TO THE EDITOR:
Population-level germ line heterogeneity, rare single nucleotide polymorphisms (SNPs), and sequence alignment to a single reference genome make it challenging for variant-calling algorithms to distinguish germ line variants from somatically acquired mutations with high throughput sequencing data sets.1 In solid tumors, blood is sequenced to facilitate the resolution of somatic mutations, distinguishing them from germ line variants through calling algorithms that consider two different sources of tissue when using the Bayesian and other statistical approaches.2-6 In myelodysplastic syndromes (MDS), SNP array karyotyping and targeted sequencing show concurrence in bone marrow (BM) and peripheral blood somatic variants.7,8 For this reason, normal skin biopsy tissue is used for germ line controls in hematologic malignancies. The feasibility of skin collection is low at many centers, so a comprehensive study of other potential normal germ line tissues is required. By using a prospective study design, we assessed the impact of quantity, quality, and hematopoietic contamination on somatic mutation detection in 4 candidate germ line tissues by using whole exome sequencing (WES) and capture-based targeted resequencing validation (supplemental Figure 1, available on the Blood Web site), which highlights the strengths and weaknesses of these germ line controls in mutation detection and establishes a new benchmark for future genomic studies in MDS.
Twenty-six patients (supplemental Table 1) who met a World Health Organization–defined MDS diagnosis were prospectively recruited from May to September 2015 at the H. Lee Moffitt Cancer Center & Research Institute and consented to collection of data and tissues under an institutional review board–approved protocol.9 Of the 26 patients enrolled, 16 were included for WES analysis and 10 were omitted because of myeloid contamination >5% in the T-cell germ line samples and for other reasons detailed in supplemental Table 3. Two rounds of negative selection or flow sorting (supplemental Figure 2A-D) were used to achieve 95% T-cell purity.
On the basis of the results in supplemental Table 2, 4 germ line control tissues were subjected to WES for comparison with BM mononuclear cells (BM-MNCs; 0.3-7.4 × 106 total cells), although the following 7 candidate germ line tissues were initially considered: a 2- to 4-mm biopsy of normal skin obtained during the BM procedure (n = 26), 12 eyebrow hair follicles (n = 26), 1.6 to 8.0 × 106 purified CD3+ T cells (n = 26), one buccal swab (n = 11), 33 to 100 mL of urine (n = 26), 10 fingernail clippings (n = 3), and 2 foam-tipped swabs for normal skin, each swiped 10 times on the forearm (n = 26) (details are provided in supplemental Data). Poor DNA quality metrics and low quantities led to discontinuing the use of urine, fingernails, and skin swabs. Buccal swabs were tested on 11 patients after the procedure was optimized for recovering epithelial cells (supplemental Methods; supplemental Figure 2E). By using an algorithm focused on genotype overlap across a subset of SNPs (>15% variant allele fraction [VAF] in 1000 Genomes), the identification of matching BM and germ line samples was validated (supplemental Figure 3).1
Although 200- to 500-ng input is now widely used for exome sequencing, whole genome amplification (WGA) was required in germ line tissue samples with ≤1.5 μg of DNA (eyebrow hair follicles and skin). WGA was also performed in all BM-MNC samples for comparison with native WES sequencing, which identified significant differences in coverage, allele dropout, WGA artifacts, and VAF (supplemental Figure 4). Targeted resequencing validation was performed with native DNA.
The median depth of coverage was 92× for WES and 404× for targeted resequencing. Despite lower DNA integrity numbers, an indicator of lower quality, postsequencing results for buccal DNA were comparable to those for other tissues, suggesting that buccal DNA is suitable for our sequencing applications (Table 1). After validation by targeted resequencing, 245 protein coding variants across all patient samples were identified in at least one BM-germ line comparison, which included 28 (10.2%) previously reported MDS-associated variants (supplemental Table 3).10 The percentage of variants identified in the respective tumor germ line pairs of the total variants detected (in any germ line pair) was assessed. The highest variant calling rate, 197 of 245 variants (80.4%; 12.3 variants per patient [range, 0-22]), was observed in the BM-T-cell comparison, followed by hair follicles (78%) and buccal swabs (71.7%) (Table 1). All BM–germ line comparisons had nonconcordant somatic variant calls. Here, variants were considered false negatives when they were missed in 1 of 4 germ line tissues or missed in 1 of 3 germ line tissues if buccal swabs were not sequenced (Figure 1; Table 1). In BM–skin biopsy analyses, 51 such false-negative somatic calls were noted, representing 92.7% (mean, 3.6 missed somatic variants per patient [range, 0-15 somatic variants]) of all such variants. BM–hair follicle and BM–T-cell comparisons missed only 1 mutation (1.8%) and BM-buccal swab pairs missed only 2 mutations (3.6%) (Table 1), indicating that hair follicle, T-cell, and buccal swab germ line tissues generated higher-confidence calls with few false-negative variants.
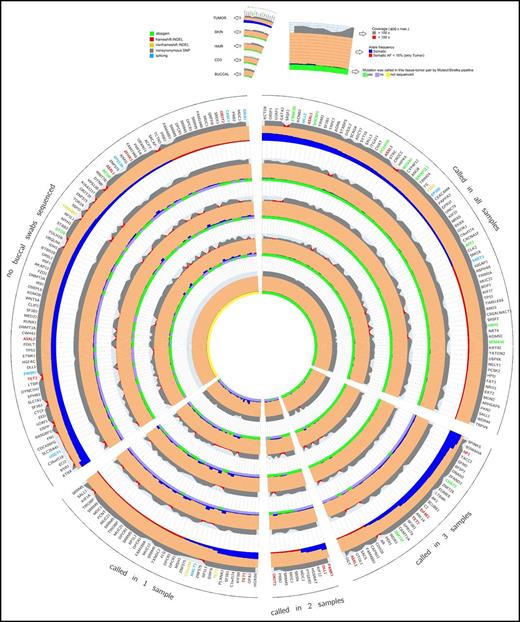
Circos diagram of tumor germ line sequencing metrics. This diagram shows a representation of capture-validated somatic mutations that occur with VAF >5% in the BM sample.11 The 5 wedges represent 4 groups of mutations called by the MuTect/Strelka pipeline and a group with missing values resulting from lack of buccal swab sample collection. The gene names orthogonal to the radius represent protein-changing somatic mutations color coded by the functional class of the mutation. The 5 main rings represent the different tissues and are further separated into 3 sections. The outer section represents the coverage at the position of the somatic mutation followed by the VAF, and the inner section represents the binary calling status (true/false) of the 4 germ line samples called by the MuTect/Strelka pipeline. Coverage, reference, VAFs, and predicted protein consequence (ANNOVAR software) across all resequenced variants are ordered by number of tumor-germ line pairs that call that variant. Each ring represents a sequenced tissue type, and those tumor–germ line pairs that called the plotted variant are denoted in green (gray if not called).
Circos diagram of tumor germ line sequencing metrics. This diagram shows a representation of capture-validated somatic mutations that occur with VAF >5% in the BM sample.11 The 5 wedges represent 4 groups of mutations called by the MuTect/Strelka pipeline and a group with missing values resulting from lack of buccal swab sample collection. The gene names orthogonal to the radius represent protein-changing somatic mutations color coded by the functional class of the mutation. The 5 main rings represent the different tissues and are further separated into 3 sections. The outer section represents the coverage at the position of the somatic mutation followed by the VAF, and the inner section represents the binary calling status (true/false) of the 4 germ line samples called by the MuTect/Strelka pipeline. Coverage, reference, VAFs, and predicted protein consequence (ANNOVAR software) across all resequenced variants are ordered by number of tumor-germ line pairs that call that variant. Each ring represents a sequenced tissue type, and those tumor–germ line pairs that called the plotted variant are denoted in green (gray if not called).
Next, we evaluated the potential for false-positive variants defined by the presence of a variant in only 1 of 4 BM–germ line analyses (1 of 3 if buccal swabs were not sequenced). A total of 77 somatic mutations were considered false positives on the basis of this criteria; 8 variants per patient (10.4%; 0.5 variants per patient [range, 0-7 variants]) were seen in BM-skin pairs, 29 (37.7%; 1.8 variants per patient [range, 0-10 variants]) in BM-hair follicle DNA pairs, 37 (48.1%; 2.3 variants per patient [range, 0-8 variants]) in BM–T-cell pairs, and 3 (3.9%; 0.2 variants per patient [range, 0-3 variants]) in BM–buccal swab pairs (Table 1).
To explore missed variant calls, a comprehensive list of sequencing metrics was assessed across BM germ line tissues by using Circos diagrams11 sorted by descending VAF in the BM and grouped by the number of BM germ line pairs that called each specific variant (Figure 1) or by individual patient samples (supplemental Figure 5). No significant differences were seen in BM-MNC coverage, BM-MNC VAF, or germ line tissue coverage at false-negative variants, indicating that lack of coverage was not responsible for these discrepant calls (Figure 1). When isolating only those variants missed by 1 germ line control, neoplastic contamination of MDS cells in the germ line samples was a likely contributor. Plotting the VAF of known MDS somatic mutations in each candidate germ line control revealed the largest degree of possible neoplastic contamination in skin biopsy samples (supplemental Figure 6),12 in agreement with the observation that a higher fraction of variants was not called in BM-skin biopsy analyses using the MuTect/Strelka pipeline.
Given the high degree of missed mutations called with BM-skin biopsy pairs, we tested VarScan,4 another variant caller that uses a distinct statistical methodology, to see if it would perform more favorably when using skin as the germ line control because its heuristic approach was hypothesized to overcome neoplastic contamination. Moreover, VarScan is one of several algorithms used in leukemia studies that incorporate skin as a germ line control.4 By using published parameters (Supplement, Variant calling, and Settings), VarScan identified variants missed by MuTect/Strelka in a subset of variants (27 of 56) from BM-skin biopsy pairs. However, 12 of 56 variants called by MuTect/Strelka were not called by VarScan against any of the tested germ line controls, suggesting that there are differences between methods (supplemental Figure 7). VarScan also called unique false positives relative to other tissues tested.
Here, skin biopsies were contaminated with neoplastic variants, which can result in missed variant identification when using our variant calling approach. Despite the hematopoietic origin of T cells, they yield sufficient DNA and high rates of somatic variant calls when highly purified (>95%). Epithelial cells derived from buccal swabs are suitable controls when procedures to minimize leukocyte contamination are implemented, and buccal swabs are superior to hair follicles because of low DNA quantity per hair. This study will be used to inform The National Myelodysplastic Syndromes (MDS) Study (NCT02775383), which will recruit 2000 MDS patients, patients with MDS/myeloproliferative neoplasms overlap disorder, and 500 patients with idiopathic cytopenia of undetermined significance through a multi-institutional, cooperative group mechanism. Given the challenge of obtaining skin biopsies across multiple enrollment sites, T cells and/or buccal swabs (from an optimized protocol) would be preferential germ line tissues for MDS genomic studies.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the individual members of the Protocol Writing Team and Steering Committee for their valuable contributions to The National MDS Study and for their help preparing this manuscript.
This work was supported by the National Institutes of Health, National Heart Lung and Blood Institute (NHLBI) contracts HHSN268201400003I and HHSN268201400002I as part of The National MDS Study (https://thenationalmdsstudy.net/), a collaboration between the NHLBI and the National Cancer Institute (NCI) being conducted in clinical sites that participate in NCI’s National Clinical Trials Network and the NCI Community Oncology Research Program, and by Shared Resources, including the Tissue Core and Genomics and Bioinformatics Core at the H. Lee Moffitt Cancer Center & Research Institute, an NCI-designated Comprehensive Cancer Center (P30-CA076292).
Authorship
Contribution: E.P. designed the research, analyzed data, and helped write the article; M.C.B. performed research, analyzed data, created display items, and helped write the article; J.K.T. performed and designed research, analyzed data, and helped write the article; J.S.P. performed bench research and tissue extractions and helped write the article; S.J.Y. analyzed data and contributed to the study design; C.Z. performed experiments; L.Z. and L.C.M. confirmed the diagnosis for study assignment; D.E.R. contributed to research design and methods; S.D.G., R.B., M.J.W., and M.A.S. contributed to the study design and helped write the article; R.S.K. served as trial leader, recruited patients, and helped write the article; and P.K.E.-B. designed research, created display items, served as project director, and helped write the article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pearlie K. Epling-Burnette, H. Lee Moffitt Cancer Center & Research Institute, 12902 Magnolia Dr, 23033 SRB, Tampa, FL 33612; e-mail: pearlie.burnette@moffitt.org.
References
Author notes
E.P. and M.C.B. contributed equally to this study.