Abstract
The National Cancer Institute Genomic Data Commons (GDC) is an information system for storing, analyzing, and sharing genomic and clinical data from patients with cancer. The recent high-throughput sequencing of cancer genomes and transcriptomes has produced a big data problem that precludes many cancer biologists and oncologists from gleaning knowledge from these data regarding the nature of malignant processes and the relationship between tumor genomic profiles and treatment response. The GDC aims to democratize access to cancer genomic data and to foster the sharing of these data to promote precision medicine approaches to the diagnosis and treatment of cancer.
Scope and vision of the National Cancer Institute Genomic Data Commons
Precision medicine, as conceived by the cancer genomics community, uses a precise knowledge of the structure and activity of a patient’s tumor genome to suggest particular therapies targeting oncogenic mechanisms in the tumor, thereby yielding meaningful therapeutic responses. The hematologic malignancies, for example, are extraordinarily diverse in clinical presentation, histology, karyotype, and genomic abnormalities. Subdivision of these cancers into molecularly defined entities that have characteristic oncogenic vulnerabilities is crucial for the development of individually tailored approaches to treatment. Significant progress toward this goal has been and is being made by exploiting the data collection and analysis efforts of large cancer genomics consortia such as The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/) and the International Cancer Genomics Consortium (https://icgc.org/). To characterize the genomes of thousands of cancers across the range of cancer histologies, these and similar programs have marshaled the expertise and resources of research institutions, cancer centers, and private companies in effective and essential team science efforts. The massive scale of these programs has been necessary to power investigations appropriately, to reveal statistically significant variability in genetic alterations, and to glean biological insights into the nature of malignant processes. These studies have implicated many new genes and molecular pathways in blood and many other cancer types by identifying recurrent genetic variants that drive cancer establishment, persistence, and growth.1
Genomic studies are beginning to influence translational cancer research. Therapeutic targets that were identified by using genomics have been defined in diffuse large B-cell lymphoma (DLBCL),2 acute myeloid leukemia,3 myelodysplastic syndrome,4 and acute lymphoblastic leukemia (ALL),5 and they are being explored with encouraging results, as we discuss below. In general, however, unlocking the knowledge within massive genomic data sets is daunting to most cancer biologists. The sheer size of the data collection makes it impractical or impossible for investigators to interrogate the data without access to high-performance computing resources. The panoply of data formats and technical details of a myriad of bioinformatics applications and analytical pipelines creates a high barrier for researchers who have creative hypotheses but do not have a dedicated team of expert bioinformaticians. These are some of the reasons why the Blue Ribbon Panel report6 for the United States Cancer Moonshot recommended the development of a national cancer data ecosystem to enable one-stop, free access for researchers, physicians, and diverse patient populations so they can share data on cancer and fuel faster progress.
The National Cancer Institute (NCI) Genomic Data Commons (GDC) is major step toward realizing that ecosystem for the benefit of both US and international cancer investigators. The GDC was launched in June 2016 as part of then President Obama’s Precision Medicine Initiative after 2 years of initial development. Initially, the GDC consolidated all clinical and genomic data from TCGA and other NCI programs, making these data available for search and download via a new portal and application program interface (API). The GDC mandate also includes the ability to accept data from any cancer genomic project worldwide that can be shared broadly with qualified researchers. The GDC will harmonize both the genomic and clinical data across programs and projects to the greatest extent possible and will enable cancer biologists and oncologists to visualize and analyze the integrated data. Ultimately, the GDC seeks to become a cancer genomics knowledge base that can inspire new avenues of integrative cancer research and provide a foundation for independent, novel bioinformatic investigation and applications.
Many worthy cancer data-sharing initiatives are underway that aspire to promote precision medicine approaches to cancer treatments (reviewed in Siu et al7 ). Rather than competing with these initiatives, the GDC is a complementary system that can play a role in their mutual integration. However, we believe the GDC is unique among these systems in that it is designed from the ground up to serve both large NCI-managed consortia and individual cancer genomics researchers. GDC exists in large part to help individual investigators and small programs by providing a permanent home for their data that also meets National Institutes of Health (NIH)8 and NCI9 genomic data sharing requirements. Laboratories without extensive bioinformatics infrastructure can take advantage of GDC’s standard processing pipeline, which generates genome alignments (hg38-based), digital gene expression, and somatic mutation calls using a suite of state-of-the-art algorithms. These bioinformatics pipelines are run on all data submitted to the GDC, and the results are made available, at no cost, to the submitters as soon as the pipeline is complete.
The GDC also acts as the long-term data steward and central source for all NCI-managed cancer genomics projects. It currently houses several large genomic projects, including TCGA, Therapeutically Applicable Research to Generate Effective Treatments (TARGET), and the Cancer Cell Line Encyclopedia. These studies serve as a starting point for many cancer investigations, and the GDC implements improved search and downloading of these data as well as in-depth browsing of derived data on the GDC Web site. Indeed, researchers are likely to prepare figures for their publications by directly using GDC’s analytic tools without ever downloading the raw genomic data to their desktop computer.
The GDC participates in and collaborates with international efforts to standardize genomic and clinical data, including the Global Alliance for Genomics and Health (http://genomicsandhealth.org/), and to adopt data formatting and interoperability standards on a rolling basis. The GDC development team consults extensively with bioinformatics, data management, and high-performance computing experts to ensure that the GDC adopts best current practices. This open and collaborative design philosophy has been instrumental in the GDC’s successful efforts to recruit important cancer data sets from outside the NCI. These include somatic mutation data for ∼18 000 cancer cases from Foundation Medicine, Inc.,10 genomic and clinical data for the Multiple Myeloma Research Foundation CoMMpass study of more than 1000 patients with multiple myeloma receiving standard-of-care treatments,11 and genomic and clinical data from ∼19 000 patients with cancer from Project GENIE.12 The bioinformatics community has also begun to embrace the GDC, developing Bioconductor13 and Python14 software packages that enable programmatic access to GDC data.
The following are major distinguishing attributes of the NCI GDC:
GDC stores raw genomic data, allowing continuous reanalysis as computation methods and genome annotations improve.
GDC uses shared bioinformatic pipelines to facilitate cross-study comparisons and integrated analysis of multiple data types.
GDC maintains clinical data in a harmonized, highly structured and extensible schema.
NCI is committed to maintaining long-term storage of cancer genomic data in the GDC with free access to researchers.
GDC enables researchers to comply with the NIH Genomic Data Sharing policy8 as well as journal requirements for data sharing.
The explanatory power of data in the GDC will grow over time as it accrues more cases, thereby promoting precision oncology.
Certain capabilities are intentionally outside the GDC’s scope of operation. For example, GDC does not provide colocated computational resources for the use of the scientific community at large. Instead, it works closely with the NCI Cloud Pilots15 that do provide such resources to supply data and support for interoperability. GDC does not provide storage or tools for collaborations working on intermediate stages of data analysis or private storage of project data. This is in keeping with the overarching GDC philosophy to promote sharing of high-quality data as broadly as possible.
Data acceptance and submission
Any investigator or consortium with cancer genomic data is welcome to apply directly to the GDC for data submission. GDC leadership reviews new project submission requests, and acceptance reflects the extent to which the data set enhances the understanding of cancer, with consideration for the number of patients in the project, the quality of the data (eg, sequence read depth and coverage), and the extent to which a data set will complement those already handled at GDC. Baseline criteria are provided on the GDC Web site,16 but these are meant to be flexible guidelines. For example, a relatively small (<100 patients) study may be accepted for submission if the molecular data are broad (cover tumor and matched normal DNA and tumor RNA sequencing [RNAseq] for all participants) and deal with a rare or understudied tumor type. Inquiries regarding data submission may be made to the GDC Support Desk (support@nci-gdc.datacommons.io).
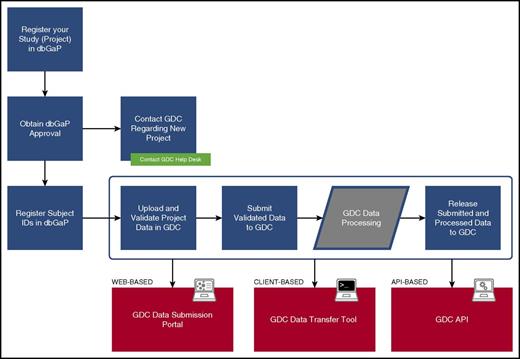
Figure 1 provides an overview of the GDC submission process. All projects housed at GDC must be registered with the National Center for Biotechnology Information database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap). This ensures that appropriate patient consent has been obtained for data sharing and that a data use committee has been duly established to handle requests for data access, as discussed below. The GDC does not participate directly in project registration. However, an introduction to the process with helpful resources is available on the GDC Web site.17
GDC data registration and submission. The diagram indicates the high-level steps an investigator takes to register and submit a cancer genomic data set to GDC. The final step “Release Submitted and Processed Data to GDC” is the point at which the investigator indicates final approval to release data to the public; data in prior steps are accessible only to GDC and the investigator.
GDC data registration and submission. The diagram indicates the high-level steps an investigator takes to register and submit a cancer genomic data set to GDC. The final step “Release Submitted and Processed Data to GDC” is the point at which the investigator indicates final approval to release data to the public; data in prior steps are accessible only to GDC and the investigator.
Data in the GDC are made publically available. Controlled access is applied to genomic sequence data as a means of protecting patient privacy, as discussed below, not to inhibit access by any legitimate researcher with appropriate data access certification. Data submitters are provided 6 months of private access in order to update data based on their own quality checks as well as on metrics generated by the GDC. After this period, the data are eligible for public release. Data submitters retain ownership of their data and may add to or update them through the GDC Data Submission Portal (https://portal.gdc.cancer.gov/submission/) at any time.
Data types accepted by GDC18 include data derived from DNA sequencing (DNAseq; exomic or genomic), RNAseq, Single Nucleotide Polymorphism 6.0 (SNP 6.0) arrays, and DNA methylation arrays, along with associated biospecimen and clinical data. Data from targeted sequencing of gene panels and derived data from mutation calling pipelines may be considered for acceptance on a case-by-case basis.
Data harmonization
Owing to the rapid expansion of next generation DNAseq and RNAseq, bioinformatics methods have been developed as needed and in parallel, leading to a vexing diversity in results derived from the same raw sequencing data. The GDC currently uses state-of-the-art bioinformatics pipelines that have been honed in the TCGA and International Cancer Genomics Consortium efforts, but over time will integrate new methods that provide a substantial improvement in either accuracy or efficiency.
Equally challenging are disparities in clinical annotations that arise from the use of different data dictionaries and clinical study designs. The GDC aims to harmonize the genomic and clinical data from independently conducted genomic studies.
Molecular harmonization
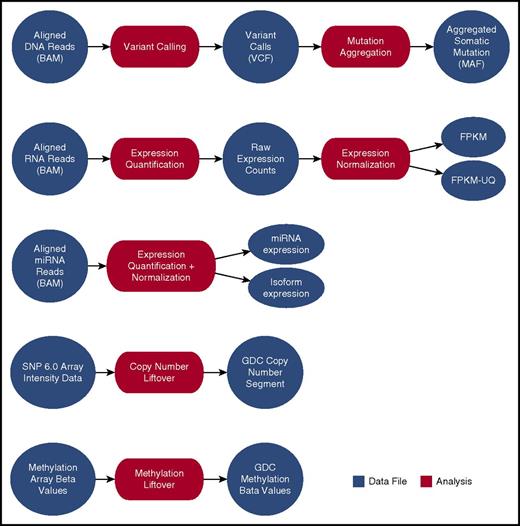
Harmonization of sequencing data (DNAseq and RNAseq) begins with a complete realignment of submitted sequencing reads to the current human genome build (GRCh38). The GDC makes the realigned sequencing data available for download by using high-performance Internet transfer protocols. The GDC uses its genome alignments to derive and distribute qualitative and quantitative descriptions of mutations and digital gene expression (Figure 2).
Current GDC bioinformatics pipelines. High-level processes indicating the GDC analysis and resulting data products created from GDC hg38-aligned data, submitted SNP 6.0 array data, or submitted methylation array data. BAM, binary alignment map; FPKM, fragments per kilobase of transcript per million mapped reads; FPKM-UQ, FPKM-upper quartile; MAF, mutation annotation format; miRNA, microRNA; VCF, variant calling format.
Current GDC bioinformatics pipelines. High-level processes indicating the GDC analysis and resulting data products created from GDC hg38-aligned data, submitted SNP 6.0 array data, or submitted methylation array data. BAM, binary alignment map; FPKM, fragments per kilobase of transcript per million mapped reads; FPKM-UQ, FPKM-upper quartile; MAF, mutation annotation format; miRNA, microRNA; VCF, variant calling format.
Since various bioinformatic methods for identifying somatic mutations differ in their sensitivity and specificity,19 the GDC currently provides the output of 4 different mutation callers: MuSE,20 Mutect2,21 SomaticSniper,22 and VarScan2.23 A fuller discussion of the rationale behind GDC mutation calling and reporting can be found on the GDC Web site.24
Other genomic data that are critical to cancer research include analyses of DNA methylation, copy number alterations (CNAs), alternative messenger RNA splicing, and fusion gene calling. Pipelines to generate derived data for methylation arrays and for SNP 6.0-array based CNA data based on human genome build hg38 are currently in place. Alternative splicing and fusion gene pipelines are under development and should be available in late 2017. Previously released versions of these derived data types based on human genome build hg19 are available for the TCGA and TARGET projects in the GDC Legacy Archive (https://portal.gdc.cancer.gov/legacy-archive/search/f).
The GDC chooses its bioinformatics pipelines in consultation with its external Bioinformatics Advisory and Steering Committees and receives ongoing input from investigators in the NCI Genomic Data Analysis Network.25 The GDC pipelines are standardized, but the GDC does not purport that its variant calls are standard. Indeed, future releases of data from the GDC may provide updated versions of the derived genomic data based on improvements in bioinformatics methods.
Clinical harmonization
High-quality clinical data associated with cancer cases is crucial to both the basic science and translational aspirations of the GDC. At the same time, the encoding of large amounts of legacy clinical data can be a burden that can discourage submission of useful and important investigations. This essential tension between the needs of data users and those of data submitters will remain with us for some time to come. The GDC attempts to chart a middle course.
Submitters are required to submit only 3 clinical fields for each case: age, sex, and diagnosis. A growing set of optional clinical fields has been cataloged by GDC, initially standardized by referencing elements in the NCI Cancer Data Standards Registry and Repository (https://cdebrowser.nci.nih.gov). New project submitters work directly with GDC user support and clinical experts to map their clinical fields and values into GDC standards. New clinical fields can be added to the GDC as needed for particular data sets. Additional clinical data can be accepted as clinical supplements, allowing bulk, unmapped information to be deposited and made available to users. Over time, GDC will map bulk clinical information to standard vocabularies and allow users to have finer control of clinical data searches and filters.
Patient privacy and controlled data
In keeping with the research-oriented mission of the GDC, it does not intend to house or distribute electronic health records or any other data that are considered personally identifiable information (PII). Submitters are informed of this policy, and the GDC takes pains to ensure that absolute dates, identifiers such as Social Security numbers, addresses, and other PII as described in the Health and Human Services Safe Harbor guidelines26 are not present in GDC clinical data.
Although genomic sequence data are not considered PII per se, the GDC nonetheless controls access to raw patient sequence (DNA and RNA) data to provide appropriate safeguards against attempts to re-identify research subjects. GDC data users interested in obtaining controlled data for research purposes must apply for access to the desired data via dbGaP. There, access requests are managed by an NIH data access committee and are not influenced by the GDC. Access to GDC data through dbGaP entails signing a Data Use Agreement that is established by the data owners in consultation with their own institutional review boards.
Aggregated analysis of controlled access data is expected and permitted in publications. Examples would include the frequency of particular genetic variants within cancer types or the relationship between genetic variants and clinical outcome. The NIH Data Access Committee that manages a particular dbGAP project can be consulted regarding acceptable presentation of GDC controlled access data in publications.
Once a user has received his or her data use certificate, the GDC provides seamless access to the relevant data. The NIH eRA Commons system (https://public.era.nih.gov/commons) is used to provide authentication and authorization.
GDC applications and user resources
The GDC provides a repository of data that users may search and access and a suite of tools that enable users to explore and analyze data in the context of their own hypotheses. As a new and evolving system, GDC currently provides a core set of data search and download capabilities; dynamic data visualization and analysis tools are under development and are targeted for release in June 2017. Graphical user interfaces to all GDC applications are available via the GDC Data Portal (https://gdc-portal.nci.nih.gov/).
All functionality available via the Web interface can be accessed programmatically by using the GDC API.27 The API enables engineering teams and technically adept users to automate access to GDC, develop novel value-added software, and obtain fine-grained control over searches and downloads. GDC Web-based interfaces themselves are built upon calls to the standard GDC API.
Data search and download
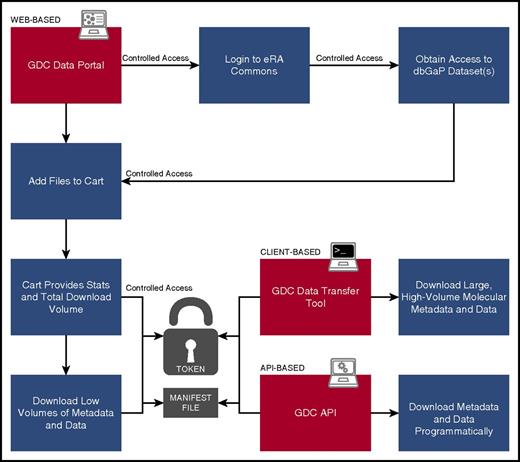
Harmonized clinical, biospecimen, and molecular data are available from the GDC Data Portal (https://gdc-portal.nci.nih.gov/search/s). Figure 3 presents an overview of typical user interaction with the Portal. A faceted search interface allows the user to filter available data files according to primary disease site, disease type, demographic metadata, analysis platform, data type, data format, and access level (ie, open or controlled access). Numbers of files that meet the search criteria are displayed either in a pie chart overview or in tabular format as desired. Charts are clickable and enable the user to further reduce the file set to desired specifications. An advanced search input allows the user to refine the search criteria manually to a very granular level. The advanced search criteria change dynamically as the user chooses facets and facet values, so the user may copy and save any query for sharing or inputting later. The search interface indicates the access level of files in the filtered set. Open access files may be downloaded by any user without logging in. Controlled access data may be downloaded by users who have received dbGaP authorization for the program under which the data were gathered, as described in the “Patient privacy and controlled data” section. Authorized users log in using their eRA Commons username and password.
User workflow. Diagram indicating user steps to authenticate and download GDC data. Red panels indicate the 3 means for accessing data: the Web-based Data Portal, the standalone Data Transfer Tools, and the programmatic API. “Token” is a short text file provided to an authenticated user that acts like a password to enable secure transfer of authorized controlled data, such as sequence alignments.
User workflow. Diagram indicating user steps to authenticate and download GDC data. Red panels indicate the 3 means for accessing data: the Web-based Data Portal, the standalone Data Transfer Tools, and the programmatic API. “Token” is a short text file provided to an authenticated user that acts like a password to enable secure transfer of authorized controlled data, such as sequence alignments.
The user downloads files by using a familiar shopping cart mechanism. Files are added to the cart while the search is underway. Users proceed to the cart screen when they are ready to download. Downloads may be performed in the browser itself or via a separate application provided by the GDC, the Data Transfer Tool (DTT).28 Browser download is impractical for large files (eg, Binary Alignment Map [BAM] alignments) or large numbers of files; in this situation, the user is directed to use the DTT. The DTT is a standalone command-line application run on the user’s own machine. It accepts GDC file identification (IDs) or a file of such IDs (called a manifest), along with a user’s authorization token (for controlled data downloads). The DTT establishes multiple parallel hypertext transfer protocol connections to download large amounts of data and is able to retry interrupted downloads automatically. Users can resume aborted connections by simply re-running the application. A graphical user interface for the DTT is nearing completion and will further simplify the download process.
Analysis and visualization
The GDC currently implements its own version of the cBioPortal29 for the exploration of somatic mutations in TCGA data. Users can examine the mutation calls in custom gene sets within each project and for each GDC mutation caller.
GDC cBioPortal will soon (around June 2017) be superseded by the GDC Data Analysis, Visualization, and Exploration (DAVE) tools, currently under development. GDC DAVE will enable users to select their own sets of cases, within or across programs or projects, to view and analyze. The case sets, referred to as “cohorts,” may be viewed in a gene context or a mutation context. In the gene context, genes that are somatically mutated may be listed and ordered by case mutation frequency. A survival plot function will enable a Kaplan-Meier comparison of survival between mutated and wild-type cases within the cohort for any mutated gene. Similar functionality for individual mutations within genes will be provided in the mutation context.
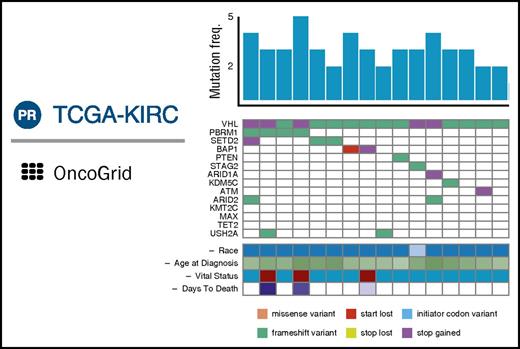
GDC DAVE will also include OncoGrid (Figure 4), an enhanced and highly interactive implementation of the cBioPortal Oncoprint graph. OncoGrid will enable users to visualize patterns in the effects of mutations in highly mutated genes over all cohort cases. Users can add to the plot clinical data and other tracks that vary by case and sort the columns by the values within tracks. By manipulating tracks and case sorting interactively, users will be able to use visual clustering to seek associations between genes and case data. GDC DAVE further allows users to visualize the position of mutations within the protein structure using standard lollipop plots. Curated Catalogue of Somatic Mutations in Cancer (COSMIC) 30 mutation annotations will be available in DAVE when it is released.
Composite of principal features of GDC OncoGrid. Each column represents a single case; the histogram indicates total number of somatic variants in the case. Rows are genes; colored cells indicate types of variants (colored according to the legend) present in these genes for the given case’s tumor sample. Clinical data for each case are presented in analogous fashion. freq., frequency; TCGA-KIRC, Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma.
Composite of principal features of GDC OncoGrid. Each column represents a single case; the histogram indicates total number of somatic variants in the case. Rows are genes; colored cells indicate types of variants (colored according to the legend) present in these genes for the given case’s tumor sample. Clinical data for each case are presented in analogous fashion. freq., frequency; TCGA-KIRC, Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma.
Documentation and support
The GDC Web site (https://gdc.cancer.gov/) is the main entry point to the GDC system. It contains a comprehensive set of GDC descriptions, overviews, how-to instructions, articles, frequently asked questions, and news items. Users can follow different tracks (data use, data submission, software development) to quickly get up to speed on their topics of interest.
The GDC documentation site (https://docs.gdc.cancer.gov/) offers in-depth user guides to the primary GDC applications, including the Data Portal, Data Submission Portal, DTT, and API. The documentation site also includes the GDC Data Dictionary viewer, which describes GDC metadata fields and values in detail and provides downloadable templates for the convenience of data submitters.
The GDC has a responsive user support team. Users who have issues with GDC applications or questions about data submission or any other aspect of the GDC should e-mail support@nci-gdc.datacommons.io for assistance.
Hematologic cancer data available in GDC
The GDC houses a growing complement of cancer genomic data related to hematologic cancers, highlighted in Table 1. Beyond these NCI-sponsored projects, the Multiple Myeloma Research Foundation recently announced its plan to deposit data from its CoMMpass11 study in the GDC. This submission includes genomic and clinical data from myeloma samples obtained at regular follow-up intervals from ∼1000 patients receiving standard-of-care treatment. In addition, exome sequencing data from more than 500 patients with various types of lymphoma and leukemia are currently accessible through the dbGaP system and could be migrated to the GDC.
Conclusion: the road ahead
In several hematologic cancers, tumor genomic analyses are beginning to yield potential precision medicine targets and associated agents with promising effects. Gene expression profiling studies of DLBCL have identified the ABC molecular subtype as responsive to the Bruton’s tyrosine kinase inhibitor ibrutinib.2 In acute myeloid leukemia and myelodysplastic syndrome, patients with IDH2 mutations may respond to a specific inhibitor of this enzyme, AG-221.4 Pediatric ALL is one of the most genomically characterized hematologic malignancies, with more than 12 molecular subtypes defined by using combined analysis of chromosomal rearrangements, CNAs, and mutations.5 Although clinical trials that aim to capitalize on this molecular taxonomy are in progress, case studies suggest that knowledge of particular oncogenic abnormalities in ALL can be used to match patients with targeted therapies. For example, a boy with B-cell ALL harboring a translocation involving the EBF1 and PDGFRB genes had a complete response to imatinib, an inhibitor of the platelet-derived growth factor receptor encoded by PDGFRB.31 An important point to emphasize is that the prevalence of each molecular subtype of ALL can range from 28% of cases to as few as 1% of cases. Moreover, it is conceivable that further molecular heterogeneity exists within the currently defined subtypes of ALL that could influence the response to therapy.
From this perspective, a national or international effort needs to be mounted to analyze each subtype of hematologic malignancy at higher case numbers and sequence depth. The number of cancer cases that need to be sequenced to identify recurrent somatic mutations with a 2% prevalence in a cancer subtype depends on the background mutation rate of the subtype.32 For chronic lymphocytic leukemia (CLL), this threshold is predicted to be ∼700 cases, meaning that the >1000 published exomes and genomes from CLL are likely to provide power to discern rare CLL subtypes.33,34 However, for DLBCL, the 2% threshold is at ∼2000 cases because of its higher background mutation rate, indicating that there are substantial discoveries of driver genetic events in DLBCL to be made with additional sequencing. Understanding the molecular consequences of a rare cancer driver event can be the most important information for a patient with that driver event because it may point to optimal targeted therapy, as in the EBF1-PDGFRB–translocated ALL example highlighted above.
We believe that the GDC can be an important tool to enable such efforts to comprehensively profile hematologic malignancies. If and when genomic analysis becomes part of routine clinical care, the GDC can also accept donations of genomic information from willing patients with cancer. In this way, the GDC will allow us to learn from each patient’s experience with cancer, ever refining the molecular taxonomy of cancer and its utility in making treatment decisions.
Acknowledgment
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and has been funded in whole, or in part, by the National Institutes of Health, National Cancer Institute (HHSN261200800001E).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Authorship
Contribution: M.A.J. and L.M.S. wrote the article; V.F. and R.L.G. edited the article; and all authors contributed to the development of the National Cancer Institute Genomic Data Commons.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Louis M. Staudt, National Cancer Institute, Building 10, Room 4N115, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: lstaudt@mail.nih.gov.