Key Points
AGGF1-PDGFRB is a novel oncogenic fusion gene in Ph-like ALL.
Genomic profiling and functional studies identified a novel PDGFRB mutation directly related to TKI resistance.
Abstract
Philadelphia chromosome (Ph)-like acute lymphoblastic leukemia (ALL) comprises ∼10% to 15% of childhood ALL cases, many of which respond exquisitely to tyrosine kinase inhibitors (TKIs), for example, imatinib in PDGFRB-rearranged ALL. However, some cases developed drug resistance to TKIs and the mechanisms are poorly understood. In this study, we identified a novel PDGFRB fusion gene, namely AGGF1-PDGFRB, and functionally characterized its oncogenic potential in vitro. Further genomic profiling of longitudinally collected samples during treatment revealed the emergence of a mutation, PDGFRBC843G, which directly conferred resistance to all generations of ABL TKIs, including imatinib, dasatinib, nilotinib, and ponatinib. PDGFRB-mutant leukemia cells are highly sensitive to multitarget kinase inhibitor CHZ868, suggesting potential therapeutic options for some patients resistant to ABL TKIs. In summary, we describe a complex clonal evolution pattern in Ph-like ALL and identified a novel PDGFRB point mutation that drives leukemia relapse after ABL TKI treatment.
Medscape Continuing Medical Education online

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and the American Society of Hematology. Medscape, LLC is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.00 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 2273.
Disclosures
CME questions author Laurie Barclay, freelance writer and reviewer, Medscape, LLC, owns stock, stock options, or bonds from Pfizer. Associate Editor Jacob M. Rowe and the authors declare no competing financial interests.
Learning objectives
Assess clinical characteristics and a novel oncogenic PDGFRB fusion gene in a patient with Philadelphia-like (Ph-like) acute lymphoblastic leukemia (ALL), based on a case report and genomic analysis.
Describe PDGFRB mutation-mediated resistance to tyrosine kinase inhibitors in a patient with Ph-like ALL.
Identify a potential therapeutic strategy to overcome such drug resistance in Ph-like ALL.
Release date: May 17, 2018; Expiration date: May 17, 2019
Introduction
Tyrosine kinase inhibitors (TKIs) such as imatinib and dasatinib are potent antileukemic agents; they inhibit aberrant kinase signaling, which is essential for leukemia survival. In fact, the addition of ABL1 inhibitors in the context of conventional chemotherapeutic regimens dramatically improved treatment outcome of Philadelphia chromosome–positive (Ph+) acute lymphoblastic leukemia (ALL) in children and adults.1-4 However, a substantial proportion of these patients develop drug resistance due to acquired mutations in the ABL1 gene.5,6 More recently, genomic studies identified a novel subtype of ALL that is characterized by a Ph-like gene expression signature and activating alterations in a variety of targetable ABL class kinases (ABL1, ABL2, PDGFRB, CSFR).7-10 In particular, Ph-like ALL cases with PDGFRB fusions (eg, EBF1-PDGFRB) have been described for exquisite clinical response to dasatinib or imatinib.7,11-15 However, there is a paucity of data on the molecular basis of relapse in Ph-like ALL patients receiving ABL TKI treatment.16,17
In this study, we identified and characterized a novel oncogenic PDGFRB fusion gene in a patient with Ph-like ALL, PDGFRB mutation–mediated TKI resistance, and also a potential therapeutic strategy for overcoming such drug resistance.
Study design
The patient was treated at Blood Diseases Hospital, Chinese Academy of Medical Sciences from November 2012 to December 2014, according to the Chinese Children’s Leukemia Group (CCLG) 2008 protocol. Bone marrow samples were collected at diagnosis, longitudinally during therapy, and at relapse after ABL TKI therapy. Saliva was used for extracting germ line DNA. The patient and carriers involved in our study signed an informed consent form approved by the institutional review board of the Institute of Hematology, Blood Diseases Hospital, Peking Union Medical College/Chinese Academy of Medical Sciences (PUMC/CAMS) (for additional details, see supplemental Methods, available on the Blood Web site).
Results and discussion
A 4-year-old boy was diagnosed with precursor B-cell ALL in November 2012 with normal karyotype. Cytogenetic analysis using fluorescence in situ hybridization suggested a possible PDGFRB rearrangement (supplemental Figure 1). The patient was refractory to conventional chemotherapeutic agents and failed to achieve complete remission even after prolonged induction therapy. Imatinib was then given as monotherapy and induced rapid leukemia clearance with decrease of blast percentage from 51% to 3% within 8 weeks. Unfortunately, the patient relapsed 5 months later with no response to further treatment with dasatinib and nilotinib, and then went on to receive bone marrow transplant therapy (Figure 1A).
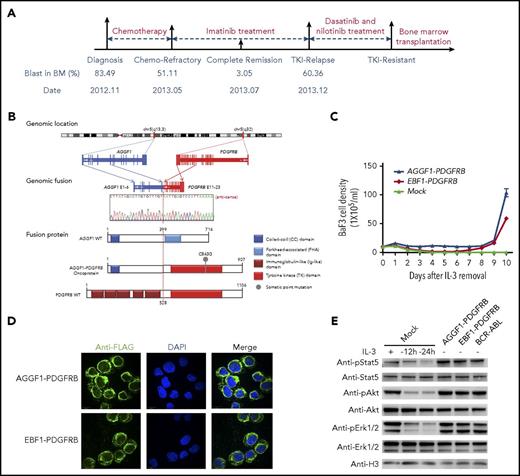
A novel fusion gene of AGGF1-PDGFRB in Ph-like ALL. (A) Schematic representation of the treatment course of the index patient with the AGGF1-PDGFRB fusion gene. Samples at diagnosis, at the end of induction (chemotherapy refractory), and at relapse were chosen for WGS and RNA-seq. (B) Schematic diagrams of AGGF1-PDGFRB based on the RNA-seq and WGS analyses. (C) Growth rate of Ba/F3 cells with AGGF1-PDGFRB, EBF1-PDGFRB, or empty lentivirus vector (mock) after removal of interleukin 3 (IL-3). (D) Localization of AGGF1-PDGFRB and EBF1-PDGFRB in Ba/F3 cells was detected by immunofluorescence (donkey anti-rabbit Alexa Fluor plus 555 and DAPI stain; original magnification ×200). (E) Lysate of Ba/F3 cells with AGGF1-PDGFRB, EBF1-PDGFRB, BCR-ABL1, and empty vector (mock) with or without IL-3 (10 ng/mL) to detect the phosphorylation of Stat5, Akt, and Erk1/2. All experiments were performed in triplicate. BM, bone marrow; DAPI, 4′,6-diamidino-2-phenylindole; pErk, phosphorylated Erk; WT, wild type.
A novel fusion gene of AGGF1-PDGFRB in Ph-like ALL. (A) Schematic representation of the treatment course of the index patient with the AGGF1-PDGFRB fusion gene. Samples at diagnosis, at the end of induction (chemotherapy refractory), and at relapse were chosen for WGS and RNA-seq. (B) Schematic diagrams of AGGF1-PDGFRB based on the RNA-seq and WGS analyses. (C) Growth rate of Ba/F3 cells with AGGF1-PDGFRB, EBF1-PDGFRB, or empty lentivirus vector (mock) after removal of interleukin 3 (IL-3). (D) Localization of AGGF1-PDGFRB and EBF1-PDGFRB in Ba/F3 cells was detected by immunofluorescence (donkey anti-rabbit Alexa Fluor plus 555 and DAPI stain; original magnification ×200). (E) Lysate of Ba/F3 cells with AGGF1-PDGFRB, EBF1-PDGFRB, BCR-ABL1, and empty vector (mock) with or without IL-3 (10 ng/mL) to detect the phosphorylation of Stat5, Akt, and Erk1/2. All experiments were performed in triplicate. BM, bone marrow; DAPI, 4′,6-diamidino-2-phenylindole; pErk, phosphorylated Erk; WT, wild type.
RNA sequencing (RNA-seq) of leukemia blasts at diagnosis, at the end of reduction therapy (refractory), and at relapse after imatinib treatment revealed a novel AGGF1-PDGFRB fusion gene: in-frame fusion of exon 11-23 of PDGFRB with exon 1-6 of the AGGF1 gene (Figure 1B). Oncogenic effects of AGGF1-PDGFRB were confirmed in mouse hematopoietic progenitor cell Ba/F3 in which ectopic expression of the fusion gene efficiently induced IL3-indepdent growth, in a fashion comparable to EBF1-PBGFRB fusion18 (Figure 1C). Similarly, AGGF1-PDGFRB expression also resulted in cytokine-independent growth of primary bone marrow cells from an Arf−/− mouse (C57/BL6) (supplemental Figure 2), suggesting that the PDGFRB rearrangement was likely the driver genomic abnormality for leukemia pathogenesis in this patient. Furthermore, in Ba/F3 cells overexpressing the fusion gene, both AGGF1-PDGFRB and EBF1-PDGFRB were mostly localized in the cytoplasm and induced constitutive activation of Stat5, Akt, and Erk1/2 signaling (Figure 1D-E).
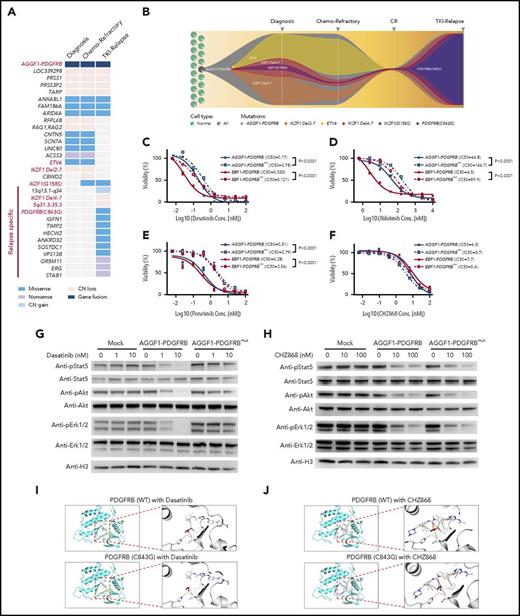
Whole-genome sequencing (WGS) was performed to search for potential genomic alteration that may confer drug resistance. Focal structural alterations and sequence mutations that were either shared or unique at various time points were identified (Figure 2A). The diagnostic blasts had 2 distinctive IKZF1 deletions both at a subclonal level, plausibly as essential but secondary events that occurred after AGGF1-PDGFRB fusion. There were very few additional genomic lesions in the refractory leukemia blasts. Interestingly, at relapse after imatinib treatment, only 1 of the 2 IKZF1 deletions remained and there was a new relapse-specific IKZF1G158S mutation. Among various mutations at this time point, most notable was a PDGFRB mutation (PDGFRBC843G) that emerged with concomitant loss of the wild-type PDGFRB allele as the result of a large deletion on 5q31.2 (Figure 2A). Droplet digital polymerase chain reaction was performed to examine the frequency of the key mutations and genomic lesions in 8 longitudinal samples obtained between diagnosis and relapse (supplemental Figure 3). The data acquired from WGS and droplet digital polymerase chain reaction were used to track the emergence of a drug-resistant leukemia clone. PDGFRBC843G was first detected 2 months prior to clinical relapse at only 0.03%, and most likely arose in the clone with IKZF1 exon4-7 deletion and IKZF1G158S. This population expanded rapidly and was clearly the major clone at relapse (Figure 2B).
Mutation of PDGFRBC843Gcaused TKI resistance in Ba/F3 cells with PDGFRB fusions. (A) Mutational landscape of leukemia cells at diagnosis, end of induction (chemotherapy refractory), and relapse. (B) A model of clonal evolution across disease progression. One subclone within the founding clone evolved to become the dominant clone at relapse, including mutations in PDGFRB. (C-F) Drug sensitivity of Ba/F3 cells with AGGF1-PDGFRB, EBF1-PDGFRB, AGGF1-PDGFRB carrying PDGFRBC843G (AGGF1-PDGFRBmut) or EBF1-PDGFRBmut upon dasatinib, nilotinib, ponatinib, and CHZ868 was detected by MTT assay. (G-H) Phosphorylation of Stat5, Akt, and Erk1/2 in Ba/F3 cells with AGGF1-PDGFRB, AGGF1-PDGFRBmut, and empty vector treated with dasatinib or CHZ868 as indicated concentration was detected by western blot. (I-J) The structure model of wild-type PDGFRB or mutant PDGFRBC843G with dasatinib or CHZ868. The cysteine (wild-type) or glycine (mutant) residue at position 843 is shown in red. All experiments were performed in triplicate. *P < .05; **P < .01; ***P < .001. CN, copy number; Conc., concentration; CR, complete remission IC50, 50% maximal inhibitory concentration.
Mutation of PDGFRBC843Gcaused TKI resistance in Ba/F3 cells with PDGFRB fusions. (A) Mutational landscape of leukemia cells at diagnosis, end of induction (chemotherapy refractory), and relapse. (B) A model of clonal evolution across disease progression. One subclone within the founding clone evolved to become the dominant clone at relapse, including mutations in PDGFRB. (C-F) Drug sensitivity of Ba/F3 cells with AGGF1-PDGFRB, EBF1-PDGFRB, AGGF1-PDGFRB carrying PDGFRBC843G (AGGF1-PDGFRBmut) or EBF1-PDGFRBmut upon dasatinib, nilotinib, ponatinib, and CHZ868 was detected by MTT assay. (G-H) Phosphorylation of Stat5, Akt, and Erk1/2 in Ba/F3 cells with AGGF1-PDGFRB, AGGF1-PDGFRBmut, and empty vector treated with dasatinib or CHZ868 as indicated concentration was detected by western blot. (I-J) The structure model of wild-type PDGFRB or mutant PDGFRBC843G with dasatinib or CHZ868. The cysteine (wild-type) or glycine (mutant) residue at position 843 is shown in red. All experiments were performed in triplicate. *P < .05; **P < .01; ***P < .001. CN, copy number; Conc., concentration; CR, complete remission IC50, 50% maximal inhibitory concentration.
Compared with IKZF1 exon4-7 deletion and IKZF1G158S, only PDGFRBC843G specifically emerged at relapse time point. Hence, we evaluated the effects of PDGFRBC843G on drug sensitivity in vitro. In Ba/F3 cells, AGGF1-PDGFRBC843G fusion efficiently induced cytokine-independent growth, in a fashion indistinguishable from the wild-type fusion gene (supplemental Figure 2). Ba/F3 cells expressing the mutant AGGF1-PDGFRB were significantly more resistant to ABL inhibitors imatinib (supplemental Figure 4), dasatinib, and nilotinib (Figure 2C-D), consistent with the patient’s clinical response to these agents at relapse. Surprisingly, ponatinib also showed significant reduced efficacy in mutant Ba/F3 cells compared with cells with a wild-type fusion gene (Figure 2E). In contrast, CHZ868, a broad-spectrum kinase inhibitor, showed specific nanomolar sensitivity in Ba/F3 cells with PDGFRB fusions (either with AGGF1 or EBF1), with minimal effects on cells expressing the BCR-ABL1 fusion (supplemental Figure 5), and CHZ868 was equally effective in mutant PDGFRB cells (Figure 2F), therefore pointing to a potential therapeutic approach to overcome drug resistance in this group of patients. Molecularly, dasatinib treatment blocked Stat5, Akt, and Erk activation in wild-type cells in a dose-dependent fashion, but this inhibitory effect was dramatically attenuated in cells expressing mutant PDGFRB fusion (Figure 2G). In contrast, CHZ868 treatment blocked the signaling pathway in a similar fashion in wild-type and mutant PDGFRB cells (Figure 2H). In fact, introduction of this mutation in the EBF1-PDGFRB fusion gene resulted in a similar pattern of drug resistance in Ba/F3 cells (Figure 2C-D), suggesting that it could be a common molecular mechanism for drug resistance in PDGFRB-rearranged leukemia. These findings were also reproduced in the Arf−/− pre-B-cell model (supplemental Figure 6).
To elucidate the structural basis of the effects of the PDGFRBC843G mutation, we modeled the conformation of the kinase domain of PDGFRB based on the crystallographic structures of c-KIT, which was most closely related to PDGFRB. With an estimated confidence level of 100% of the homology modeling, we observed that the C843 residue was located in a flexible loop within the binding pocket that directly interacts with kinase inhibitors (shown as dasatinib). The substitution of cysteine by glycine led to increased hydrophobicity and flexibility of the binding pocket causing a loss of van der Waals interaction with dasatinib (Figure 2I), plausibly resulting in lower affinity of dasatinib toward the kinase domain and thus drug resistance. In contrast, the CHZ868 compound is predicted to have 443 proximal interactions with the wild-type PDGFRB kinase domain (compared with 335 contacts predicted with dasatinib), many of which involve residues other than C843. In particular, the benzimidazole moiety of CHZ868 forms potentially essential interactions that are preserved in the C843G mutant protein (Figure 2J). Although our structural modeling analyses provided hints of how this mutation might alter PDGFRB interactions with various inhibitors, future studies are warranted to experimentally characterize the exact molecular mechanisms of TKI resistance in this group of patients.
In our study, a novel fusion gene of AGGF1-PDGFRB was identified and functionally characterized in Ph-like ALL. More importantly, this is the first report of the mutation of PDGFRBC843G, which caused resistance to ABL TKIs, including imatinib, dasatinib, and ponatinib, but responded to multitarget kinase inhibitor CHZ868. Our findings provide new insights into the TKI-resistance mechanism in Ph-like ALL, which may also apply to other hematopoietic malignancies with PDGFRB fusion genes.19-21
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patient in this study.
This work was supported by grants from the Ministry of Science and Technology of China (2016YFA0100600 [T.C.]), the CAMS Innovation Fund for Medical Sciences (2016-I2M-1-002 [X.Z.], 2017-I2M-1-015 [Y.Z.], and 2016-I2M-1-017 [T.C.]), the National Nature Science Foundation of China (81421002 [T.C., X.Z.]; 81470339 [X.Z.]; 81400137 and 81770175 [Y.Z.]; 81300401 [H.Z.]; 81570173 [Xin Liu]), the Hundred Talents Program of the Chinese Academy of Sciences (Xin Liu), the Nature Science Fund of Tianjin Municipal Science and Technology Commission (12ZCDZSY18100 [X.Z.]), the PUMC Youth Fund (3332016091 [Y.Z.]), the Czech Science Foundation (15-06582S [A.H.]), the Social Development Project of Jiangsu Province (CXTDA 2017014 [Q.W.]), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (F.H.), St. Baldrick’s Foundation (522589 [H.Z.]), US National Institutes of Health (NIH), National Cancer Institute grant CA21765 and NIH National Institute of General Medical Sciences grant GM115279 (J.J.Y.), and American Lebanese Syrian Associated Charities (H.Z., C.-H.P., J.J.Y.).
Authorship
Contribution: Xin Liu, X.Z., J.J.Y., T.C., and C.-H.P. designed the research; Y.Z., H.Z., Y. Gao, and J.Z. performed experiments; Xiaoming Liu and J.Z. recruited and followed up the patient; Y. Gocho, M.Q., A.H., F.H., and Q.W. analyzed results; and Y.Z., Y. Gao, H.Z., Q.W., C.-H.P., and J.J.Y. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jun J. Yang, St. Jude Children’s Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105; e-mail: jun.yang@stjude.org; Xiaofan Zhu, Institute of Hematology and Blood Diseases Hospital, 288 Nanjing Rd, Tianjin, 300020 China; e-mail: xfzhu@ihcams.ac.cn; or Xin Liu, Beijing Institute of Genomics, Chinese Academy of Sciences, 1 Beichen West Road, Beijing 100101, China; e-mail: liuxin@big.ac.cn.
REFERENCES
Author notes
Y.Z., Y. Gao, and H.Z. contributed equally to this work.