Key Points
The sphingosine-1-phosphate receptor 2 is a bona fide tumor suppressor and transcriptionally regulated by the TGF-β/TGF-βR2/SMAD1 axis.
The aberrant loss of SMAD1 expression is very common in DLBCL and provides a proliferative advantage to B cells in vitro and in vivo.
Abstract
The sphingosine-1-phosphate receptor S1PR2 and its downstream signaling pathway are commonly silenced in diffuse large B-cell lymphoma (DLBCL), either by mutational inactivation or through negative regulation by the oncogenic transcription factor FOXP1. In this study, we examined the upstream regulators of S1PR2 expression and have newly identified the transforming growth factor-β (TGF-β)/TGF-βR2/SMAD1 axis as critically involved in S1PR2 transcriptional activation. Phosphorylated SMAD1 directly binds to regulatory elements in the S1PR2 locus as assessed by chromatin immunoprecipitation, and the CRISPR-mediated genomic editing of S1PR2, SMAD1, or TGFBR2 in DLBCL cell lines renders cells unresponsive to TGF-β–induced apoptosis. DLBCL clones lacking any 1 of the 3 factors have a clear growth advantage in vitro, as well as in subcutaneous xenotransplantation models, and in a novel model of orthotopic growth of DLBCL cells in the spleens and bone marrow of MISTRG mice expressing various human cytokines. The loss of S1pr2 induces hyperproliferation of the germinal center (GC) B-cell compartment of immunized mice and accelerates MYC-driven lymphomagenesis in spontaneous and serial transplantation models. The specific loss of Tgfbr2 in murine GC B-cell phenocopies the effects of S1pr2 loss on GC B-cell hyperproliferation. Finally, we show that SMAD1 expression is aberrantly downregulated in >85% of analyzed DLBCL patients. The combined results uncover an important novel tumor suppressive function of the TGF-β/TGF-βR2/SMAD1/S1PR2 axis in DLBCL, and show that DLBCL cells have evolved to inactivate the pathway at the level of SMAD1 expression.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most commonly diagnosed lymphoma in adults. It arises de novo at nodal or extranodal sites or as a consequence of high-grade transformation of indolent lymphomas or leukemias such as follicular lymphoma, chronic lymphocytic leukemia, and marginal zone lymphoma.1-3 DLBCL represents a heterogeneous disease with molecular subtypes characterized by distinct gene expression profiles, specific sets of somatic mutations, and differentially active intracellular signaling pathways.4 Three subtypes of DLBCL can be distinguished based on their gene expression similarities to their presumed normal B-cell counterparts, with activated B-cell–like (ABC) DLBCL resembling the postgerminal center plasmablast, germinal center B-cell–like (GCB) DLBCL deriving from GC B cells, and primary mediastinal B-cell lymphoma arising from a rare subset of thymic B cells.5,6 One of the biomarkers that is used to discriminate between GCB and ABC DLBCL is the Forkhead Box Protein 1 (FOXP1), a transcription factor that is highly expressed in the ABC subtype and is associated with poor prognosis.7-9 FOXP1 predominantly functions as a repressor of protein-coding genes, several of which have documented tumor suppressive activities in B cells.10-12 We have recently identified the sphingosine 1-phosphate receptor 2 (S1PR2) as being negatively regulated by FOXP1 in ABC-DLBCL; its expression was inversely correlated with FOXP1 expression in a large cohort of DLBCL patients and was restored in DLBCL cell lines upon depletion of FOXP1.10 Thus, S1PR2 is aberrantly silenced because of FOXP1 overexpression in ABC-DLBCL; conversely, in GCB-DLBCL, the S1PR2 locus is subject to recurrent mutations that typically affect 1 of the 2 alleles.13 Therefore, the 2 main DLBCL subtypes have evolved distinct mechanisms of silencing S1PR2 expression, both of which result in inactivation of the downstream signaling pathway involving the small G-protein Gα13 (encoded by the GNA13 gene), ρ, and possibly the kinase AKT. The S1PR2/Gα13/AKT signaling axis has been implicated in the regulation of GC B-cell growth and dissemination.13 Several pieces of evidence indicate that S1PR2 serves as a bona fide tumor suppressor in DLBCL: (1) the forced, doxycycline-induced expression of S1PR2 in DLBCL xenografts strongly delays tumor outgrowth, (2) the loss of 1 S1PR2 allele is sufficient to predispose mice to c-MYC–driven lymphomagenesis, and (3) low S1PR2 expression, independently and in conjunction with high FOXP1 expression, represents a strong negative prognosticator of survival in patients with either subtype of DLBCL.10
In this study, we have addressed the consequences of S1PR2 inactivation for normal and malignant B-cell fate and have investigated the mechanisms of S1PR2 regulation in normal B cells and in DLBCL. We show that the mutational inactivation of the S1PR2 locus by CRISPR/Cas9 provides a major proliferative advantage to DLBCL cell lines in vitro and in xenotransplantation models, and that the loss of 1 or both alleles of S1pr2 in murine GC B cells promotes hyperproliferation of the GC compartment upon sheep red blood cell immunization. We further provide evidence in various cell cultures, xenotransplantation, and genetically manipulated mouse models of the tumor suppressive activity of a newly identified transforming growth factor-β receptor II (TGF-βR2)/SMAD1/S1PR2 axis in DLBCL.
Methods
Cell culture
The panel of DLBCL cell lines used here includes 4 of GCB DLBCL subtype (SU-DHL-6, SU-DHL-16, SU-DHL5, and RC-K8) and 2 of the ABC DLBCL subtype (OCI-Ly3, OCI-Ly10). Cell lines were subjected to human TGF-β-1 (referred to as TGF-β) (PreproTech) treatment at various concentrations and analyzed with respect to cell viability, apoptosis, S1PR2 expression by quantitative reverse transcription polymerase chain reaction (qRT-PCR), transcription factor binding to the S1PR2 promoter region by chromatin immunoprecipitation (ChIP) and protein expression by western blot. Culture conditions, TGF-β treatment conditions, cell viability, proliferation and apoptosis assays, RNA extraction and qRT-PCR, small interfering RNA (siRNA) transfections, CRISPR manipulations, ChIP-PCR, and western blotting techniques are described in the supplemental Material and methods, available on the Blood Web site.
Animal experimentation
Nonobese diabetic/severe combined immunodeficiency/IL2Rγ−/− (NSG) mice and macrophage colony-stimulating factorh (CSFh);interleukin-3 (IL-3)/granulocyte-macrophage CSFh;hSIRPAtg;TPOh;Rag2−;γc− (MISTRG) mice14 were obtained from a local repository. Tgfbr2fl/fl mice (B6;129-Tgfbr2tm1Karl/J) were crossed with AID-Cre mice (B6;FVB-Tg[Aicda-cre]1Rcas/J, both from the Jackson Laboratories). S1pr2−/− mice15 were crossed with Emu-MYC mice expressing the c-MYC oncogene under the control of the immunoglobulin heavy chain enhancer (B6.Cg-Tg[IghMyc]22Bri/J, also from Jackson Laboratories)16 to obtain composite strains. For induction of germinal centers, 6- to 8-week-old mice were intraperitoneally immunized twice with a 10-day interval with 200 µL of 10% sheep red blood cells (Innovative Research, Michigan) and euthanized 10 days after the last immunization. Spleens were processed and subjected to flow cytometric analysis. All flow cytometry and staining procedures are described in the supplemental Material and methods. MYCtg mice were examined and palpated 2 to 3 times weekly to detect signs of lymph node enlargement. Mice were euthanized within 1 week of developing palpable tumors. Tumor cells pooled from the axillary and inguinal lymph nodes were cryopreserved in fetal calf serum with 10% dimethyl sulfoxide. For serial transplantation studies, 1 × 106 cells were injected IV into wild-type BL6 mice in a volume of 100 µL. Mice were palpated every other day for signs of lymph node engraftment. For xenotransplantation studies, CRISPR clones or wild-type RC-K8 or SU-DHL-6 (10 × 106 cells in 200 µL phosphate-buffered saline [PBS]) were injected subcutaneously into both flanks of 6- to 8-week-old NSG mice, or IV in a volume of 100 μL into 6- to 8-week-old MISTRG mice for orthotopic growth. Once palpable tumors had formed in the subcutaneous model, the volume of the tumors was measured by calipers and calculated using the equation (A2 × B)/2, where A is the shorter and B the longer tumor dimension. Once palpable, wild-type SU-DHL-6 tumors were treated every 72 hours with 0.2 μg human TGF-β (PreproTech) in a volume of 50 μL reconstituted in 10 mM citric acid and diluted in 0.1% bovine serum albumin in 1x PBS. Control tumors were treated with 50 μL 10 mM citric acid/0.1% bovine serum albumin diluted in 1x PBS. IV-injected mice were monitored 3 times per week for weight loss and other symptoms of disease. All animal studies were reviewed and approved by the Zürich Cantonal Veterinary Office (licenses 224/2014, 227/2015, 235/2015).
Patient cohorts and SMAD1 immunohistochemistry
Expression of SMAD1 was studied by immunohistochemistry on a phenotypically and genotypically well characterized collective of 76 patients uniformly treated with rituximab, cyclophosphamide, doxorubicin hydrochloride, vincristine sulfate, and prednisone every 14 days (R-CHOP-14) and prospectively followed in the Swiss Group for Clinical Cancer Research 38/07 clinical trial (ClinicalTrials.gov NCT00544219)17 and on a retrospective collective of 184 primary DLBCL treated with rituximab-free regimens (mainly CHOP),18 of which 74 patients had complete (and compatible with the former patients’) follow-up data. The primary polyclonal antibody (CellSignaling cs9743) was diluted 1:40 and incubated for 20 minutes in an automated immunostainer (Benchmark, Ventana/Roche) after heat-induced antigen retrieval with the CC1 buffer for 40 minutes.
Statistics
All statistical analyses were performed using Graph Pad Prism software. Graphs represent means plus standard error of the mean (SEM) of at least 2 independent experiments for cell culture work and medians for mouse experiments unless otherwise indicated in the figure legend. Statistical analysis was performed using 2-tailed Student t test for in vitro assays and 2-tailed Mann-Whitney U test for in vivo studies as well as 2-tailed Fisher’s exact test for correlations of mutational status and SMAD1 expression in the patient cohort.
Results
Deletion of S1PR2 by CRISPR/Cas9 confers a strong growth advantage to DLBCL cell lines in vitro and in vivo
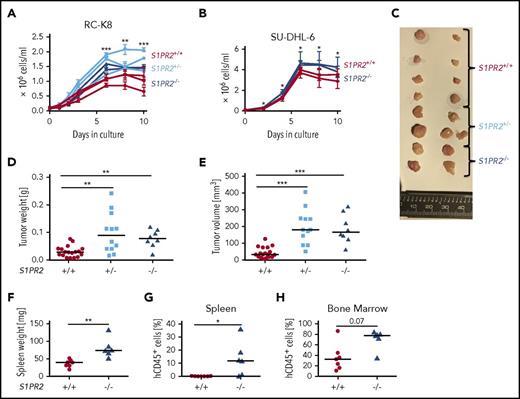
To confirm the putative tumor suppressive properties of S1PR2 in DLBCL, we used CRISPR to delete 1 or both alleles of the S1PR2 locus in a cell line that is particularly suitable for this purpose because of its low FOXP1 and corresponding high S1PR2 expression (RC-K8; supplemental Figure 1A-B). Of the 5 S1PRs (S1PR1-5), only S1PR2 is expressed in DLBCL cells and controlled by FOXP1 expression (supplemental Figure 1C). Sequencing of the S1PR2 and the GNA13 genomic loci confirmed that both loci carry the wild-type sequence in RC-K8 cells (supplemental Figure 1D). A CRISPR strategy (supplemental Figure 1E) was then used to delete the single S1PR2 exon in 1 or both alleles of the locus (supplemental Figure 1F). Three S1PR2+/− and 2 S1PR2−/− clones were grown from fluorescence-activated cell sorted (FACS) single cells and compared with respect to their in vitro growth to 3 wild-type clones (S1PR2+/+) that had been subjected to the same electroporation and sorting procedures. Mono- and biallelically mutated clones exhibited a robust growth advantage over wild-type clones in vitro (Figure 1A); the same observation was made with S1PR2−/− clones generated using a second cell line (SU-DHL-6, Figure 1B). To address whether the growth advantage of S1PR2−/− clones was due to reduced apoptosis or to enhanced proliferation, we flow cytometrically quantified the surface Annexin V and nuclear Ki67 expression at various time points of the growth curve. Whereas apoptosis rates were similar across clones as determined by Annexin V staining, S1PR2−/− clones showed significantly stronger Ki67 staining than S1PR2+/+ clones (supplemental Figure 1G-H). S1PR2+/− and S1PR2−/− clones further also grew faster and formed larger tumors at the study end point compared with wild-type clones in a subcutaneous xenograft model of RC-K8 (Figure 1C-E). The NSG mice used for subcutaneous xenotransplantation do not support orthotopic growth of DLBCL cell lines upon IV injection. We instead turned to an alternative immunodeficient mouse developed on the Rag2−/−IL2Rγ−/− background, termed MISTRG, which expresses the human cytokines macrophage-CSF, IL-3, thrombopoietin, granulocyte-macrophage CSF as well as SIRP1α, from the respective murine loci19 and is known to support improved (orthotopic) growth of various myeloid neoplasias and solid tumors.14,20,21 MISTRG mice supported the efficient engraftment of SU-DHL-6 cells, which grew in bone marrow and spleen; interestingly, S1PR2-mutant SU-DHL-6 clones grew more rapidly in both bone marrow and spleen than wild-type clones, accounting for the higher frequencies of human cells identified by FACS in these organs (Figure 1F-H; supplemental Figure 1I). The combined data indicate that S1PR2 expression restricts tumor cell proliferation in a cell-autonomous manner, further supporting the concept that S1PR2 is a bona fide tumor suppressor in DLBCL.
Genomic editing of the S1PR2 locus provides a growth advantage to DLBCL cell lines in vitro and in vivo. The DLBCL cell lines (A) RC-K8 and (B) SU-DHL-6 were subjected to S1PR2 inactivation using CRISPR/Cas9 editing. Absolute cell counts of 2 to 3 independent clones derived from FACS single cells of the indicated genotypes were compared under standard cell culture conditions over 10 days without medium change. Pooled results from (A) 2 of a total of 4 independent experiments and (B) of 4 independent experiments are shown. P values were calculated using the Student t test on the average value for each genotype pooling S1PR2+/− and S1PR2−/− clones. (C-E) Ten million cells each of 5 S1PR2+/+ clones (red), 4 S1PR2+/− clones (light blue), and 2 S1PR2−/− clones (all in the RC-K8 cell line; blue) were injected subcutaneously into the flanks of NSG mice. (C) Tumors were excised, representative macroscopic images were taken, and (D) tumor weights and (E) volumes were determined at the study end point 40 days post injection. Every dot represents 1 tumor, and plots show pooled data from 2 independent experiments. (F-H) Ten million cells per mouse of 2 to 3 replicates each of 3 independent S1PR2+/+ clones (red) and 3 S1PR2−/− clones (all in the SU-DHL-6 cell line; blue) were injected IV into MISTRG mice. Mice were euthanized 35 days after tumor cell injection, (F) their spleens were weighed, and the frequencies of hCD45+ cells in the (G) spleen and (H) bone marrow was determined by flow cytometry. Every dot represents 1 mouse; graphs represent pooled data from 2 independent experiments. (D-H) Horizontal lines indicate medians; P values were calculated using the Mann-Whitney U test. *P < .05; **P < .01; ***P < .001.
Genomic editing of the S1PR2 locus provides a growth advantage to DLBCL cell lines in vitro and in vivo. The DLBCL cell lines (A) RC-K8 and (B) SU-DHL-6 were subjected to S1PR2 inactivation using CRISPR/Cas9 editing. Absolute cell counts of 2 to 3 independent clones derived from FACS single cells of the indicated genotypes were compared under standard cell culture conditions over 10 days without medium change. Pooled results from (A) 2 of a total of 4 independent experiments and (B) of 4 independent experiments are shown. P values were calculated using the Student t test on the average value for each genotype pooling S1PR2+/− and S1PR2−/− clones. (C-E) Ten million cells each of 5 S1PR2+/+ clones (red), 4 S1PR2+/− clones (light blue), and 2 S1PR2−/− clones (all in the RC-K8 cell line; blue) were injected subcutaneously into the flanks of NSG mice. (C) Tumors were excised, representative macroscopic images were taken, and (D) tumor weights and (E) volumes were determined at the study end point 40 days post injection. Every dot represents 1 tumor, and plots show pooled data from 2 independent experiments. (F-H) Ten million cells per mouse of 2 to 3 replicates each of 3 independent S1PR2+/+ clones (red) and 3 S1PR2−/− clones (all in the SU-DHL-6 cell line; blue) were injected IV into MISTRG mice. Mice were euthanized 35 days after tumor cell injection, (F) their spleens were weighed, and the frequencies of hCD45+ cells in the (G) spleen and (H) bone marrow was determined by flow cytometry. Every dot represents 1 mouse; graphs represent pooled data from 2 independent experiments. (D-H) Horizontal lines indicate medians; P values were calculated using the Mann-Whitney U test. *P < .05; **P < .01; ***P < .001.
Loss of S1pr2 promotes hyperproliferation of the GC B-cell compartment and increases lymphoma incidence in a spontaneous and a serial transplantation mouse model of DLBCL
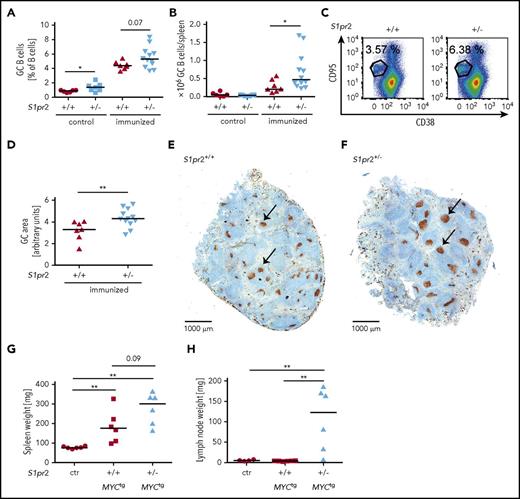
DLBCL arises from germinal center and post-GC B cells that reside in secondary lymphoid organs or in lymphoid tissues at extranodal sites. To address whether the expression of S1PR2 affects GC formation and GC B-cell proliferation, we immunized wild-type or S1pr2+/− mice with 2 doses of sheep red blood cells (SRBCs) and flow cytometrically quantified GC B cells, centrocytes, and centroblasts in the spleen at the study end point. The frequencies and absolute numbers of centrocytes, centroblasts, and of all GC B cells were increased in S1pr2+/− mice (Figure 2A-C; supplemental Figure 2A-D). The increase in GC B cells could be attributed to an increase in the size of individual GCs, but not their multiplicity per spleen, as determined by Ki67 staining of spleen sections (Figure 2D-F; supplemental Figure 2E). As previously observed in a small preliminary cohort,10 we confirmed that mice harboring a heterozygous S1pr2 mutation with an additional c-MYC transgene under the B-cell–specific immunoglobulin heavy chain enhancer showed accelerated lymphoma development compared with S1pr2-proficient mice harboring the c-MYC transgene (supplemental Figure 2F). When lymph node cells from S1pr2+/− or S1pr2+/+c-MYC transgenic mice were serially transplanted into wild-type mice, we observed faster lymphoma development with transplanted S1pr2+/− cells relative to wild-type cells in recipient mice (Figure 2G-H; supplemental Figure 2G-H). These results suggest that S1PR2 controls GC B-cell proliferation and that the hyperproliferation of GC B cells from loss of S1pr2 may represent an early, initiating event in lymphomagenesis.
The monoallelic loss of S1pr2 promotes hyperproliferation of the GC B-cell compartment and increases the lymphoma burden in a spontaneous and a serial transplantation model of MYC-driven lymphomagenesis. (A-F) S1pr2+/+ and S1pr2+/− mice on the BL/6 background were immunized twice intraperitoneally with 200 μL 10% SRBC, with a 10-day interval between the 2 immunizations. (A-C) Mice were euthanized 10 days after the last immunization and GC B cells were flow cytometrically identified as CD95hi CD38lo in the CD19+ B-cell compartment. GC B-cell frequencies in % of all CD19+ B cells as well as absolute numbers per spleen are shown alongside representative FACS plots. Nonimmunized littermates are shown as control. (D-F) The GC area (arbitrary units) of immunized mice was determined by quantifying 3 Ki-67-stained spleen sections per mouse (D) using ImageJ. (E-F) Representative pictures of spleens of immunized S1pr2+/+ and S1pr2+/− mice are shown. Size bar represents 1000 μm; arrows point to GCs. (A-B,D) Every dot represents 1 mouse and data from 5 pooled experiments are shown. (G-H) One million lymph node cells per mouse, harvested from 3 S1pr2+/+ and 3 S1pr2+/−MYCtg donor mice of the cohorts shown in supplemental Figure 2F were injected IV into wild-type BL/6 recipients. Mice were palpated every other day for enlarged lymph nodes and euthanized after 20 days (ie, when the first mice showed disease symptoms). (G) Spleen and (H) lymph node weights were determined. Lymph node weights represent the average of 2 inguinal and 2 axillary lymph nodes. Control mice were not injected with tumor cells. (A-B,D,G-H) Horizontal lines indicate medians; P values were calculated using the Mann-Whitney U test. *P < .05; **P < .01.
The monoallelic loss of S1pr2 promotes hyperproliferation of the GC B-cell compartment and increases the lymphoma burden in a spontaneous and a serial transplantation model of MYC-driven lymphomagenesis. (A-F) S1pr2+/+ and S1pr2+/− mice on the BL/6 background were immunized twice intraperitoneally with 200 μL 10% SRBC, with a 10-day interval between the 2 immunizations. (A-C) Mice were euthanized 10 days after the last immunization and GC B cells were flow cytometrically identified as CD95hi CD38lo in the CD19+ B-cell compartment. GC B-cell frequencies in % of all CD19+ B cells as well as absolute numbers per spleen are shown alongside representative FACS plots. Nonimmunized littermates are shown as control. (D-F) The GC area (arbitrary units) of immunized mice was determined by quantifying 3 Ki-67-stained spleen sections per mouse (D) using ImageJ. (E-F) Representative pictures of spleens of immunized S1pr2+/+ and S1pr2+/− mice are shown. Size bar represents 1000 μm; arrows point to GCs. (A-B,D) Every dot represents 1 mouse and data from 5 pooled experiments are shown. (G-H) One million lymph node cells per mouse, harvested from 3 S1pr2+/+ and 3 S1pr2+/−MYCtg donor mice of the cohorts shown in supplemental Figure 2F were injected IV into wild-type BL/6 recipients. Mice were palpated every other day for enlarged lymph nodes and euthanized after 20 days (ie, when the first mice showed disease symptoms). (G) Spleen and (H) lymph node weights were determined. Lymph node weights represent the average of 2 inguinal and 2 axillary lymph nodes. Control mice were not injected with tumor cells. (A-B,D,G-H) Horizontal lines indicate medians; P values were calculated using the Mann-Whitney U test. *P < .05; **P < .01.
S1PR2 expression is regulated by TGF-β and SMAD signaling
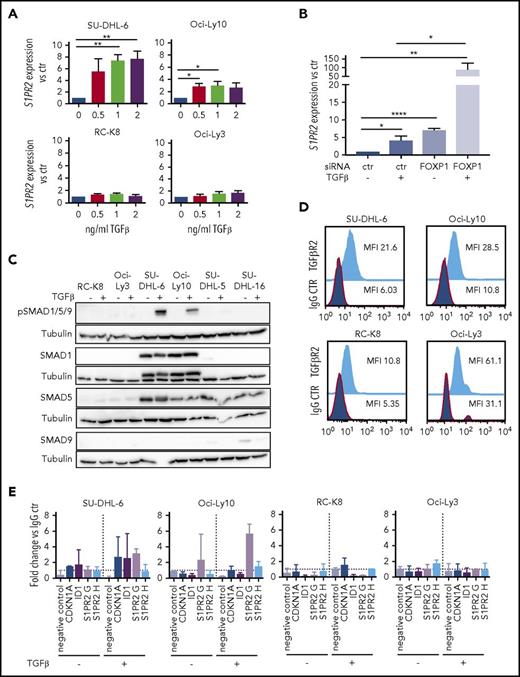
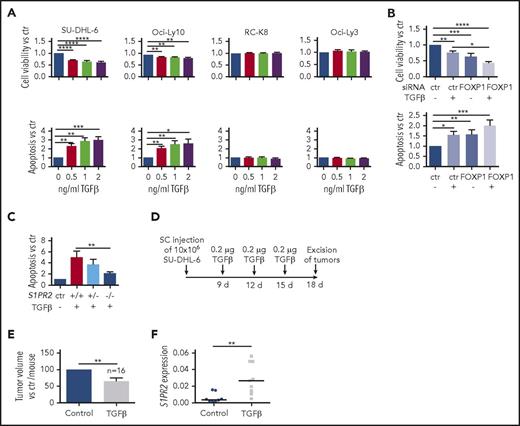
We have previously shown that FOXP1 represses transcription at the S1PR2 locus by binding to 2 regulatory elements located 2.5 (referred to as S1PR2 H) and 5 kb (S1PR2 G) upstream of the transcription start site.10 However, little is known regarding activating transcription factors that regulate S1PR2 expression. We therefore screened several cytokines known to affect normal B-cell biology for their effects on S1PR2 expression and found that the addition of TGF-β, but not of IL-3, IL-5, or IL-6, dose-dependently induced the expression of S1PR2 in a subset of cell lines analyzed (Figure 3A; supplemental Figure 3A-B), with SU-DHL-6 and Oci-Ly10 showing the strongest response, RC-K8 and Oci-Ly3 being completely unresponsive to TGF-β, and SU-DHL-16 and SU-DHL-5 showing a partial response. The effect of TGF-β was particularly strong when FOXP1 was simultaneously depleted by siRNAs (Figure 3B; supplemental Figure 3C); of note, TGF-β exposure had no effect on FOXP1 levels (supplemental Figure 3D), ruling out an indirect effect of TGF-β on S1PR2 expression via regulation of FOXP1. To examine which components of the TGF-β signaling pathway control the differential susceptibility of DLBCL cell lines to TGF-β treatment, we examined the expression of various SMAD transcription factors that were previously found to transactivate target genes of the pathway in DLBCL.22 Although SMAD9 was not expressed in any of the cell lines and SMAD5 was expressed in both highly and moderately responsive cell lines, the expression of SMAD1 was selectively detected only in those cell lines that showed strong responses to TGF-β and strong SMAD1/5/9 phosphorylation (Figure 3C; supplemental Figure 3E). We further examined the expression of the TGF-β receptor II (TGF-βR2) and found it to be universally expressed in all cell lines (Figure 3D; supplemental Figure 3F).
S1PR2 expression is regulated by the TGF-β/SMAD signaling pathway. (A) S1PR2 expression after 24 hours of treatment with the indicated increasing doses of TGF-β, as assessed in the SU-DHL-6, Oci-Ly10, RC-K8, and Oci-Ly3 DLBCL cell lines by qRT-PCR. (B) The DLBCL cell line SU-DHL-6 was treated with FOXP1 targeting siRNA for 48 hours and subjected to treatment with 2 ng/mL TGF-β for an additional 24 hours. (A-B) Data are pooled from 3 or more independent experiments. Graphs show mean ± SEM; P values were calculated using the Student t test. (C) The indicated DLBCL cell lines were treated with 2 ng/mL TGF-β for 1 hour and subjected to immunoblotting with antibodies against the indicated SMAD proteins, p-SMAD1/5/9 and tubulin. Representative immunoblots of at least 2 independent experiments are shown. (D) TGF-βR2 surface expression of the indicated DLBCL cell lines, as assessed by flow cytometry. The plots are representative for 2 independent experiments. (E) pSMAD1/5/9 ChIP of cells treated or not with 5 ng/mL TGF-β for 4 hours; an unspecific rbIgG antibody was used as control. Eluted DNA was subjected to PCR using primers amplifying 2 regions 2.5 and 5 kb upstream of the S1PR2 TSS. MyoD was amplified as negative control; CDKN1A and ID1 were used as positive controls for canonical TGF-β and BMP signaling. Graphs represent the fold change of the yield relative to 1% input of the pSMAD1/5/9 sample vs rbIgG; means ± SD of 2 independent experiments are shown. *P < .05; **P < .01; ***P < .001; ****P < .0001.
S1PR2 expression is regulated by the TGF-β/SMAD signaling pathway. (A) S1PR2 expression after 24 hours of treatment with the indicated increasing doses of TGF-β, as assessed in the SU-DHL-6, Oci-Ly10, RC-K8, and Oci-Ly3 DLBCL cell lines by qRT-PCR. (B) The DLBCL cell line SU-DHL-6 was treated with FOXP1 targeting siRNA for 48 hours and subjected to treatment with 2 ng/mL TGF-β for an additional 24 hours. (A-B) Data are pooled from 3 or more independent experiments. Graphs show mean ± SEM; P values were calculated using the Student t test. (C) The indicated DLBCL cell lines were treated with 2 ng/mL TGF-β for 1 hour and subjected to immunoblotting with antibodies against the indicated SMAD proteins, p-SMAD1/5/9 and tubulin. Representative immunoblots of at least 2 independent experiments are shown. (D) TGF-βR2 surface expression of the indicated DLBCL cell lines, as assessed by flow cytometry. The plots are representative for 2 independent experiments. (E) pSMAD1/5/9 ChIP of cells treated or not with 5 ng/mL TGF-β for 4 hours; an unspecific rbIgG antibody was used as control. Eluted DNA was subjected to PCR using primers amplifying 2 regions 2.5 and 5 kb upstream of the S1PR2 TSS. MyoD was amplified as negative control; CDKN1A and ID1 were used as positive controls for canonical TGF-β and BMP signaling. Graphs represent the fold change of the yield relative to 1% input of the pSMAD1/5/9 sample vs rbIgG; means ± SD of 2 independent experiments are shown. *P < .05; **P < .01; ***P < .001; ****P < .0001.
To address whether the SMAD proteins bind to regulatory elements of the S1PR2 locus, we performed ChIP followed by quantitative PCR specific for the regions 2.5 (H) and 5 kb (G) upstream of the S1PR2 transcription start site. SMADs 1, 5, and 9 are all phosphorylated on serine residues by the active (ligand-bound) TGF-β receptor complex (consisting of type I and II receptor heterotetramer), which promotes the nuclear translocation of the complex and binding to chromatin. We therefore used a pSMAD1/5/9-specific antibody for ChIP, and found that pSMAD1/5/9 indeed precipitated with S1PR2 genomic DNA, especially when cells were exposed to TGF-β for 4 hours before ChIP; this effect was observed in the TGF-β–responsive cell lines SU-DHL-6 and Oci-Ly10, but not the resistant cell lines RC-K8 and Oci-Ly3 (Figure 3E). Speculating that FOXP1 and SMAD1 interact directly at the S1PR2 promoter, we immunoprecipitated FOXP1 and checked for a possible coprecipitation of SMAD1; however, no evidence could be found to that end in a cell line that coexpresses both proteins (supplemental Figure 3G).
Because S1PR2 signaling has previously been shown to inhibit AKT phosphorylation and AKT-driven migration,13 we speculated that TGF-β/SMAD1-induced S1PR2 expression might impair DLBCL cell survival by preventing AKT-mediated survival signaling. However, TGF-β treatment failed to affect AKT activation, as assessed by its autophosphorylation on serine 473, and also did not change overall AKT levels (supplemental Figure 3D) and thus does not appear to act through this pathway. The combined results indicate that TGF-β activates S1PR2 expression, especially if the repressor FOXP1 is removed, and implicate the noncanonical TGF-βR2/SMAD1/5/9 pathway, and in particular SMAD1, in the transcriptional activation of S1PR2.
TGF-β induces S1PR2-dependent apoptosis in DLBCL cell lines in vitro and in vivo
We have shown in a previous study that forced S1PR2 expression kills DLBCL cell lines both in culture and in xenotransplantation models.10 To address whether TGF-β exposure and the concomitant endogenous S1PR2 upregulation is toxic to DLBCL cell lines, we treated cell lines showing differential responses to TGF-β (Figure 3A,C) with increasing doses of the cytokine. TGF-β exposure efficiently reduced cell viability and increased apoptosis dose-dependently; this effect was restricted to cell lines that upregulate S1PR2 upon TGF-β exposure and that are positive for SMAD1 (Figure 4A; supplemental Figure 4A-B). The effect was again particularly strong when FOXP1 was simultaneously depleted by siRNAs (Figure 4B). SU-DHL-6 clones that had been subjected to S1PR2 deletion failed to undergo apoptosis upon TGF-β exposure, suggesting a critical role of S1PR2 in TGF-β–driven apoptosis (Figure 4C). The TGF-β–sensitive cell line SU-DHL-6 was further assessed with respect to its susceptibility to TGF-β treatment in a xenograft model. Regular doses of intratumorally administered TGF-β resulted in lower tumor volumes and increased S1PR2 expression at the study end point relative to the vehicle control (Figure 4D-F; supplemental Figure 4C). The combined results indicate that TGF-β signaling is cytotoxic for DLBCL cells in vitro and in vivo, and suggest that the apoptosis-promoting activity of TGF-β requires S1PR2.
TGF-β induces S1PR2-dependent apoptosis in DLBCL cell lines in vitro and in vivo. (A) Cell viability and apoptosis, as determined by Cell Titer Blue assay and Annexin V staining, of the indicated cell lines after 24 hours of exposure to increasing concentrations of TGF-β; values are normalized to the untreated control sample. (B) The DLBCL cell line SU-DHL-6 was treated with FOXP1 targeting siRNA for 48 hours, subjected to 2 ng/mL TGF-β for additional 24 hours, and analyzed as shown in panel A. (A-B) Data are pooled from 3 or more independent experiments. Graphs show means ± SEM; P values were calculated using the Student t test. (C) Three S1PR2+/+, 1 S1PR2+/−, and 3 S1PR2−/− clones generated in the SU-DHL-6 cell line were treated with 2 ng/mL TGF-β and analyzed for apoptosis by Annexin V staining. Bars represent pooled data for each genotype relative to the untreated control of each clone. Each clone was analyzed 3 to 6 times. Graphs represent means ± SEM; P values were calculated using the Student t test. (D-F) Ten million SU-DHL-6 cells were injected subcutaneously into both flanks of NSG mice. (D) One tumor per mouse was injected intratumorally with TGF-β at the depicted intervals; the other received vehicle only. Tumor volumes were measured (E) after excision and (F) RNA was extracted and qRT-PCR for S1PR2 was performed on excised tumor tissue. (F) Each dot represents 1 tumor and results are pooled from 2 independent experiments. S1PR2 expression analysis was performed in only 1 of the 2 studies with n = 10 per group. Two control and 1 TGF-β–treated tumor had to be excluded because of insufficient RNA quality. TGF-β–treated and control tumors are compared for each mouse. P values were calculated using the Mann-Whitney U test. *P < .05; **P < .01; ***P < .001; ****P < .0001.
TGF-β induces S1PR2-dependent apoptosis in DLBCL cell lines in vitro and in vivo. (A) Cell viability and apoptosis, as determined by Cell Titer Blue assay and Annexin V staining, of the indicated cell lines after 24 hours of exposure to increasing concentrations of TGF-β; values are normalized to the untreated control sample. (B) The DLBCL cell line SU-DHL-6 was treated with FOXP1 targeting siRNA for 48 hours, subjected to 2 ng/mL TGF-β for additional 24 hours, and analyzed as shown in panel A. (A-B) Data are pooled from 3 or more independent experiments. Graphs show means ± SEM; P values were calculated using the Student t test. (C) Three S1PR2+/+, 1 S1PR2+/−, and 3 S1PR2−/− clones generated in the SU-DHL-6 cell line were treated with 2 ng/mL TGF-β and analyzed for apoptosis by Annexin V staining. Bars represent pooled data for each genotype relative to the untreated control of each clone. Each clone was analyzed 3 to 6 times. Graphs represent means ± SEM; P values were calculated using the Student t test. (D-F) Ten million SU-DHL-6 cells were injected subcutaneously into both flanks of NSG mice. (D) One tumor per mouse was injected intratumorally with TGF-β at the depicted intervals; the other received vehicle only. Tumor volumes were measured (E) after excision and (F) RNA was extracted and qRT-PCR for S1PR2 was performed on excised tumor tissue. (F) Each dot represents 1 tumor and results are pooled from 2 independent experiments. S1PR2 expression analysis was performed in only 1 of the 2 studies with n = 10 per group. Two control and 1 TGF-β–treated tumor had to be excluded because of insufficient RNA quality. TGF-β–treated and control tumors are compared for each mouse. P values were calculated using the Mann-Whitney U test. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Loss of TGF-β signaling in the GC compartment induces GC B-cell hyperproliferation
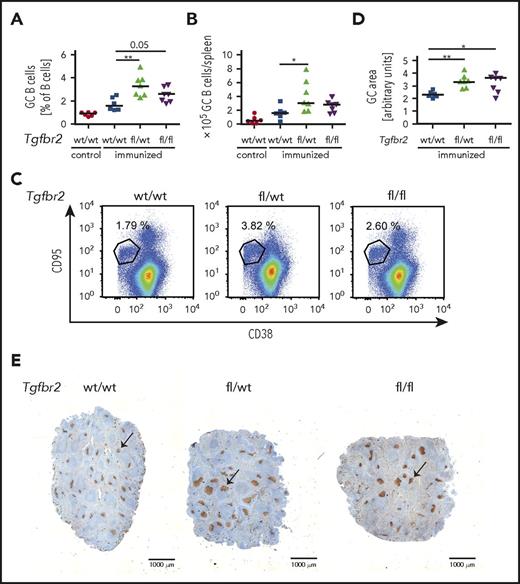
To address in a genetic model whether the cell-intrinsic loss of TGF-β signaling in the GC B-cell compartment affects the GC reaction, we crossed Tgfbr2fl/fl mice with animals expressing Cre recombinase under the control of the GC-specific activation-induced cytidine deaminase promoter (AID-Cre). Mice lacking TGF-βR2 specifically in GC B cells exhibited hyperproliferation of their GC B cells, centroblasts, and centrocytes upon 1 round of SRBC immunization (Figure 5A-C; supplemental Figure 5A-C) and thus phenocopied the loss of S1PR2 in this compartment. The overall increase in GC cells could be attributed to an increase in the size of individual GCs, but not of their multiplicity per spleen, as determined by Ki67 staining of spleen sections (Figure 5D-E; supplemental Figure 5D). Together, the results demonstrate a critical role of the TGF-βR2/SMAD1/S1PR2 axis in the physiological control of the GC reaction and show for the first time that GC B-cell intrinsic TGF-β signaling is required for GC confinement.
Loss of TGF-β signaling in the GC compartment induces GC B-cell hyperproliferation. (A-E) Tgfbr2fl/fl mice were crossed with AID-Cre mice; Tgfbr2wt/wt, Tgfbr2fl/wt, and Tgfbr2fl/fl x AID-Cre mice were immunized IV with 200 μL 10% SRBCs, euthanized 10 days after immunization, and GC cells were analyzed by flow cytometry as described in Figure 2. (A-C) GC B-cell frequencies in % of all CD19+ B cells as well as absolute numbers per spleen are shown alongside representative FACS plots. Nonimmunized littermates are shown as control. (D-E) The GC area (arbitrary units) of immunized mice was determined by quantifying 3 Ki-67-stained spleen sections per mouse (D) using ImageJ. (E) Representative pictures of spleens of immunized mice of the indicated genotypes. Size bar represents 1000 μm; arrows point to GCs. (A-B,D) Every dot represents 1 mouse, and data from 3 pooled experiments are shown. Graphs show medians. *P < .05; **P < .01.
Loss of TGF-β signaling in the GC compartment induces GC B-cell hyperproliferation. (A-E) Tgfbr2fl/fl mice were crossed with AID-Cre mice; Tgfbr2wt/wt, Tgfbr2fl/wt, and Tgfbr2fl/fl x AID-Cre mice were immunized IV with 200 μL 10% SRBCs, euthanized 10 days after immunization, and GC cells were analyzed by flow cytometry as described in Figure 2. (A-C) GC B-cell frequencies in % of all CD19+ B cells as well as absolute numbers per spleen are shown alongside representative FACS plots. Nonimmunized littermates are shown as control. (D-E) The GC area (arbitrary units) of immunized mice was determined by quantifying 3 Ki-67-stained spleen sections per mouse (D) using ImageJ. (E) Representative pictures of spleens of immunized mice of the indicated genotypes. Size bar represents 1000 μm; arrows point to GCs. (A-B,D) Every dot represents 1 mouse, and data from 3 pooled experiments are shown. Graphs show medians. *P < .05; **P < .01.
TGF-β signaling via TGF-βR2 and SMAD1 controls the proliferation of DLBCL cells
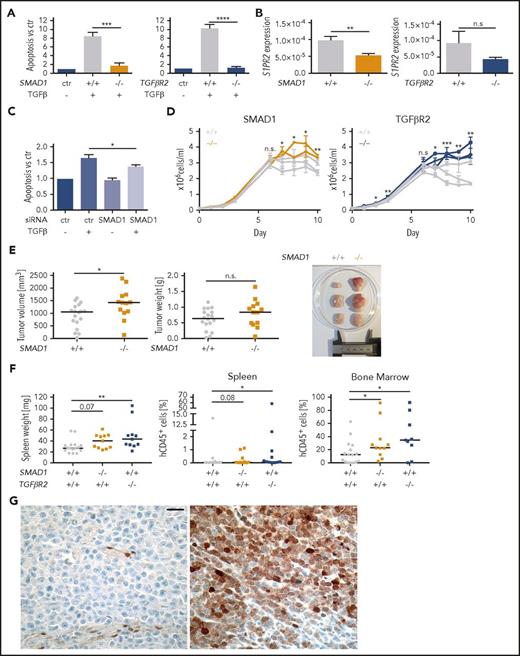
To examine the contribution of the TGF-βR2/SMAD1 axis to cell death signaling in DLBCL in more detail, we inactivated SMAD1 and TGFβR2 in the SU-DHL-6 cell line by editing their first exons (supplemental Figure 6A-B) and exposing several clones each to TGF-β. The loss of SMAD1 and TGFβR2 rendered clones resistant to TGF-β–induced apoptosis and reduced their S1PR2 expression (Figure 6A-B). The contribution of SMAD1 to TGF-β–induced apoptosis could further be confirmed by a SMAD1-specific siRNA (Figure 6C; supplemental Figure 6C-D). Clones lacking either SMAD1 or TGFβR2 grew faster than wild-type clones and thus phenocopied the effect of S1PR2 inactivation in terms of their growth advantage in vitro (Figure 6D). Clones lacking SMAD1, but not those lacking TGFβR2, also grew faster and formed larger tumors when growing as subcutaneous xenografts on the flanks of NSG mice (Figure 6E; supplemental Figure 6E) and both SMAD1 and TGFβR2 knockout clones engrafted more readily in the spleen and the bone marrow when injected IV into MISTRG mice (Figure 6F). Furthermore, analysis of SMAD1 expression in a panel of 11 DLBCL cell lines confirmed its expression only in 2 cell lines (supplemental Figure 6F). Correlation analysis in the cell line panel confirmed the negative correlation between FOXP1 and S1PR2 expression and showed a trend toward a positive correlation between SMAD1 and TGF-βR2 vs S1PR2 expression (supplemental Figure 6G). The combined results indicate that TGF-β signals via TGF-βR2 and SMAD1 to control S1PR2 expression and DLBCL survival in vitro and restrict tumor growth in vivo.
TGF-β signaling via TGF-βR2 and SMAD1 activates S1PR2 expression and induces apoptosis of DLBCL cells and SMAD1 expression is downregulated in DLBCL patients. (A) Three SMAD1+/+ and 2 SMAD1−/− as well as 3 TGFβR2+/+ and 3 TGFβR2−/− clones (all generated in the SU-DHL-6 cell line) were treated with 2 ng/mL TGF-β for 24 hours and analyzed for apoptosis with Annexin V staining. Bars represent pooled data for each genotype relative to the untreated control of each clone. (B) The same clones as in panel A were subjected to RNA extraction and S1PR2-specific qRT-PCR. (A-B) Each clone was analyzed twice; graphs represent means ± SEM; P values were calculated using the Student t test. (C) The DLBCL cell line SU-DHL-6 was treated with SMAD1-targeting siRNA for 48 hours and subjected to 2 ng/mL TGF-β for additional 24 hours. Cells were analyzed for apoptosis by Annexin V staining. Graphs show pooled results from 6 independent experiments. Means ± SEM are represented. P values were calculated using the Student t test. (D) Absolute cell counts of 2 to 3 independent clones derived from FACS single cells of the indicated genotypes were compared under standard cell culture conditions over 10 days without medium change. Two experimental replicates are shown. P values were calculated using the Student t test on the average value for each genotype. (E) Ten million cells each of 3 SMAD1+/+ clones (gray) and 2 SMAD1−/− clones (orange, in SU-DHL-6) were injected subcutaneously into the flanks of NSG mice. Tumors were excised and tumor weights and tumor volumes were determined at the study end point 24 days after injection. Every dot represents 1 tumor; plots show pooled data from 2 independent experiments. (F) Ten million cells of 6 SMAD1/TGFβR2+/+ clones (gray) and 2 SMAD1−/− (orange) or 3 TGFβR2−/− (blue, all in SU-DHL-6) clones were injected IV into MISTRG mice. Mice were euthanized 35 days postinjection, their spleens were weighed, and the frequencies of hCD45+ cells in the spleens and bone marrow was determined by flow cytometry. Every dot represents 1 mouse; graphs represent data from 3 experiments. (E-F) Horizontal lines indicate medians; P values were calculated using the Mann-Whitney U test. (G) Negative (left) and positive (right) SMAD1 immunohistochemical staining of DLBCL patient samples. Size bar represents 20 μm. *P < .05; **P < .01; ***P < .001; ****P < .0001. n.s., not significant.
TGF-β signaling via TGF-βR2 and SMAD1 activates S1PR2 expression and induces apoptosis of DLBCL cells and SMAD1 expression is downregulated in DLBCL patients. (A) Three SMAD1+/+ and 2 SMAD1−/− as well as 3 TGFβR2+/+ and 3 TGFβR2−/− clones (all generated in the SU-DHL-6 cell line) were treated with 2 ng/mL TGF-β for 24 hours and analyzed for apoptosis with Annexin V staining. Bars represent pooled data for each genotype relative to the untreated control of each clone. (B) The same clones as in panel A were subjected to RNA extraction and S1PR2-specific qRT-PCR. (A-B) Each clone was analyzed twice; graphs represent means ± SEM; P values were calculated using the Student t test. (C) The DLBCL cell line SU-DHL-6 was treated with SMAD1-targeting siRNA for 48 hours and subjected to 2 ng/mL TGF-β for additional 24 hours. Cells were analyzed for apoptosis by Annexin V staining. Graphs show pooled results from 6 independent experiments. Means ± SEM are represented. P values were calculated using the Student t test. (D) Absolute cell counts of 2 to 3 independent clones derived from FACS single cells of the indicated genotypes were compared under standard cell culture conditions over 10 days without medium change. Two experimental replicates are shown. P values were calculated using the Student t test on the average value for each genotype. (E) Ten million cells each of 3 SMAD1+/+ clones (gray) and 2 SMAD1−/− clones (orange, in SU-DHL-6) were injected subcutaneously into the flanks of NSG mice. Tumors were excised and tumor weights and tumor volumes were determined at the study end point 24 days after injection. Every dot represents 1 tumor; plots show pooled data from 2 independent experiments. (F) Ten million cells of 6 SMAD1/TGFβR2+/+ clones (gray) and 2 SMAD1−/− (orange) or 3 TGFβR2−/− (blue, all in SU-DHL-6) clones were injected IV into MISTRG mice. Mice were euthanized 35 days postinjection, their spleens were weighed, and the frequencies of hCD45+ cells in the spleens and bone marrow was determined by flow cytometry. Every dot represents 1 mouse; graphs represent data from 3 experiments. (E-F) Horizontal lines indicate medians; P values were calculated using the Mann-Whitney U test. (G) Negative (left) and positive (right) SMAD1 immunohistochemical staining of DLBCL patient samples. Size bar represents 20 μm. *P < .05; **P < .01; ***P < .001; ****P < .0001. n.s., not significant.
The expression of SMAD1 is aberrantly downregulated in DLBCL patients
To test the clinical relevance of our observations in 2 Swiss patient cohorts, we studied the expression of SMAD1 by immunohistochemistry performed on tissue microarrays. Seventy-five patient samples of a uniformly R-CHOP–treated collective and 184 patients of a CHOP-treated collective were evaluable. Only 7 of the 75 (9.3%) and 29 of the 184 patients (15.7%), respectively, were found to express SMAD1 in 10% to 90% of tumor cells (Figure 6G); in all other (negative) cases, internal positive controls (vessels) stained as expected, but the lymphoma cells did not show SMAD1 positivity. In contrast, SMAD1 staining of normal tonsil samples showed uniformly positive SMAD1 expression in centrocytes, but not centroblasts (supplemental Figure 6H). In both cohorts, the fraction of SMAD1-positive cases was equally low in patients with GCB (15%) and non-GCB (ABC; 13%) DLBCL subtypes. The results indicate that SMAD1 is selectively downregulated in DLBCL, irrespective of the subtype. Further analyses of SMAD1 expression in relation to clinical characteristic of the patients, and the mutational profiles of the tumors as assessed by targeted high-throughput sequencing of all exons or hotspots of 68 frequently mutated genes, revealed that SMAD1 was more commonly expressed in CD79b mutant cases (2/4 vs 2/70, P = .041), in female patients (in the respective collectives: 6/34 vs 1/41, P = .030 and 14/60 vs 16/124, P = .043), and, not statistically significantly, in MYD88 mutant cases (P = .066). We also examined whether aberrant silencing of SMAD1 was more common in patients with wild-type GNA13 than in patients with mutations in this important downstream mediator of S1PR2 signaling. Thirteen of 75 analyzed patients (17.3%) exhibited GNA13 mutations. Among GNA13 wild-type tumors, 7 of 62 were positive for SMAD1 (11.2%), whereas 0 of 13 GNA13-mutant tumors were SMAD1-positive (0%). The difference was not statistically significant (P = .25). In conclusion, SMAD1 expression is aberrantly silenced in a large majority of GCB and non-GCB DLBCL cases, which does not appear to be (inversely) correlated with inactivating mutations in the S1PR2/Gα13 signaling pathway.
Discussion
A growing body of evidence implicates S1PR2 as a novel, and uniformly important, tumor suppressor that is mutationally or transcriptionally inactivated in both major subtypes of DLBCL. In the GC B-cell subtype, the S1PR2 locus, as well as genes encoding downstream components of the signaling pathway such as GNA13, are subject to recurrent mutational inactivation.13 In the ABC subtype arising from post-GC plasmablasts, S1PR2 is repressed because of high FOXP1 expression (Figure 7), with FOXP1 acting as a direct negative regulator of S1PR2 expression by binding to 2 elements in the promoter region of the S1PR2 gene.10 In this study, we uncover a third and very common mechanism of S1PR2 downregulation that affects >85% of our patient cohort and the majority of examined DLBCL cell lines and involves the aberrant silencing of SMAD1. In SMAD1-expressing DLBCL cells, SMAD1 functions alongside other components of the noncanonical SMAD1/5/9 signaling complex in relaying signals from the type II TGF-β receptor upon binding of its ligand TGF-β (Figure 7). Whereas the receptor itself is universally expressed on all examined cell lines in our panel and appears to not be the limiting factor restricting signaling activity of the pathway, SMAD1 expression is absent in the majority of our cell lines (and of our cohorts), possibly reflecting its transcriptional silencing by DNA methylation. The knockdown of SMAD1 by RNA interference, or the CRISPR-mediated editing and inactivation of the SMAD1 gene in SMAD1-positive DLBCL cells, both inhibit TGF-β−dependent S1PR2 expression, indicating that SMAD1-negative cases of DLBCL likely are unresponsive to the pro-apoptotic activities of this cytokine in vivo. We indeed found in various xenotransplantation settings and under cell culture conditions, that SMAD1 and TGF-βR2 are not only required for S1PR2 expression, but also for the induction of apoptosis by TGF-β, which in turn was dependent on S1PR2 as judged by the pro-apoptotic response of S1PR2-proficient but not S1PR2-deficient DLBCL clones to TGF-β. DLBCL clones lacking SMAD1, or TGF-βR2, or S1PR2 expression all have a growth advantage over wild-type clones in vitro and in vivo. This effect was particularly evident in a novel model of orthotopic DLBCL growth that allows for efficient engraftment of various cell lines, including the SU-DHL-6 used here, in the spleen and bone marrow of MISTRG mice. The new model overcomes the current limitations in DLBCL research by providing a versatile, practical, fast, and readily available model for in vivo work that, especially in combination with CRISPR manipulation of the transplanted cell lines, lends itself to multiple applications, including drug testing.
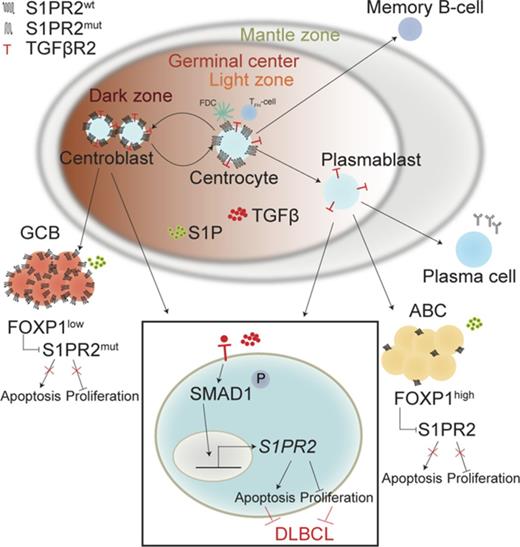
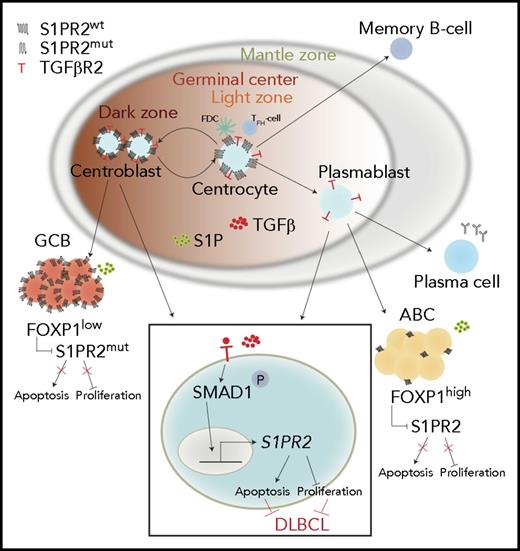
Schematic summarizing the tumor-suppressive properties of the TGF-β/SMAD1/S1PR2 axis in DLBCL. Under physiological conditions, centrocytes and centroblasts express large amounts of S1PR2, which promotes GC confinement because of a gradient of S1P that increases in concentration toward the borders of the GC and leads to apoptosis in GC cells that attempt to exit the GC. In DLBCL, S1PR2 is either mutated (in the GCB subtype) or transcriptionally downregulated by FOXP1 (in the ABC subtype). Loss of S1PR2 thus is an early initiating event in both major subtypes of DLBCL. The expression of S1PR2 is further regulated by TGF-β, which binds to its receptor TGF-βR2 and activates SMAD1 phosphorylation and nuclear translocation. p-SMAD1 binds directly to regulatory elements in the S1PR2 promoter and activates S1PR2 expression; most cases of DLBCL exhibit aberrantly low or absent expression of SMAD1.
Schematic summarizing the tumor-suppressive properties of the TGF-β/SMAD1/S1PR2 axis in DLBCL. Under physiological conditions, centrocytes and centroblasts express large amounts of S1PR2, which promotes GC confinement because of a gradient of S1P that increases in concentration toward the borders of the GC and leads to apoptosis in GC cells that attempt to exit the GC. In DLBCL, S1PR2 is either mutated (in the GCB subtype) or transcriptionally downregulated by FOXP1 (in the ABC subtype). Loss of S1PR2 thus is an early initiating event in both major subtypes of DLBCL. The expression of S1PR2 is further regulated by TGF-β, which binds to its receptor TGF-βR2 and activates SMAD1 phosphorylation and nuclear translocation. p-SMAD1 binds directly to regulatory elements in the S1PR2 promoter and activates S1PR2 expression; most cases of DLBCL exhibit aberrantly low or absent expression of SMAD1.
The observation that TGF-β activates noncanonical SMAD1/5/9 signaling via TGF-βR2 in normal B cells and in various other cell types is not new,23-27 and some evidence is available for the concept that this process is impaired in malignant B cells. For example, in Burkitt lymphoma and in Epstein-Barr virus–transformed B-lymphoblastoid cell lines, reduced TGF-βR2 expression renders cells partially resistant to TGF-β.28 Point mutations in TGF-βR2 were found to inhibit its tumor suppressive activities in cutaneous T-cell lymphoma.29 Furthermore, TGF-βR2 expression has been proposed as a positive prognostic factor in DLBCL patients,30 an observation that fits very well with our model. Large-scale screening and drug testing efforts have identified the TGF-β signaling pathway as a promising target in DLBCL that may be exploited for chemosensitization purposes.31-33 Our results thus provide a mechanistic link between 2 previously known or suspected tumor suppressive pathways frequently inactivated in DLBCL: 1 involving aberrant inactivation of S1PR2 signaling and its pro-apoptotic activities and the other involving the TGF-β signaling pathway. SMAD1 links the 2 pathways by directly binding to the S1PR2 promoter once it is phosphorylated by ligand-bound TGF-βR2 and has translocated to the nucleus.
Mechanistic evidence from animal models, presented here and in 2 previous studies,34,35 suggests that S1PR2 inactivation represents an early event in DLBCL initiation. The loss of only 1 functional allele of S1pr2 was sufficient to predispose mice to hyperproliferation of the GC compartment upon immunization, a phenotype that was recapitulated by the loss of 1 or both copies of Tgfbr2. Both centrocytes and centroblasts were more numerous in the absence of S1pr2 or Tgfbr2 and their overrepresentation in splenocyte preparations could be linked to larger rather than more numerous GCs. The fact that there is no reliable S1PR2 antibody available did not allow us to confirm the decreased S1PR2 expression in heterozygous knockout mice; nevertheless, the clear phenotype of these mice strongly suggests that the mice have a lower receptor expression upon loss of 1 copy of S1pr2. That S1PR2 plays an important role in GC confinement has been shown previously35 and has been linked to a gradient of S1P within the GC that prevents GC cells from prematurely exiting the compartment. Our data suggest that an additional layer of regulation exists that involves TGF-β acting further upstream within GC B cells to control S1PR2 expression. This model is supported by the observations that tonsillar centrocytes, but not centroblasts, express high levels of SMAD1, and that the S1PR2 repressor FOXP1 is downregulated in the GC B-cell compartment.10
Several lines of evidence suggest that S1PR2 acts cell-intrinsically in tumor B cells, rather than in tumor-infiltrating nonmalignant cells to promote cell death (for example, in response to TGF-β) and/or to limit the proliferative capacities of normal and malignant B cells. This model is supported by the hyperproliferation of S1PR2-deficient DLBCL cells in vitro and in vivo, by the fact that MYC-expressing murine lymphoma cells, injected IV into wild-type recipient mice, form larger tumors in lymph nodes and spleen if they lack 1 copy of S1pr2, and by our previous observation that forced expression of S1PR2 reduces tumor size in established lymphomas.10 Patients with low S1PR2 expression have a significantly worse prognosis than patients with high S1PR2 expression, and the benefits of high S1PR2 expression are especially obvious in combination with low FOXP1 expression.10 In summary, we show here for the first time that 2 major tumor suppressive pathways in DLBCL intersect at the level of the transcription factor SMAD1, which mediates pro-apoptotic TGF-β signaling by binding directly to the S1PR2 promoter and thereby activating S1PR2 expression, a process that is particularly efficient in the absence of the repressor FOXP1 (Figure 7). In accordance with its critical role in TGF-β–driven apoptosis, SMAD1 expression is aberrantly low or absent in a large majority of DLBCL patients. The combined results reveal an important novel tumor suppressive function of the TGF-β/TGF-βR2/SMAD1/S1PR2 axis in DLBCL and demonstrate for the first time that DLBCL cells have evolved to inactivate this cell death-promoting pathway at the level of SMAD1 expression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Michael Flori, Cheuk Ting Wu, Maries van den Broek, and all consortium members of the Clinical Research Priority Program for support and helpful discussions.
This study was funded by the Swiss Cancer Leagues grants KLS-3612-02-2015 and KFS-4120-02-2017 (A.M.) and the Clinical Research Priority Program “Human Hemato-lymphatic Diseases” of the University of Zurich.
Authorship
Contribution: A.S. designed, performed, and analyzed most of the experiments, and cowrote the manuscript; H.H. and K.B. helped with experiments and provided critical tools and advice; A.T. analyzed patient cohorts; M.G.M. provided critical tools and intellectual input; and A.M. supervised the study and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anne Müller, Institute of Molecular Cancer Research, University of Zürich, Winterthurerstr 180, 8057 Zürich, Switzerland; e-mail: mueller@imcr.uzh.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal