Key Points
B-cell–specific overexpression of SOX11 promotes oncogenic proliferation of B1a B cells and drives an MCL-like phenotype.
SOX11 overexpression is associated with increased signaling through the BCR pathway that can be reversed by pharmacological BTK inhibition.
Abstract
Mantle cell lymphoma (MCL) is characterized by increased B-cell receptor (BCR) signaling, and BTK inhibition is an effective therapeutic intervention in MCL patients. The mechanisms leading to increased BCR signaling in MCL are poorly understood, as mutations in upstream regulators of BCR signaling such as CD79A, commonly observed in other lymphomas, are rare in MCL. The transcription factor SOX11 is overexpressed in the majority (78% to 93%) of MCL patients and is considered an MCL-specific oncogene. So far, attempts to understand SOX11 function in vivo have been hampered by the lack of appropriate animal models, because germline deletion of SOX11 is embryonically lethal. We have developed a transgenic mouse model (Eμ-SOX11-EGFP) in the C57BL/6 background expressing murine SOX11 and EGFP under the control of a B-cell–specific IgH-Eμ enhancer. The overexpression of SOX11 exclusively in B cells exhibits oligoclonal B-cell hyperplasia in the spleen, bone marrow, and peripheral blood, with an immunophenotype (CD5+CD19+CD23−) identical to human MCL. Furthermore, phosphocytometric time-of-flight analysis of the splenocytes from these mice shows hyperactivation of pBTK and other molecules in the BCR signaling pathway, and serial bone marrow transplant from transgenic donors produces lethality with decreasing latency. We report here that overexpression of SOX11 in B cells promotes BCR signaling and a disease phenotype that mimics human MCL.
Introduction
Mantle cell lymphoma (MCL) is an aggressive, typically fatal subtype of B-cell lymphoma comprising ∼6% of all non-Hodgkin lymphoma (NHL), which is the most common hematological malignancy worldwide.1 MCL is characterized by increased B-cell–receptor (BCR) signaling, and BTK inhibition is an effective therapeutic intervention in MCL, inducing remission in 60% of relapsed patients.2-4 The mechanisms leading to increased BCR signaling in MCL are poorly understood, because mutations in upstream regulators of BCR signaling, such as CD79A, commonly observed in other lymphomas,5 are rare in MCL.3
The transcription factor SOX11 is overexpressed in the majority (78% to 93%) of MCL patients and is considered an MCL-specific oncogene,1,6-10 regulating numerous oncogenic pathways.7 Elucidation of SOX11 function in vivo and its cooperation with a canonical MCL oncogene, CCND1, have been limited by a lack of animal models because germline deletion of SOX11 is embryonically lethal.11 We have developed a transgenic (Tg) mouse model (Eμ-SOX11-EGFP) in the C57BL/6 background expressing murine SOX11 and EGFP under the control of a B-cell–specific immunoglobulin H (IgH)-Eμ enhancer. We report here that overexpression of SOX11 in B cells promotes BCR signaling and a disease phenotype that mimics human MCL.
Methods
Mouse modeling
All animal experiments were carried out under protocols approved by the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee.
Eμ-SOX11 Tg mice were generated by injecting the BstBI/BciVI digested, pEμSV-SOX11-IRES-eGFP vector containing mouse SOX11 full-length sequence into fertilized oocytes from B6 animals. pEμSV-SOX11-IRES-eGFP was generated by inserting the IgH-μ enhancer and SV40 promoter (EμSV) amplified from pUEμSV-M13R upstream of the murine SOX11 sequence on EX-Mm05367-M61 Vector (Genecopoeia) using BstBI and KpnI. Mice were screened for the presence of the SOX11-IRES-eGFP transgene by polymerase chain reaction (PCR) analysis on tail DNA using the primer pairs from the list below. The Eμ-SOX11 mouse colony was then maintained by crossing transgene-carrying, heterozygote founders with C57Bl/6 (Jackson Laboratory). Genotyping was performed on tail DNA by quantitative PCR (Transnetyx). Transgenic heterozygote mice were studied and compared with nontransgenic littermates (wild-type, WT) reared under identical conditions.
EμSV 2 F: 5′-ggccgcTTCGAAGCAATTCTAAATACATTTTAGAAGTCG-3′
EμSV 2 R: 5′-ccaagcGGTACCtCTAGAGCTTTTTGCAAAAGCCT-3′
mSOX11-Scr-F: 5′-TCATGTTCGACCTGAGCTTG-3′
eGFP-Scr-R: 5′-GCTGAACTTGTGGCCGTTT-3′
mSOX11-Scr-R: 5′-GCTGTCCTTCAGCATCTTCC-3′
Cytometric time-of-flight (CyTOF) sample preparation
Antibodies were either purchased preconjugated from Fluidigm (formerly DVS Sciences) or purchased, purified, and conjugated in house using MaxPar X8 Polymer Kits (Fluidigm) according to the manufacturer’s instructions. Cells were washed and stained with cell surface antibodies for 30 minutes on ice, before being fixed, permeabilized (BD Biosciences), and incubated in 0.125 nM iridium intercalator (Fluidigm) diluted in phosphate-buffered saline (PBS) containing 2% formaldehyde; cells were then stored at 4°C until acquisition.
For signaling experiments, samples were stimulated with biotinylated anti-IgM and then fixed with 2% formaldehyde immediately following stimulation. The samples were bar-coded using a commercial Pd Barcoding Kit (Fluidigm) and pooled for combined analysis to minimize any technical variation during sample processing. The pooled samples were stained with cell surface antibodies for 30 minutes on ice and then permeabilized with ice-cold methanol and stained with antibodies against intracellular phophoprotein targets. The samples were then washed and incubated in 0.125 nM iridium intercalator (Fluidigm) diluted in PBS containing 2% formaldehyde and stored at 4°C until acquisition. Supplemental Tables 1 and 3, available on the Blood Web site, supply complete CyTOF panels used to determine the phenotype and BCR signaling status of mouse splenocytes.
CyTOF data acquisition.
Immediately prior to acquisition, samples were washed once with PBS, once with deionized water, and then resuspended at a concentration of 1 million cells per milliliter in deionized water containing a 1/20 dilution of EQ 4 Element Beads (Fluidigm). The samples were acquired on a CyTOF2 (Fluidigm) equipped with a SuperSampler fluidics system (Victorian Airships) at an event rate of <500 events per second. After acquisition, the data were normalized using bead-based normalization in the CyTOF software. Bar codes were deconvoluted using the Fluidigm de–bar-coding software, or by manual Boolean gating in the case of CD45–bar-coded samples. The data were gated to exclude residual normalization beads, debris, dead cells, and doublets for subsequent clustering and high dimensional analyses.
CyTOF clustering by viSNE and SPADE.
CyTOF data were visualized using Spanning Tree Progression of Density Normalized Events (SPADE)12 and viSNE,13 as implemented in Cytobank14 to facilitate visualization and identification of populations on the basis of multiple markers. SPADE was performed on total viable cells using all surface markers as clustering parameters. Major immune populations were identified on the basis of canonical marker expression patterns, and change in relative frequency of populations between WT and Tg-SOX11 mice were visualized on the tree. viSNE analysis was performed on pregated B cells using only B-cell–relevant surface markers as clustering parameters. All samples for paired comparisons were included as part of the same run. For signaling experiments, phosphoproteins were not included as clustering parameters, but their expression was visualized on SPADE and viSNE maps to resolve signaling across cell subsets.
CyTOF statistical analysis.
Heat maps of normalized marker expression, relative marker expression, and relative difference of population frequency were generated by GENE-E or Morpheus from the Broad Institute (https://software.broadinstitute.org/morpheus/).
Cell sorting
B1a cells from both Tg-SOX11 and WT mice were sorted from red blood cell (RBC)–depleted, B-cell–enriched splenocytes using Aria II (BD) sorter. In brief, spleens were homogenized using forceps to make single-cell suspensions. RBCs were lysed using 1× BD Pharm Lyse (BD) at room temperature for 10 minutes and centrifuged at 300g. Splenic B cells were enriched using the pan B-cell isolation kit II (Miltenyi) and stained with anti-mouse CD19-e405, anti-mouse CD3-PE-Dazzle549, anti-mouse B220-PE-Cy7, and anti-mouse CD5-APC (Biolegend) and resuspended in PBS containing 2% fetal bovine serum and 10 µg/mL propidium iodide (Invitrogen). Stained B cells were stored on ice in the dark until being sorted.
Flow cytometry analysis
Single-cell suspensions from murine spleen, peripheral blood, bone marrow, or lymph node were collected. Cells were stained with antibodies specific for murine CD5 (Biolegend, Clone 53-7.3), CD19 (Biolegend, Clone 6D5), CD3 (Biolegend, Clone 17A2), B220 (Biolegend, Clone RA3-6B2), CD23 (Biolegend, Clone B3B4), BP-1 (Miltenyi, Clone 6C3), TdT (eBioscience, Clone 19-3), and CD43 (Miltenyi, Clone REA80). Flow cytometry was carried out on a BD LSRFortessa. Flow data were then analyzed using FlowJo and Cytobank.
PhosphoFlow
Spleens were homogenized using forceps to make single-cell suspensions. RBCs were lysed using 1× BD Pharm Lyse (BD) at room temperature for 10 minutes and centrifuged at 300g. Splenic B cells were enriched using the pan B-cell isolation kit II, mouse (Miltenyi). B-cell–enriched splenocytes (2 × 106) were then resuspended in 300 µL of RPMI 1640 10% fetal bovine serum and stimulated with 10 µg/mL anti-mouse IgM F(ab′)2 at 37°C for 10 to 15 minutes. After incubation, cells were put on ice for 10 minutes, centrifuged, and stained with anti-mouse CD19-BV711, anti-mouse CD3-PE-Dazzle549, anti-mouse CD5-PE-Cy7, and anti-mouse B220-AF700 (Biolegend). Stained cells were then washed and fixed using the FoxP3/transcription factor staining buffer set (eBioscience) following the manufacturer’s instructions. Fixed cells were stained with anti-phospho PLCγ2 (pY759) and with anti-phospho Btk (pY223)/Itk (pY180) (BD) and analyzed with BD LSRFortessa cell analyzer. Plots were done using CytoBank.
Bone marrow transfer
Two million RBC-lysed bone marrow cells from tumor-bearing Tg-SOX11 mice or healthy non-Tg littermate controls (WT) were injected into 10-week-old lethally irradiated CD45.1 hosts (B6.SJL-Ptprca Pepcb/BoyJ; Jackson Laboratory). Mice receiving bone marrow were fed with Neomycin water at 20 mg/mL for 14 days. Blood was withdrawn 40 days after bone marrow transplant and stained with anti-mouse CD45.1 (Biolegend, Clone A20), anti-mouse CD45.2 (Biolegend, Clone 104), anti-mouse CD19, anti-mouse CD3, anti-mouse CD5, for flow cytometer analysis to monitor bone marrow engraftment. Mice were monitored weekly.
Immunohistochemistry
Mouse organs were fixed in 4% formaldehyde and embedded in paraffin. Four-micron sections were deparaffinized and heat antigen–retrieved in citrate buffer pH 6.4, and endogenous peroxidase activity was blocked by treating the sections with 3% hydrogen peroxide in methanol. Indirect immunohistochemistry was performed with antispecies-specific biotinylated secondary antibodies followed by avidin–horseradish peroxidase or avidin–alkaline phosphatase and developed by Vector Blue or DAB color substrates (Vector Laboratories). Sections were counterstained with hematoxylin if necessary. The following primary antibodies were used: anti-B220 (BD Bioscience, 550286), anti-CD3 (Abcam, ab16669), anti-BCL6 (Santa Cruz Biotechnology, sc-858), and anti-CD34 (Abcam, ab8158).
IgH sequencing and analysis
Sample preparation.
Genomic DNA was prepared from sorted B1a splenocytes of 2 WT and 4 Tg-SOX11 mice using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. Samples were quantified using NanoDrop (ThermoFisher) and sent to Adaptive Biotechnologies for IgH gene survey.
Library preparation and sequencing.
Sample data were generated using the immunoSEQ Assay (Adaptive Biotechnologies). The somatically rearranged murine IgH locus CDR3 was amplified from genomic DNA using a 2-step, amplification bias-controlled multiplex PCR approach.15,16 The first PCR consists of forward and reverse amplification primers specific for every V and J gene segment and amplifies the hypervariable complementarity-determining region 3 (CDR3) of the immune receptor locus. The second PCR adds a proprietary bar-code sequence and Illumina adapter sequences.17 CDR3 libraries were sequenced on an Illumina instrument according to the manufacturer’s instructions.
Data analysis.
Raw sequence reads were demultiplexed according to Adaptive’s proprietary bar-code sequences. Demultiplexed reads were then further processed to remove adapter and primer sequences and to remove primer dimer and other contaminant sequences. The data are filtered and clustered using both the relative frequency ratio between similar clones and a modified nearest-neighbor algorithm to merge closely related sequences. The resulting sequences were sufficient to allow annotation of the V(N)D(N)J genes constituting each unique CDR3 and the translation of the encoded CDR3 amino acid sequence. V, D, and J gene definitions were based on annotation in accordance with the IMGT database (http://www.imgt.org/).
The set of observed biological IgH CDR3 sequences were quantified and normalized to correct for residual multiplex PCR amplification bias, using a set of synthetic IgH CDR3 sequence analogs that are included in each PCR reaction.16 Data were analyzed, and plots were generated using the immunoSEQ Analyzer toolset.
Karyotyping
Metaphase chromosomes were prepared from mouse splenocytes after Colcemid arrest (1 hour; final concentration 0.1 μg/mL), and standard hypotonic treatment and fixation in methanol/acetic acid. SKY was performed essentially as described before.18 Briefly, mouse SKY Paint Probes (Applied Spectral Imaging) were hybridized to metaphase slides aged at 37°C for 24 hours. After in situ hybridization, slides were counterstained with 4′,6-diamidino-2-phenylindole, and images were acquired using an epifluorescence microscope (Olympus BX61) connected to an imaging interferometer (SD200; Applied Spectral Imaging, Migdal HaEmek, Israel). Chromosomes were unambiguously identified using a spectral classification algorithm that results in the assignment of a separate classification color to all pixels with identical spectra. Chromosome aberrations were defined using the nomenclature rules from the International Committee on Standardized Genetic Nomenclature for Mice.19 Ten to 12 metaphases were analyzed for each sample.
Results
Tg overexpression of murine SOX11 in B cells phenocopies human MCL
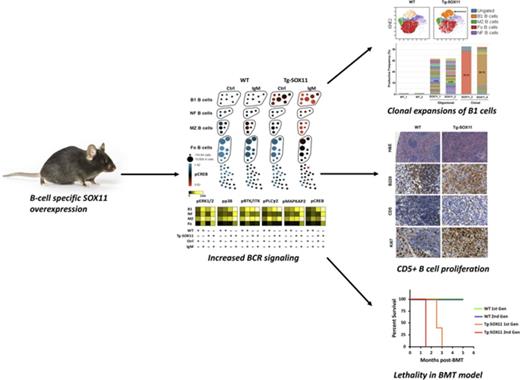
We generated a Tg mouse line cooverexpressing murine SOX11 and EGFP under the control of the B-cell–specific Eµ-SV enhancer (Figure 1A). As early as 1.5 months of age, Tg-SOX11 mice show a significant increase in a subset of B cells compared with age- and sex-matched (WT) littermate controls. Comprehensive analysis of all splenic immune subsets using a 30-parameter single-cell mass CyTOF analysis confirmed that this abnormal increase is restricted specifically to a subset of CD19+CD5+CD21−CD23− cells, which are phenotypically characteristic of B1a B cells20,21 (Figure 1B-C; complete CyTOF panel shown in supplemental Table 1). We validated these results using conventional flow cytometry, showing an increased frequency of CD19+CD5+CD23− B cells in the blood, spleens, and lymph nodes (P < .05), and also in the bone marrow of Tg-SOX11 mice when compared with their WT littermate controls (Figure 1D). We observed complete penetrance of this immunophenotype in the peripheral blood of all Tg-SOX11 mice we tested (N = 36/group; age range: 1.5-24 months; Figure 1E). The clinical presentation of MCL includes splenomegaly, and we observed enlarged spleens in Tg-SOX11 mice but not in their WT littermate controls (Figure 1F). The histopathological analysis of the spleens from Tg-SOX11 mice shows a disruption of splenic architecture in H&E staining, by B220+ CD3−CD34− B cells replacing the germinal center (BCL6 staining) as is expected in human MCL (Figure 1G).
B-cell–specific SOX11 expression produces MCL phenotype. (A) Schematic representation of the gene cassette for the coexpression of SOX11 and EGFP in the B cells of a C57BL/6 mouse model. (B) Single-cell mass cytometry (CyTOF) data analyzed using SPADE to comprehensively characterize splenocytes from WT vs age-matched Tg-SOX11 mice. Major immune subsets are annotated based on canonical marker expression patterns, and the SPADE tree is colored by [log10([% of total in SOX11]/[% of total in WT])], highlighting selective expansion of a specific B-cell subset in Tg-SOX11 mice. (C) Further analysis of B-cell subsets by viSNE highlights the phenotype of the expanded population of cells as CD19+CD21−CD23−CD5+ (black arrows). (D) Frequency of CD5+CD19+CD23− B1 cells in peripheral blood, spleens, lymph nodes, and bone marrow of Tg-SOX11 mice vs WT littermates analyzed by flow cytometry (N = 3/group). (E) Frequency of CD5+CD19+CD23− B1 cells in the peripheral blood of 1.5- to 36-month-old Tg-SOX11 mice vs WT littermates analyzed by flow cytometry (N = 3/group). (F) Spleens from 6-month-old mice showing splenomegaly in Tg-SO11 mice compared with WT littermate controls (N = 3/group). (G) Pathological analysis of spleen sections from a Tg-SOX11 mouse confirming a neoplastic B220+CD3−CD34−BCL6− immunophenotype. Fo, follicular; GC, germinal center; H&E, hematoxylin and eosin; Mono/Mac, monocytes/macrophages; MZ, marginal zone; NF, newly formed transitional; NK, natural killer; pDC, plasmacytoid dendritic cells.
B-cell–specific SOX11 expression produces MCL phenotype. (A) Schematic representation of the gene cassette for the coexpression of SOX11 and EGFP in the B cells of a C57BL/6 mouse model. (B) Single-cell mass cytometry (CyTOF) data analyzed using SPADE to comprehensively characterize splenocytes from WT vs age-matched Tg-SOX11 mice. Major immune subsets are annotated based on canonical marker expression patterns, and the SPADE tree is colored by [log10([% of total in SOX11]/[% of total in WT])], highlighting selective expansion of a specific B-cell subset in Tg-SOX11 mice. (C) Further analysis of B-cell subsets by viSNE highlights the phenotype of the expanded population of cells as CD19+CD21−CD23−CD5+ (black arrows). (D) Frequency of CD5+CD19+CD23− B1 cells in peripheral blood, spleens, lymph nodes, and bone marrow of Tg-SOX11 mice vs WT littermates analyzed by flow cytometry (N = 3/group). (E) Frequency of CD5+CD19+CD23− B1 cells in the peripheral blood of 1.5- to 36-month-old Tg-SOX11 mice vs WT littermates analyzed by flow cytometry (N = 3/group). (F) Spleens from 6-month-old mice showing splenomegaly in Tg-SO11 mice compared with WT littermate controls (N = 3/group). (G) Pathological analysis of spleen sections from a Tg-SOX11 mouse confirming a neoplastic B220+CD3−CD34−BCL6− immunophenotype. Fo, follicular; GC, germinal center; H&E, hematoxylin and eosin; Mono/Mac, monocytes/macrophages; MZ, marginal zone; NF, newly formed transitional; NK, natural killer; pDC, plasmacytoid dendritic cells.
SOX11 overexpression is associated with increased BCR signaling
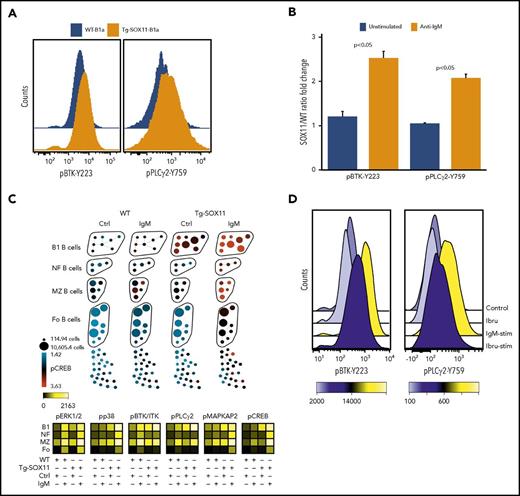
To characterize the BCR signaling status of the aberrant B-cell population in Tg-SOX11 mice, we assayed critical BCR pathway members, including BTK and PLCγ by flow cytometry. We saw increased levels of phospho-BTK(Y223) and phospho-PLCγ(Y759) by phospho-flow cytometry (Figure 2A; supplemental Table 2A) in unstimulated SOX11-overexpressing B1a cells as compared with WT B1a controls. We also examined dynamic BCR signaling in response to IgM stimulation in these mice and found that phospho-BTK and phospho-PLCγ levels were significantly increased in Tg-SOX11-B1a splenocytes when compared with WT-B1a splenocytes after anti-IgM stimulation (Figure 2B) in a dose- and time-dependent manner (supplemental Figure 1). In addition, we used phospho-CyTOF to more comprehensively analyze the activation of major phosphoproteins in the BCR signaling pathway in response to anti-IgM stimulation (10 µg/mL). SPADE analysis was used to visualize differences in phosphoprotein levels with the various B-cell subsets in Tg-SOX11 and WT splenocytes. Figure 2C upper panel shows elevated pCREB levels in Tg-SOX11-B1a splenocytes as compared with WT-B1a splenocytes. Similar differences between Tg-SOX11 and WT B1a splenocytes were also observed for increased levels of phosphorylated BTK, PLCγ2, ERK1/2, p38, and MAPKAP2. The heat map in Figure 2C lower panel summarizes these results for all 6 members of the BCR signaling pathway (complete CyTOF panel shown in supplemental Table 3). We also observed signaling differences in other B-cell subsets, but the differences were most pronounced in the B1 population. Our data suggest that the BCR signaling cascade is increased at baseline levels and further amplified by antigenic stimulation in SOX11 overexpressing cells. We found this increased BCR signaling to be reversible, because treatment of Tg-SOX11-B1a splenocytes with 100 nM ibrutinib prior to anti-IgM stimulation decreased BTK phosphorylation to near prestimulation levels (Figure 2D; supplemental Table 2B).
SOX11 expression promotes BCR signaling in MCL cells. (A) p-BTK-Y223 and p-PLCγ-Y759 levels in CD5+CD19+CD23− (B1a) splenocytes from Tg-SOX11 mice as compared with WT controls. (B) Elevated response to anti-IgM stimulation of BCR signaling seen by increased p-BTK(Y223) and p-PLCγ(Y759) levels in Tg-SOX11-B1a splenocytes as compared with WT-B1a splenocytes assayed by flow cytometry. (C) SPADE analysis (upper panel) showing increased p-CREB levels in various B-cell subsets of Tg-SOX11 and WT splenocytes in anti-IgM–stimulated B cells. The heat map (lower panel) summarizes findings for all the key members of the BCR signaling pathway showing elevated expression of p-BTK, p-PLCγ2, p-ERK, p-p38, p-MAPKAP2, and p-CREB across B-cell subsets in Tg-SOX11 overexpressing and WT splenocytes in response to IgM stimulation. (D) Treatment of fluorescence-activated cell–sorted SOX11-overexpressing CD5+CD19+ splenocytes with 100 nM of ibrutinib (Ibru) prior to anti-IgM stimulation showing reversibility of elevated BCR signaling. Ctrl, control.
SOX11 expression promotes BCR signaling in MCL cells. (A) p-BTK-Y223 and p-PLCγ-Y759 levels in CD5+CD19+CD23− (B1a) splenocytes from Tg-SOX11 mice as compared with WT controls. (B) Elevated response to anti-IgM stimulation of BCR signaling seen by increased p-BTK(Y223) and p-PLCγ(Y759) levels in Tg-SOX11-B1a splenocytes as compared with WT-B1a splenocytes assayed by flow cytometry. (C) SPADE analysis (upper panel) showing increased p-CREB levels in various B-cell subsets of Tg-SOX11 and WT splenocytes in anti-IgM–stimulated B cells. The heat map (lower panel) summarizes findings for all the key members of the BCR signaling pathway showing elevated expression of p-BTK, p-PLCγ2, p-ERK, p-p38, p-MAPKAP2, and p-CREB across B-cell subsets in Tg-SOX11 overexpressing and WT splenocytes in response to IgM stimulation. (D) Treatment of fluorescence-activated cell–sorted SOX11-overexpressing CD5+CD19+ splenocytes with 100 nM of ibrutinib (Ibru) prior to anti-IgM stimulation showing reversibility of elevated BCR signaling. Ctrl, control.
Next-generation sequencing of the IgH locus reveals MCL-specific clonal expansions of B cells in Tg-SOX11 mice
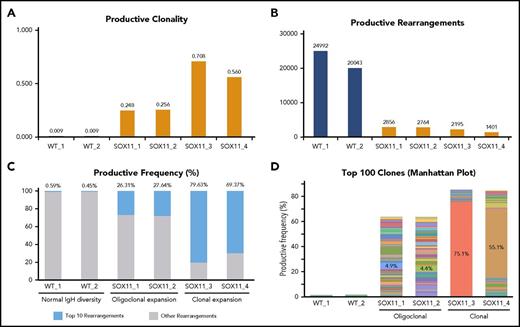
MCL is typified by reduced IgH gene diversity and clonal expansions of B-cell subpopulations. We conducted next-generation deep sequencing of the genomic DNA of CD19+/CD5+/CD23− (B1a) splenocytes to study their IgH gene repertoire (Adaptive Biotechnologies). In 4 of 4 Tg-SOX11 mice, we found a 25- to 75-fold increase in clonality compared with their WT-B1a littermates (Figure 3A). This was reflected in a corresponding decrease in the total number of productive rearrangements in Tg-SOX11 mice vs WT littermates (Figure 3B). When we compared the frequency of the top 10 rearrangements to all other rearrangements of the IgH locus, we observed near clonality in 2 Tg-SOX11 mice and oligoclonality in the other 2, whereas the WT mice displayed normal IgH diversity (Figure 3C). These (oligo)clonal expansions in the Tg-SOX11 mice became even more apparent when the relative abundance of each of the top 100 rearrangements was visualized in the Manhattan plot shown in Figure 3D. The sum of the frequencies of these top 100 clones comprise ∼65% and 85% of the repertoire of the oligoclonal and near clonal expansions, respectively, which is in sharp contrast to the <2% observed for their WT littermates. The numerical value for the top clone frequency is annotated for each of these Tg-SOX11 mice (also see supplemental Figure 2A for the maximum productive frequency of the top clone in each of the mice, including WT). Biased usage of certain VH3 and VH4 alleles is well established in human MCL.22 In the Eµ-SOX11-EGFP mice, we observed a biased usage of specific alleles of the IgHV gene families corresponding to the human VH3 and VH4 (supplemental Figure 2B). The karyotype of the splenocytes from these Tg-SOX11 mice was found to be normal, showing no significant translocation or aneuploidy (supplemental Figure 3).
SOX11 expression leads to clonal expansion of B cells. (A) Deep sequencing of the genomic DNA from B1a cells showing increased IgH gene clonality in Tg-SOX11 mice vs WT mice. (B) Decreased total productive rearrangements in the BCR repertoire of B1a cells of Tg-SOX11 mice vs WT mice showing a decrease in IgH gene diversity. (C) Frequency of the top 10 rearrangements compared with all other rearrangements of the IgH locus in B1a cells showing clonality in 2 Tg-SOX11 mice and oligoclonality in 2 Tg-SOX11 mice vs normal IgH diversity in the 2 WT mice. (D) Productive frequency of the top 100 IgH gene rearrangements shows loss of IgH diversity in all Tg-SOX11 mice (N = 4) as compared with WT mice (N = 2). In the histogram, each color represents a unique IgH clone, whereas the numerals represent the frequency of top clone in each mouse.
SOX11 expression leads to clonal expansion of B cells. (A) Deep sequencing of the genomic DNA from B1a cells showing increased IgH gene clonality in Tg-SOX11 mice vs WT mice. (B) Decreased total productive rearrangements in the BCR repertoire of B1a cells of Tg-SOX11 mice vs WT mice showing a decrease in IgH gene diversity. (C) Frequency of the top 10 rearrangements compared with all other rearrangements of the IgH locus in B1a cells showing clonality in 2 Tg-SOX11 mice and oligoclonality in 2 Tg-SOX11 mice vs normal IgH diversity in the 2 WT mice. (D) Productive frequency of the top 100 IgH gene rearrangements shows loss of IgH diversity in all Tg-SOX11 mice (N = 4) as compared with WT mice (N = 2). In the histogram, each color represents a unique IgH clone, whereas the numerals represent the frequency of top clone in each mouse.
Tg-SOX11 cells can propagate disease in isogenic animals with short latency
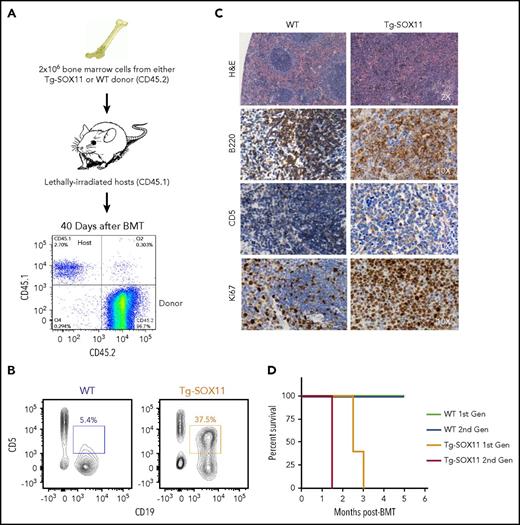
Next, we set up bone marrow transplantation (BMT) experiments to determine whether the bone marrow compartment in Eμ-SOX11-GFP mice contains disease-propagating activity. Two million RBC-depleted bone marrow cells from Tg-SOX11 or WT donors (both expressing CD45.2) were transplanted via retroorbital injection into lethally irradiated CD45.1 congenic C57BL/6 recipients. Engraftment of CD45.2 donor bone marrow was tracked in the peripheral blood of recipient mice (Figure 4A). We observed a significant increase in CD5+CD19+CD23− cells in the peripheral blood of recipients engrafted with bone marrow from Tg-SOX11 mice (Figure 4B) when compared with WT mice. The normal architecture of the spleen was completely disrupted by rapidly proliferating (high Ki-67) CD5+B220+ atypical B cells (Figure 4C) in the Tg-SOX11 BMT recipients. Finally, mice receiving marrow from SOX11 donors died in 3 months, whereas WT bone marrow recipients had normal survival (Figure 4D). When bone marrow was serially transplanted from BMT recipients (first generation) into congenic recipients (second generation), Tg-SOX11 recipients died in 1.5 months, whereas the WT recipients once again showed normal survival (Figure 4D). Thus, BM cells from Tg-SOX11 donor mice confer rapid lethality characterized by tumor infiltration mimicking human MCL and can serve as a model for preclinical drug development.
BMT from Tg-SOX11 donors models lethal MCL. (A) Flow cytometry tracking engraftment of CD45.2 donor bone marrow in the peripheral blood of CD45.1-recipient mice. (B) CD5+CD19+ cell fraction in splenocytes of sublethally irradiated recipients of bone marrow from Eμ-SOX11-EGFP donors as compared with recipients of bone marrow from WT donors. (C) Pathological analysis of the spleen in recipient mice transplanted with BM from Eμ-SOX11-EGFP donors. (D) Kaplan-Meier survival curves for recipient mice (1st gen) transplanted once with bone marrow from Tg-SOX11 vs WT donors and upon second serial BMT (2nd gen); N = 5/group.
BMT from Tg-SOX11 donors models lethal MCL. (A) Flow cytometry tracking engraftment of CD45.2 donor bone marrow in the peripheral blood of CD45.1-recipient mice. (B) CD5+CD19+ cell fraction in splenocytes of sublethally irradiated recipients of bone marrow from Eμ-SOX11-EGFP donors as compared with recipients of bone marrow from WT donors. (C) Pathological analysis of the spleen in recipient mice transplanted with BM from Eμ-SOX11-EGFP donors. (D) Kaplan-Meier survival curves for recipient mice (1st gen) transplanted once with bone marrow from Tg-SOX11 vs WT donors and upon second serial BMT (2nd gen); N = 5/group.
Discussion
The pathogenesis of MCL is characterized by cell-cycle dysregulation and Cyclin D1 (CCND1) overexpression as a result of t(11;14).23 However, CCND1 overexpression is not universal in this disease, and mouse models overexpressing CCND1 do not recapitulate the phenotype of the disease.24 Whole-exome sequencing of primary MCL has shown recurrent mutations inactivating tumor suppressors P53, CDKN2A, and CDKN2C25 in subsets of MCL patients, and additional genetic lesions and epigenetic alterations continue to be characterized.
SOX11 belongs to the SOXC family of high-mobility-group transcription factors, which consists of SOX4, SOX11, and SOX12.26 High-mobility-group transcription factors bind to DNA and facilitate conformational changes that allow binding of other transcription factors, usually leading to activation or repression of downstream genes. In early B cells, SOX4, a closely related SOX11 family member with identical target binding motifs, can inhibit differentiation via direct inhibition of WNT signaling.27 SOXC transcription factors have overlapping roles in central and peripheral nerve development, and it is likely that these factors may play a role in B-cell development as well, because SOX11-null mice do not form spleens, and SOX4-deficient mice do not form B lymphocytes.11,26 SOX11 expression is specific for MCL as compared with other NHLs1,6,28 and present in premalignant lymph nodes examined in serial biopsies from MCL patients, suggesting that the expression of this protein is an early event in the malignant transformation of lymphocytes in MCL.29 SOX11 is expressed in ∼90% of MCL patients, including 5% to 10% of those who are CCND1 negative.9 Microarray studies performed on primary MCL samples by us previously confirm SOX11 expression to be significantly higher in CD19+ tumor cells as compared with naive B cells from healthy controls, which are the cell of origin for MCL.10 We have developed a SOX11 overexpressing mouse model under a B-cell–specific (Eμ-SV) enhancer to understand the role of SOX11 in MCL pathogenesis. Our experiments implicate SOX11 as a novel upstream regulator of BCR and demonstrate increased BCR signaling as a novel mechanism of SOX11-driven lymphomagenesis. We see the hallmarks of MCL in Eμ-SOX11-EGFP mice from an early age. CD5 is specifically expressed on B1 cells, the cell of origin for MCL,30 and CD23 negativity accompanied with the absence of CD34, CD3, and BCL6 (Figure 1G) expression distinguishes these cells from those found in other T- and B-cell lymphomas and leukemias (negative for acute lymphoblastic leukemia marker, terminal deoxynucleotidyl transferase, and immature B-cell markers BP-1/CD43, as shown in supplemental Figures 4-6). The data generated from our Tg-SOX11 model are consistent with human MCL. We acknowledge the use of a single Tg line, and our results must be interpreted with that caveat in mind.
Future research will focus on the SOX11-regulated transcription program that links aberrant SOX11 expression to increased BCR signaling in MCL. SOX11 expression has been associated with activating histone marks (H3K9/14Ac and H3K4me3), hypomethylation of its promoter, and the de novo looping of a distant enhancer (marked by H3K27ac) in both MCL cell lines and patient samples.31 Directed sequencing of DNA has not revealed any mutations in SOX11, consistent with an active role of epigenetic changes to the regulation of SOX11 expression in MCL. In addition, it will be necessary to identify the drivers of the upstream epigenetic changes leading to SOX11 overexpression.
Previous in vivo MCL mouse models have relied on xenografting human MCL cell line–derived and patient-derived cells in immunodeficient mice32 or Tg expression of oncogenes implicated in MCL. Although useful for efficacy studies, xenografts have the obvious limitations of a lacking immune system and an incorrect microenvironment.33 Tg models employing Eµ-driven B-cell–specific expression of Cyclin D1 alone have failed to develop lymphoid tumors.24,34,35 Double Tgs coexpressing Cyclin D1 and a MYC oncogene rapidly develop pre–B- and B-cell lymphomas, but these are more characteristic of Myc-driven tumors,24,33 representing only a minority of clinical MCL cases and phenotypically corresponding to the blastoid variant of the disease.36 Another double Tg model overexpresses Cyclin D1 in Bim-deficient B cells but requires exogenous immune stimulation with sheep RBCs and pristane to induce MCL.34 In contrast, Tg-SOX11 (EμSOX11::GFP) mice show 100% penetrance of a disease phenotype reminiscent of classical disseminated human MCL with a leukemic component not requiring exogenous stimuli for MCL induction. As evidenced by the full repertoire of immune cells (Figure 1B), Tg-SOX11 mice are immunocompetent, making our model amenable to investigating immunotherapeutic strategies for MCL therapy.37 It will be interesting to investigate whether double Tg EμCycD1 × EμSOX11 mice show an accelerated disease progression, akin to our isogenic BMT model, and whether they retain the phenotype of classical MCL.
We also propose that Eµ-Sox11-EGFP mice can serve as a relevant model for studying MCL disease biology, for identifying novel therapeutic opportunities, and for preclinical drug development. Despite advances in chemotherapy and immunotherapy, the majority of MCL patients relapse and die of their disease, and advances in therapeutics are urgently needed.38 Most patients treated with ibrutinib relapse with a median survival of 3 months on progression. Ibrutinib resistance is a significant challenge and occurs due to a variety of mechanisms.39 We recently described that hyperactivation of ERK and JAK/STAT signaling correlated with insensitivity to BTK inhibition and poor clinical outcomes.40 Mechanistically distinct therapeutic strategies are therefore urgently needed for relapsed MCL patients. Our studies suggest targeting SOX11 as an upstream inducer of the BCR signaling pathway in MCL as a strategy distinct from direct BTK inhibition may hold merit.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are thankful to the immune core (HIMC) for their assistance with CyTOF.
This work was funded by an ASH Bridge grant and Chemotherapy Foundation Research grants. Instrumentation at the Human Immune Monitoring Center was supported by National Institutes of Health, NIH Office of the Director grant S10 OD023547-01.
Authorship
Contribution: P.-Y.K., S.S.J., and S.P. designed the study, had access to raw data, analyzed the data, and wrote the manuscript; A.H.R. developed, designed, and analyzed the CyTOF experiments; D.E. and Z.J. maintained small animal colonies, harvested tissue, and prepared samples; K.A., D.P., and V.V.L. contributed to manuscript preparation; R.S. processed and analyzed histopathological samples; B.H.Y. provided the gene transfer vector and contributed to the design of the Tg mouse model; and S.P., J.B., and S.S.J. had full access to all the data and had final responsibility for the decision to submit for publication.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Samir Parekh, Mount Sinai Hospital, Icahn School of Medicine, 1 Gustave L. Levy Pl, Box 1185, New York, NY 10029; e-mail: samir.parekh@mssm.edu.
REFERENCES
Author notes
P.-Y.K. and S.S.J. are joint first authors.

![Figure 1. B-cell–specific SOX11 expression produces MCL phenotype. (A) Schematic representation of the gene cassette for the coexpression of SOX11 and EGFP in the B cells of a C57BL/6 mouse model. (B) Single-cell mass cytometry (CyTOF) data analyzed using SPADE to comprehensively characterize splenocytes from WT vs age-matched Tg-SOX11 mice. Major immune subsets are annotated based on canonical marker expression patterns, and the SPADE tree is colored by [log10([% of total in SOX11]/[% of total in WT])], highlighting selective expansion of a specific B-cell subset in Tg-SOX11 mice. (C) Further analysis of B-cell subsets by viSNE highlights the phenotype of the expanded population of cells as CD19+CD21−CD23−CD5+ (black arrows). (D) Frequency of CD5+CD19+CD23− B1 cells in peripheral blood, spleens, lymph nodes, and bone marrow of Tg-SOX11 mice vs WT littermates analyzed by flow cytometry (N = 3/group). (E) Frequency of CD5+CD19+CD23− B1 cells in the peripheral blood of 1.5- to 36-month-old Tg-SOX11 mice vs WT littermates analyzed by flow cytometry (N = 3/group). (F) Spleens from 6-month-old mice showing splenomegaly in Tg-SO11 mice compared with WT littermate controls (N = 3/group). (G) Pathological analysis of spleen sections from a Tg-SOX11 mouse confirming a neoplastic B220+CD3−CD34−BCL6− immunophenotype. Fo, follicular; GC, germinal center; H&E, hematoxylin and eosin; Mono/Mac, monocytes/macrophages; MZ, marginal zone; NF, newly formed transitional; NK, natural killer; pDC, plasmacytoid dendritic cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/20/10.1182_blood-2018-02-832535/10/m_blood832535f1.jpeg?Expires=1769105789&Signature=MmjTNRckIeykO2MN9x8c3Bk0K1-qIeOSkohc8TUrlFtQ8DnZDEOMvJbapnNOip6XcMEY-CWKe~LvC-WWsUpOfdUz~UWA1wLD3uI58KTATASJ3xuBwtsYW79KKpI4ZgptRr1EB32a8BBT1gmxzT866gQt5xE~FrxGwwgXd20-AecKAgHJEkAhlNN5I55Sm8KpQTqlOH-QnHI~QVRqAzfRIdIX8ur7SWF9dkAV3hp9G9nQ0n~~5xuLz-xPNztVXUf6i3ANEw7ZP2Fth-QEKoUdaQ5o8VDx-iSKNAYEdhKmCagOidiLTUNBfC2HTwtMWPruMIXPoYZxlapt4ZPqdVfb5w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal