. The mean ± SD is shown. All data were analyzed with Wilcoxon rank test, *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Transient hematopoietic expansion is followed by HSC exhaustion and BM failure in CUX1-knockdown BM transplant recipients. (A) Schematic of the BM transplant (BMT) time course. BM from Cux1mid, Cux1low, or Ren mice was transplanted into lethally irradiated C57BL/6 wild-type recipients. Four weeks after transplant, recipient mice were treated continuously with dox. Most mice were monitored long-term, up to 350 to 370 days of dox or time of euthanization because of disease. A cohort of mice was euthanization after 3 months of dox for analysis and to harvest BM for secondary transplantation. Secondary transplant recipients were given dox starting on the day of transplantation. (B) Kaplan-Meier plot shows decreased survival of Cux1mid and Cux1low BM transplant recipients (log-rank test). (C) Complete blood count analysis shows a transient WBC and platelet count expansion in Cux1 knockdown BM transplant recipients, whereas WBC, platelets, and RBCs were decreased by the time of euthanization. The mean ± SEM is shown. (D) Blood cell indices at the time of euthanization show a macrocytic anemia. The mean ± SD is shown. (E) BM from hind legs and hips, and spleen leukocyte cellularity at the indicated time points. Cell counts for Cux1mid spleens were not determined for the 3-month time point. (F) Cux1low BM transplant recipients have greater numbers of HSPC after 3 months of dox, as determined by flow cytometry. The mean ± SD is shown. (G) At the time that diseased mice were euthanized because of anemia and weakness, HSPC were decreased in Cux1 knockdown BM transplant recipients compared with Ren BM transplant recipients. The mean ± SD is shown. (H) Absolute LT-HSC from the BM of secondary BMT recipients is shown at the time of euthanization for disease in Cux1low recipients and after 350 days of dox for Cux1mid recipients. (I) Competitive BM transplantation was performed with Cux1low, Cux1mid, or Ren BM (CD45.2) at a 1:10 ratio with competitor BM (CD45.1). Dox was given on the day of transplant. One replicate of 3 biological replicates is shown, n = 6 mice per experimental group. Flow cytometry of the peripheral blood shows that Cux1mid and Cux1low HSPCs outcompete controls in the total WBC population and myeloid lineages. The mean ± SD is shown. All data were analyzed with Wilcoxon rank test, *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Both Cux1mid and Cux1low BMTs had transient WBC expansions, whereas Cux1low BMTs also had transient increased platelet counts (Figure 4C). In contrast, RBC counts were decreased in Cux1mid and Cux1low BMTs during the time course (Figure 4C). Mice were euthanized because of weakness and/or anemia, at which time Cux1low BM recipients had pancytopenia with a macrocytic anemia (Figure 4C-D). Of the 4 Cux1mid BM recipients that became diseased, 2 were anemic, all were thrombocytopenic, and none had an elevated WBC count (Figure 4C).
At 3 months posttransplant, Cux1low and Cux1mid BM and spleen cellularity was normal or increased, similar to nontransplanted mice (Figure 4E). In contrast, the BM and spleens tended to be hypocellular in Cux1low and Cux1mid BMTs at the time of euthanization (Figure 4E). Both Cux1low and Cux1mid BMT recipients had remarkable dysplasia in megakaryocyte, erythroid, and granulocyte lineages (supplemental Figure 7). In sum, the cytopenias and trilineage dysplasia in Cux1mid and Cux1low BMT recipients are consistent with MDS, akin to RCMD.
At 3 months of dox, Cux1low BMT recipients had increased LSK, ST-HSC, MP, and MPP (Figure 4F). In stark contrast, most stem and myeloid progenitor populations were significantly depleted in Cux1low BMT at euthanization (Figure 4G). Cux1mid BMT had decreased myeloid progenitors at euthanization. Decreased HSPC numbers may partly explain the BM failure in these mice. Indeed, secondary Cux1mid and Cux1low BMT recipients had a significant reduction in LT-HSC, consistent with impaired HSC self-renewal (Figure 4H). Although Cux1low secondary BMT exhibited pancytopenia, Cux1mid secondary BMT did not within 350 days of dox (supplemental Figure 8E-F). The finding that Cux1mid secondary BMT had decreased LT-HSC suggests that these mice may ultimately develop anemia over longer times. These data indicate that CUX1 knockdown leads to a transient HSPC expansion followed by depletion, via a hematopoietic-intrinsic mechanism.
These findings led us to test the hypothesis that CUX1-knockdown HSPCs outcompete control cells in short-term competitive BMT. Because CUX1 is associated with CHIP, we transplanted Cux1low, Cux1mid, or Ren BM (CD45.2) at a 1:10 ratio with competitor BM (CD45.1) to model low-frequency clonal hematopoiesis.18-21 In these experiments, dox was given on the day of transplant to assess HSPC engraftment. Within 2 months, both Cux1mid and Cux1low outcompeted control cells, specifically in the myeloid lineages (Figure 4I); thus, CUX1 knockdown provides a myeloid clonal advantage for HSPCs.
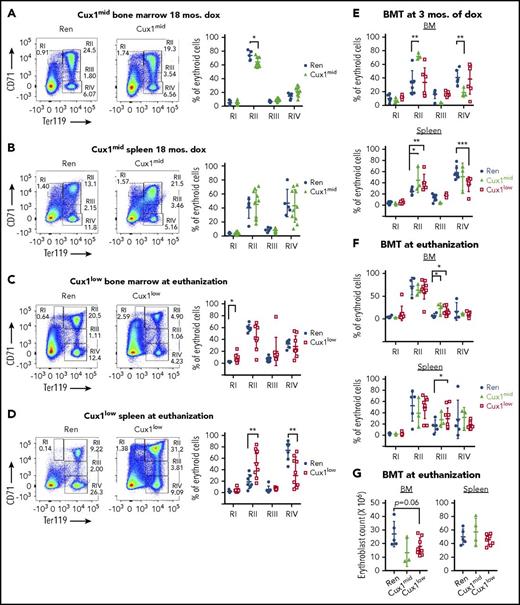
CUX1 knockdown leads to ineffective erythropoiesis
To better understand the cause of the anemia, we explored erythropoiesis in the mouse models. During erythroid maturation, CD71+/Ter119med (RI) pro-erythroblasts gradually lose CD71 expression as they progress through basophilic (RII), polychromatophilic (RIII), and ultimately orthochromatophilic (RIV) stages followed by enucleation.40 We gated on all erythroid cells and assessed the proportion of erythroblasts within each of these stages. Aged Cux1mid mice had a mild decrease in the BM RII population and normal splenic RI-RIV frequencies (Figure 5A-B). In contrast, Cux1low splenic erythroblasts had an increased frequency in RII and a decrease in RIV, with relatively normal BM populations (Figure 5C-D). The splenic RII expansion and RIV reduction was recapitulated in Cux1mid and Cux1low BMTs 3 months after transplant (Figure 5E-F). Thus, in nontransplanted Cux1low, as well as in Cux1mid or Cux1low BM recipients, there is a partial block in progression to the RIV orthochromatophilic erythroblast stage.
CUX1 knockdown leads to a partial block in erythropoiesis. Flow cytometric analysis of BM and spleen erythroblast populations was performed for: (A-B) Cux1mid mice treated with dox for 18 months; (C-D) Cux1low mice at the time of euthanization for disease; (E) primary BMT recipients 3 months after transplantation; and (F) primary BMT recipients at the time of euthanization. (G) Absolute count of all RI through RIV erythroblasts in the BM and spleens of BMT recipients at the time of euthanization is provided. Age-matched, littermate control Ren mice were used for all experiments. The erythroid cell populations from least to most differentiated are indicated as RI through RIV. RI, pro-erythroblasts; RII, basophilic erythroblasts; RIII, polychromatophilic erythroblasts; RIV, orthochromatophilic erythroblasts. (A-D) Representative flow analysis. The mean ± SD is shown. All data were analyzed with Wilcoxon rank test, *P ≤ .05, **P ≤ .01, ***P ≤ .001.
CUX1 knockdown leads to a partial block in erythropoiesis. Flow cytometric analysis of BM and spleen erythroblast populations was performed for: (A-B) Cux1mid mice treated with dox for 18 months; (C-D) Cux1low mice at the time of euthanization for disease; (E) primary BMT recipients 3 months after transplantation; and (F) primary BMT recipients at the time of euthanization. (G) Absolute count of all RI through RIV erythroblasts in the BM and spleens of BMT recipients at the time of euthanization is provided. Age-matched, littermate control Ren mice were used for all experiments. The erythroid cell populations from least to most differentiated are indicated as RI through RIV. RI, pro-erythroblasts; RII, basophilic erythroblasts; RIII, polychromatophilic erythroblasts; RIV, orthochromatophilic erythroblasts. (A-D) Representative flow analysis. The mean ± SD is shown. All data were analyzed with Wilcoxon rank test, *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Interestingly, at the time of disease in BMT recipients, the RII and RIV frequencies normalized (Figure 5F). At this time, the total erythroblast cell count was decreased in the BM of transplant recipients (Figure 5G). Because HSC exhibited evidence of exhaustion by this time (Figure 4), decreased erythropoiesis at euthanization may be largely due to upstream HSC defects, masking downstream erythroblast maturation defects.
We next determined if ineffective erythropoiesis was due to erythroblast apoptosis. After 1 month of dox, the frequency of Annexin V+ cells was similar in Cux1low and Ren erythroblast populations, suggesting the anemia is not due to decreased erythroblast survival (supplemental Figure 5G). In summary, these data indicate that CUX1 is novel transcriptional regulator of red cell development regulating at least 2 stages of erythropoiesis: (1) maintenance of HSCs and (2) progression into the orthochromatophilic erythroblast stage.
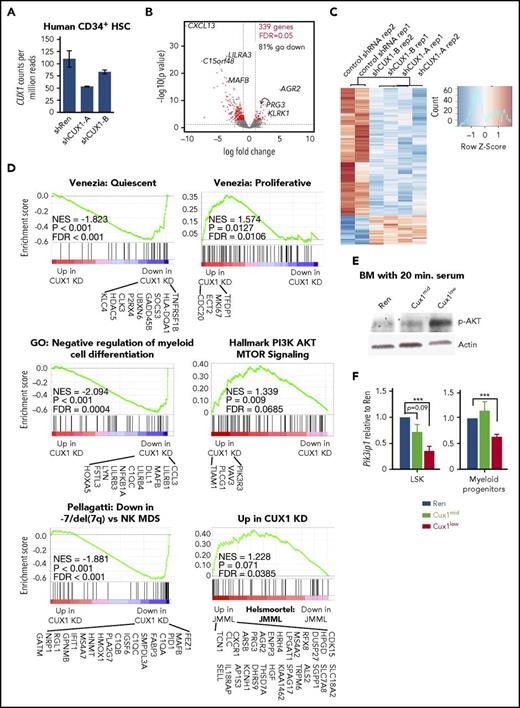
CUX1 regulates proliferative and phosphatidylinositol 3-kinase (PI3K) pathways and JMML and −7/del(7q) MDS gene signatures
These findings led to us to determine the genes regulated by CUX1 in primary human HSPC, the normal counterparts thought to give rise to myeloid malignancies.41 We transduced human CD34+ cells with shRNA targeting CUX1 or a control shRNA,7 resulting in approximately 50% and 80% residual CUX1 transcripts (Figure 6A). RNA-sequencing analysis identified 339 differentially expressed genes (5% false discovery rate), 81% of which decreased, indicating that CUX1 is primarily at transcriptional activator in this cell type (Figure 6B-C).
CUX1 regulates genes associated with MDS and JMML gene signatures and PI3K/AKT signaling. Primary human CD34+ HSPC were transduced with shCUX1-A, shCUX1-B, or nonspecific control shRNA. After 7 days, GFP+ CD34+ cells were sorted and RNA collected for RNA-sequencing analysis, with 2 biological replicates. (A) CUX1 transcript levels after CUX1 knockdown as determined by RNA-sequencing. (B) The volcano plot shows gene expression changes after CUX1 knockdown. Red indicates differentially expressed genes with a 5% false discovery rate (FDR). The horizontal dashed line represents a nominal P value = .05. Vertical dashed lines indicate ± one-fold change. (C) Heat map of the 339 differentially expressed genes identified after CUX1 knockdown, with a 5% FDR. (D) Gene set enrichment analysis of RNA-sequencing data. Gene sets used were: “Quiescent” and “Proliferative”43 ; GO gene set from the MSig Database,42 −7/del(7q) MDS vs normal karyotype MDS,33 and JMML.34 Genes listed are those that had nominal P values of <.05 (CUX1 knockdown vs control) and were within the GSEA “Core enrichment” results. (E) Whole BM was collected from Cux1mid, Cux1low, or Ren control mice after 5 days of dox, cultured in serum-free media for 2 hours, and stimulated with serum for 20 minutes. Whole cell lysates were probed for phospho-AKT by western blot. One representative blot of 3 biological replicates is shown. (F) qRT-PCR results from LSK and myeloid progenitors sorted from the indicated mice. One of 3 biological replicates is shown.
CUX1 regulates genes associated with MDS and JMML gene signatures and PI3K/AKT signaling. Primary human CD34+ HSPC were transduced with shCUX1-A, shCUX1-B, or nonspecific control shRNA. After 7 days, GFP+ CD34+ cells were sorted and RNA collected for RNA-sequencing analysis, with 2 biological replicates. (A) CUX1 transcript levels after CUX1 knockdown as determined by RNA-sequencing. (B) The volcano plot shows gene expression changes after CUX1 knockdown. Red indicates differentially expressed genes with a 5% false discovery rate (FDR). The horizontal dashed line represents a nominal P value = .05. Vertical dashed lines indicate ± one-fold change. (C) Heat map of the 339 differentially expressed genes identified after CUX1 knockdown, with a 5% FDR. (D) Gene set enrichment analysis of RNA-sequencing data. Gene sets used were: “Quiescent” and “Proliferative”43 ; GO gene set from the MSig Database,42 −7/del(7q) MDS vs normal karyotype MDS,33 and JMML.34 Genes listed are those that had nominal P values of <.05 (CUX1 knockdown vs control) and were within the GSEA “Core enrichment” results. (E) Whole BM was collected from Cux1mid, Cux1low, or Ren control mice after 5 days of dox, cultured in serum-free media for 2 hours, and stimulated with serum for 20 minutes. Whole cell lysates were probed for phospho-AKT by western blot. One representative blot of 3 biological replicates is shown. (F) qRT-PCR results from LSK and myeloid progenitors sorted from the indicated mice. One of 3 biological replicates is shown.
Gene set enrichment analysis42 determined that genes preferentially expressed in quiescent HSPC43 were downregulated after CUX1 knockdown (Figure 6D). Conversely, genes preferentially expressed in proliferative HSPC43 were upregulated. This suggests a transcriptional role for CUX1 in regulating HSPC quiescent vs proliferative states. Genes upregulated by activation of the PI3K/AKT/mTOR pathway (ie, the “Hallmark PI3K AKT MTOR Signaling” gene set), were enriched in upregulated genes, which is notable because increased PI3K signaling is associated with HSC exit from quiescence and proliferation.44 The Gene Ontology pathway, “Negative regulation of myeloid cell differentiation” was downregulated, indicating CUX1 represses myeloid differentiation. This is in accordance with the myeloid expansion we observed after CUX1 knockdown in vivo.
To test whether CUX1-regulated genes are associated with human disease, we compared the transcriptional profile of CUX1-knockdown HSPC to gene signatures from MDS patients.33 Strikingly, genes that are significantly decreased in −7/del(7q) MDS compared with normal karyotype MDS were also decreased after CUX1 knockdown in human HSPCs (Figure 6D). Because of the myelomonocytic phenotype of the MPD in Cux1low mice, we next assessed if CUX1-regulated genes are enriched for a JMML gene signature. Using microarray data from JMML patients and healthy controls, we found that genes upregulated in JMML are enriched for genes upregulated after CUX1 knockdown (Figure 6D). Thus, the CUX1 gene signature is associated with JMML and −7/del(7q) MDS disease states, consistent with the JMML and MDS phenotypes of the CUX1 knockdown mice.
CUX1 knockdown induced genes upregulated by PI3K signaling (Figure 6D), implying increased PI3K activity. Indeed, Cux1low BM cells have increased phosphorylation of the PIK3 substrate, AKT (Figure 6E). These data align with reports that CUX1/Cut suppresses PI3K activity,17,45 by transcriptionally activating expression of the PI3K inhibitor, PIK3IP1.17 In agreement, we determined CUX1 knockdown leads to decreased Pik3ip1 in Cux1low LSK and MP (Figure 6F). Although Cux1mid LSKs had a slight decrease in Pik3ip1, we did not reliably detect increased phospho-AKT in Cux1mid whole BM. Perhaps BM heterogeneity masks putatively increased phospho-AKT in rare HSPC in Cux1mid mice. Overall, these findings fit a model wherein CUX1 knockdown leads to decreased PI3KIP1, increased PI3K activity, and resulting in increased stem/progenitor cell-cycle entry.
CUX1 regulates HSPC homeostasis
To further understand the role of CUX1 in HSPC differentiation and self-renewal, we performed colony-forming unit (CFU) and serial replating assays. Because CUX1 knockdown alters HSPC populations, we used BM from dox-naïve mice and provided dox only during the assay. Cux1low BM had a significant increase in CFU on passage 1 (Figure 7A; supplemental Figure 9). CUX1 knockdown colonies were larger than Ren, particularly Cux1low at passage 1 and Cux1mid at passage 2 (supplemental Figure 9). Indeed, the number of cells enumerated at the end of most passages was greater for Cux1low and Cux1mid, consistent with increased progenitor proliferation (Figure 7B). Both Cux1low and Cux1mid BM had increased CFU in passages 2 and 3 and extinguished by passage 4 (Figure 7A). It is unclear why Cux1mid CFU increased only after the first passage. Because of the decreased potency of Cux1mid shRNA, perhaps Cux1mid lagged Cux1low in achieving sufficient CUX1 knockdown, resulting in a delayed phenotype. Overall, the CFU results are in agreement with the in vivo data demonstrating temporarily increased HSPC number and proliferation but decreased long-term self-renewal (Figure 4). In addition, these in vitro assays further confirm the cell-intrinsic role for CUX1 regulation of HSPCs.
CUX1 regulates HSPC numbers, proliferation, and lineage specification. Total BM cells isolated from dox-naïve Cux1mid, Cux1low, or littermate Ren mice were mixed with M3434 methylcellulose medium (25 000 cells per well) and dox for CFU assays. At each passage (P1-P4), cells were collected and 25 000 cells were replated, with 7 to 11 days between each passage. (A) Total colony numbers and (B) cell counts were quantified at the end of each passage (n = 3). (C) Specific colonies (CFU-GM, BFU-E, and CFU granulocyte, erythrocyte, monocyte, and megakaryocyte [CFU-GEMM]) were identified by morphology days 10 through 12 in a separate CFU assay. (D) BM cells were collected from Cux1mid, Cux1low, and Ren littermate controls after 3 days of dox. Lin− cells were cultured in StemSpan SFEM medium (Stemcell Technologies) supplemented with 100 ng/mL of murine stem cell factor, throbopoietin, and Flt3 and dox for 3 days before flow analysis. Representative flow cytometric analysis of HSPC cell-cycle analysis is shown. (E) The bar graph represents the mean of 3 biological replicates ± SEM. All data were analyzed with Student t test, *P ≤ .05, **P ≤ .01, ***P ≤ .001.
CUX1 regulates HSPC numbers, proliferation, and lineage specification. Total BM cells isolated from dox-naïve Cux1mid, Cux1low, or littermate Ren mice were mixed with M3434 methylcellulose medium (25 000 cells per well) and dox for CFU assays. At each passage (P1-P4), cells were collected and 25 000 cells were replated, with 7 to 11 days between each passage. (A) Total colony numbers and (B) cell counts were quantified at the end of each passage (n = 3). (C) Specific colonies (CFU-GM, BFU-E, and CFU granulocyte, erythrocyte, monocyte, and megakaryocyte [CFU-GEMM]) were identified by morphology days 10 through 12 in a separate CFU assay. (D) BM cells were collected from Cux1mid, Cux1low, and Ren littermate controls after 3 days of dox. Lin− cells were cultured in StemSpan SFEM medium (Stemcell Technologies) supplemented with 100 ng/mL of murine stem cell factor, throbopoietin, and Flt3 and dox for 3 days before flow analysis. Representative flow cytometric analysis of HSPC cell-cycle analysis is shown. (E) The bar graph represents the mean of 3 biological replicates ± SEM. All data were analyzed with Student t test, *P ≤ .05, **P ≤ .01, ***P ≤ .001.
To test if CUX1 is acting at an early stem/progenitor stage to regulate lineage specification, we identified colony morphologies. We consistently observed that Cux1low led to an excess of CFU-granulocyte/monocyte (CFU-GM) and a paucity of burst-forming units (BFU-Es, erythroid; Figure 7C). Cux1mid also had decreased BFU-Es. Although Cux1mid BM did not demonstrate increased CFU-GM in this assay, we note that the majority of increased CFU in passages 2 and 3 of serial replating of Cux1mid BM were CFU-GM (Figure 7A). These data concur with the in vivo myelomonocytic expansion and anemia of Cux1low and Cux1mid mice and indicate that CUX1 levels control cell fate of early stem and/or progenitor cells.
The elevated PI3K activity upon CUX1 knockdown led us to test the hypothesis that CUX1 knockdown promotes HSC exit from quiescence.44 Cell-cycle analyses showed a significant reduction in the fraction of Cux1mid and Cux1low LT-HSC in the quiescent, G0, state and a concordant increase in the fraction of cells in S/G2 (Figure 7D-E). ST-HSC, MPP, and GMP also had an increase in frequency of cycling cells in S/G2 (Figure 7D-E). These findings show that CUX1 knockdown promotes LT-HSC exit from quiescence and cell cycling and are in agreement with the expansion of downstream ST-HSC in vivo and the quiescent and proliferative gene signatures in human HSPC.
Discussion
In our studies, all transplant and nontransplant CUX1 knockdown mouse models had anemia and multilineage dysplasia in common, demonstrating a hematopoietic-intrinsic role for CUX1 reduction in MDS (Table 1). The nontransplanted Cux1low mice additionally exhibit myelomonocytic expansion, comparable to MDS/MPN. In many ways, the Cux1low phenotypes were stronger than those of Cux1mid; for example, BMT survival, AKT phosphorylation, and fur loss, indicating a dosage effect. A prior study of mice homozygous for deletion of the CUX1 homeodomain (Cux1HD/HD) reported only 5.4% were viable and few survived beyond 4 week of age, precluding long-term analysis.12 However, as with Cux1low mice, Cux1HD/HD mice had hair loss, cachexia, and myeloid hyperplasia; elevated tumor necrosis factor was thought to explain some of the extrahematopoietic effects.12 We speculate that the myelomonocytic expansion in Cux1low mice is also due to hematopoietic-extrinsic inflammatory cytokines stemming from ubiquitous, low CUX1 levels.
Hematopoietic abnormalities have not been reported for Cux1HD/wt heterozygous mice.12 However, based on our work, the phenotype would not be apparent until 18 months; it is unclear how long heterozygous mice were aged in the prior study. In a second Cux1 knock-out mouse model, with deletion of only the first cut repeat domain, no hematopoietic abnormalities were found in heterozygous or homozygous mice at 2-3 weeks of age.28 This may be due to the young age of the mice and/or residual hypomorphic CUX1 protein in that model.
Our studies show that CUX1 is essential for maintaining HSC quiescence, suppressing HSPC proliferation and self-renewal, and regulating lineage specification and differentiation. We propose a model wherein at the top of the hematopoietic hierarchy, CUX1 promotes LT-HSC quiescence through transcriptional activation of Pik3ip1, thereby limiting growth factor–induced signaling pathways. CUX1 knockdown leads to increased PI3K activity, a central activator of the HSC cell cycle,44 promoting LT-HSC exit from quiescence and proliferation. These LT-HSCs give rise to ST-HSC, expanding the ST-HSC pool and downstream progenitors. Eventually, the HSC exhaust and are depleted, leading to BM failure. These findings are consistent with prior reports that increased PI3K signaling leads to transient HSC proliferative expansion followed by exhaustion.46-48
In MPP, loss of CUX1 causes increased proliferation and a myeloid differentiation bias, increasing downstream myeloid progenitor numbers, and ultimately myelomonocytic expansion. CUX1 is also necessary for normal erythroid and megakaryocyte development because CUX1 deficiency led to dysplasia and cytopenia in both lineages. Acting at multiple stages of hematopoiesis, CUX1 reduction generally leads to the mobilization of myeloid progenitors with differentiation defects. Increased proliferation and aberrant differentiation are the cardinal features of myeloid malignancies, and CUX1 deficiency drives both.
Our results shed light on how del(7q)49,50 and somatic CUX1 inactivating mutations18,19 give a clonal advantage in CHIP. CUX1 haploinsufficiency may provide human HSC with a proliferative advantage that enables mutant HSC to outcompete wild-type counterparts, resulting in clonal expansion, as we observed in competitive transplantation assays. That CUX1 knockdown eventually causes HSC exhaustion provides an intriguing biological explanation for the transient −7 or del(7q) that has been observed in some children and adults.51,52 A second genetic alteration may be required to increase the long-term survival and/or self-renewal of −7/del(7q) clones, thereby enabling transformation.
We reported that in one-half of −7/del(7q) cases, the “second hit” is a mutation that results in Ras pathway activation.53 Oncogenic Ras signaling may provide the enhanced self-renewal capacity necessary for sustained CUX1-deficient HSC clonal expansion.54 Of note, effectors of Ras include the PI3K/AKT/mTOR and Raf/MEK/ERK axes. Our finding that CUX1 knockdown enhances PI3K signaling raises the prospect that CUX1 haploinsufficiency cooperates with Ras by potentiating Ras signaling via increased PI3K activity.
There is evidence for other pathogenic genes on 7q,55-61 provoking the notion of a contiguous gene syndrome, wherein haploinsufficiency of multiple TSGs contributes to disease.62 Nonetheless, our data provide strong evidence that CUX1 knockdown alone is sufficient for MDS and MDS/MPN, especially because the disease was also reversible upon CUX1 restoration. It will be critical to identify how combined haploinsufficiency of 7q genes cooperate in neoplasia, however.
Cut, in Drosophila, is exquisitely dose sensitive and changes in cut levels can result in dramatically different cellular programs, including proliferation, differentiation, and apoptosis, even within the same cell type.24-26 Transcription factor dosage can be critical in myeloid differentiation and transformation, as evidenced, for example, by Pu.1.63,64 In future work, it will be important to determine the threshold of CUX1 necessary for maintaining myeloid TSG activity and if changes in CUX1 levels also dictate hematopoietic cell fate, as described for cut in other models.25,26
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Angela Stoddart, Jeffrey Kurkewich, and Kevin Shannon for critical reading of the manuscript. The authors also thank Christine Labno and The University of Chicago’s Integrated Light Microscopy Core Facility, Pieter Faber and the High Throughput Genomics Core Facility, and David Leclerc in the Flow Cytometry Facility for special assistance and services. Linda Degenstein and the Transgenic Mouse and Embryonic Stem Cell Facility helped generate the Cux1mid mice. Lentivirus was packaged and concentrated at the Northwestern University RNA/DNA Delivery Core with special assistance from Alex Yemelyanov.
This work was supported by grants from the National Institutes of Health (NIH) National Cancer Institute (K08CA181254) (M.E.M.), The University of Chicago Medicine Comprehensive Cancer Center Support Grant (P30CA014599), NIH National Institute of General Medicine Sciences (T32GM007281) (M.K.I.), NIH National Institute of Allergy and Infectious Diseases (T32AI007090) (S.N.K.), and NIH National Center for Advancing Translational Sciences (UL1 TR000430); V Foundation for Cancer Research V Foundation Scholar Award; American Cancer Society Institutional Research Grant (IRG-58-004); Cancer Research Foundation Young Investigator Award; and the University of Chicago Cancer Research Foundation Auxiliary Board.
Authorship
Contribution: M.E.M. conceived the experiments, analyzed the RNA-seq data, and wrote the manuscript. N.A. performed the mouse experiments and in vitro assays, analyzed data, and edited the manuscript. S.K. performed human hematopoietic stem and progenitor cell experiments, western blots, and mouse genotyping. M.K.I. performed quantitative reverse transcription polymerase chain reaction. S.N.K. performed apoptosis assays. S.K.G. performed hematopathologic analyses and edited the manuscript. M.R.B. generated the shCux1.581 LMNcherry construct.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Megan E. McNerney, Department of Pathology, The University of Chicago, Knapp Center for Biological Discovery Room 5128, 900 E. 57th St, Chicago, IL, 60637; e-mail: megan.mcnerney@uchospitals.edu.





![Figure 7. CUX1 regulates HSPC numbers, proliferation, and lineage specification. Total BM cells isolated from dox-naïve Cux1mid, Cux1low, or littermate Ren mice were mixed with M3434 methylcellulose medium (25 000 cells per well) and dox for CFU assays. At each passage (P1-P4), cells were collected and 25 000 cells were replated, with 7 to 11 days between each passage. (A) Total colony numbers and (B) cell counts were quantified at the end of each passage (n = 3). (C) Specific colonies (CFU-GM, BFU-E, and CFU granulocyte, erythrocyte, monocyte, and megakaryocyte [CFU-GEMM]) were identified by morphology days 10 through 12 in a separate CFU assay. (D) BM cells were collected from Cux1mid, Cux1low, and Ren littermate controls after 3 days of dox. Lin− cells were cultured in StemSpan SFEM medium (Stemcell Technologies) supplemented with 100 ng/mL of murine stem cell factor, throbopoietin, and Flt3 and dox for 3 days before flow analysis. Representative flow cytometric analysis of HSPC cell-cycle analysis is shown. (E) The bar graph represents the mean of 3 biological replicates ± SEM. All data were analyzed with Student t test, *P ≤ .05, **P ≤ .01, ***P ≤ .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/24/10.1182_blood-2017-10-810028/5/m_blood810028f7.jpeg?Expires=1766393423&Signature=XtnZCPgJmqwrl~gIkOQsMqWfgJT1FR6JjCiP7UL2sFkg5EusogEEmMWs1QeTowmKSM5HN~cba4USPBeqhbuQMj2FSKCm9Rfo0fWhc4Si1bv1TWfpoiabBKFG9Zw3tTcUxgKFwooU6Gxbeahk9zZVMMEoIkA3Xlzxx1Fgh4Ltdap8J8YCQmg2uDcYtT6IKLdZvIAzU8iZt5~u~Y1BoVmdElAiUALoAltV7wgZK9dgDfDDbdZingFozlqfQbzMekEbcWmh4jjfWQwctmvyewWe8tb-RJA1HzLfbOFS2ecdKUYIu18CbeEqupc39PLryDNPi4yLb~qVOrkL9M6zxMo5IA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
