Key Points
Critical roles for dynamic translational control during terminal erythroid differentiation.
RBM38 can regulate translation during terminal erythropoiesis.
Abstract
Cell development requires tight yet dynamic control of protein production. Here, we use parallel RNA and ribosome profiling to study translational regulatory dynamics during murine terminal erythropoiesis. Our results uncover pervasive translational control of protein synthesis, with widespread alternative translation initiation and termination, robust discrimination of long noncoding from micropeptide-encoding RNAs, and dynamic use of upstream open reading frames. Further, we identify hundreds of messenger RNAs (mRNAs) whose translation efficiency is dynamically controlled during erythropoiesis and that enrich for target sites of RNA-binding proteins that are specific to hematopoietic cells, thus unraveling potential regulators of erythroid translational programs. A major such program involves enhanced decoding of specific mRNAs that are depleted in terminally differentiating/enucleating cells with decreasing transcriptional capacity. We find that RBM38, an erythroid-specific RNA-binding protein previously implicated in splicing, interacts with the general translation initiation factor eIF4G and promotes translation of a subset of these irreplaceable mRNAs. Inhibition of RBM38 compromises translation in erythroblasts and impairs their maturation, highlighting a key function for this protein during erythropoiesis. These findings thus reveal critical roles for dynamic translational control in supporting specialized mammalian cell formation.
Introduction
Decoding of transcriptome information by ribosomes is a key step in controlling cell differentiation.1 Translation is tightly regulated in response to developmental and environmental cues, and the rate of translation initiation, elongation, and termination at individual messenger RNAs (mRNAs) can be tuned to control protein synthesis, folding, and localization.2,3 Ribosome profiling, the sequencing of ribosome-protected mRNA fragments, enables systematic analysis of the complexity and regulation of ribosome decoding.4 Ribosome profiling studies have documented widespread translation of micropeptides and unanticipated protein isoforms, as well as extensive variation in mRNA translation efficiencies. However, how these processes respond to transcriptome dynamics during cell differentiation is poorly understood.
Erythropoiesis represents an attractive model for the study of translational regulatory dynamics during cell differentiation. Erythroid cells are particularly sensitive to disturbances in translational mechanisms; for example, mutations affecting the production of various ribosomal proteins underlie anemias that cause bone marrow failure.5,6 Moreover, translational control is uniquely vital in enucleated reticulocytes, as they require ongoing protein synthesis but are transcriptionally inactive.
Here, we use parallel RNA and ribosome profiling to comprehensively characterize translational control during mouse fetal liver erythroid differentiation. The resulting translational landscape of erythropoiesis reveals precise yet dynamic translational control of protein synthesis. Ribosomes accurately distinguish between noncoding and micropeptide-encoding long RNAs and enhance proteome diversity via alternative translation initiation and termination, while upstream open reading frames (uORFs) function dynamically to lessen translation of developmentally regulated factors such as TAL1 and BCL11A. We further uncover hundreds of mRNAs with dynamic translation efficiencies during erythropoiesis. The untranslated regions (UTRs) of these mRNAs enrich for target sites of RNA-binding proteins that are specifically enriched in hematopoietic cells, thus implicating these proteins in erythroid translational regulatory programs. We functionally characterize one such protein, RBM38, which is specifically induced in late-differentiating erythroblasts by GATA1/TAL1 and has been linked to splicing during late erythropoiesis. We find that RBM38 associates with the translation initiation factor eIF4G and can promote translation of select mRNAs with decreasing mRNA levels in terminally differentiating/enucleating cells. Inhibiting Rbm38 confers a translation defect and blocks reticulocyte generation, arguing for a critical role of RBM38 during erythropoiesis. Together, these findings illustrate how developing cells exploit translational control to expand and remodel their proteomes and reveal how tissue-specific factors can tune translation to support the formation of functionally specialized cells.
Methods
Cell isolation, culture, and flow cytometry
RNA and ribosome profiling
Luciferase assays
The Dual-Luciferase Reporter Assay System (Promega) was used by following the provided protocol. Plasmids were transfected into K562 cells by using Lipofectamine LTX (Life Technologies), and the ratio of firefly to Renilla luciferase activity was measured 30 hours after transfection. Tethering experiments were performed as described previously.8
Polysome assays
Polysome analysis and RNA quantification were conducted as described previously.8
Protein assays
Antibodies against the proteins RBM38 (Santa Cruz sc-365898), GAPDH (Santa Cruz sc-32233), eIF4G (Santa Cruz sc-11373), eIF4E (Santa Cruz sc-9976), and HA (Sigma H9658) were used. Immunoprecipitation experiments were performed as described previously.8
Data analysis
Data analysis details can be found in the supplemental Methods (available on the Blood Web site). RNA and ribosome profiling data have been deposited in the Gene Expression Omnibus (accession number GSE83823).
Results
Global translation profiling during red blood cell development
We investigated translational dynamics by using terminal differentiation of primary erythroid progenitors in culture as a model. Erythroid progenitors were purified from E14.5 mouse fetal livers and cultured 48 hours in erythropoietin-containing media to induce terminal proliferation and differentiation,12 modeling terminal in vivo erythropoiesis.8,13 We collected cells at 0, 24, 33, and 48 hours after differentiation, as these represent cells at different stages of late erythropoiesis, with colony-forming unit and proerythroblasts constituting more than 95% of cells at 0 hours, and with enucleated reticulocytes and orthochromatic erythroblasts composing more than 80% of cells at 48 hours (supplemental Figure 1).12,13
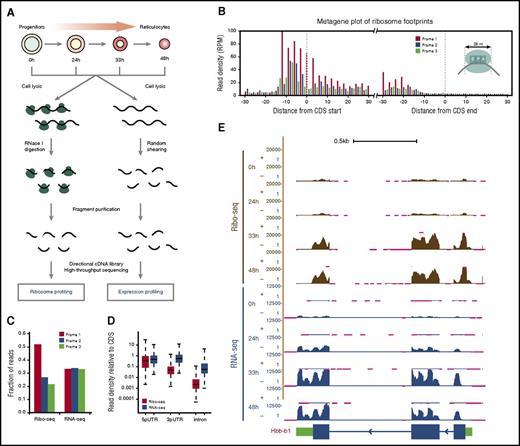
To profile gene expression and translation in parallel, we split samples and subjected each fraction to either RNA profiling (RNA-seq) or ribosome profiling (Ribo-seq). For RNA-seq, directional sequencing libraries were generated from randomly fragmented RNA, whereas for Ribo-seq they were generated from RNA fragments protected from nuclease digestion by ribosomes (Figure 1A). The resulting strand-specific RNA-seq and Ribo-seq reads were then aligned to a comprehensive erythroid differentiation transcriptome,10 with more than 0.5 billion reads mapped in total (supplemental Table 1). Our ribosome profiling data sets are highly reproducible (Pearson r = 0.87-0.99; P < 10−15 for all comparisons, Fisher’s exact test) and capture elemental features of translation with single-nucleotide resolution (supplemental Figure 2A). Ribosome footprints (RFPs) accurately delineate known coding sequences (CDSs) and their exons, with 12-nt and 15-nt offsets upstream of CDS starts and ends reflecting known distances from RFP 5′ termini to the P- and A-site codons, respectively, and their density is not biased by CDS length (Figure 1B; supplemental Figure 2B-D).14 Crucially, Ribo-seq but not RNA-seq reads show 3-nt codon periodicity and are highly specific to CDS regions (Figure 1C-D). These data enable global analysis of translation during erythropoiesis.
Global translation profiling during red blood cell development. (A) Workflow for parallel ribosome and RNA profiling during erythropoiesis. (B) Ribosome footprints (RFPs) delineate known CDSs. Metagene plot shows the rise and fall in 28-nt RFP density (reads per million mapped reads, RPM) near starts and ends of annotated CDSs, respectively. The 12-nt and 15-nt offsets from starts and ends reflect distances from RFP 5′ termini to the ribosome P- and A-site codons at translation initiation and termination, respectively (see inset). (C) Subcodon resolution of ribosome footprints. Note that 3-nt codon periodicity relative to the known CDS is seen for 28-nt RFPs but not RNA-seq reads. (D) Ribosome footprints are highly specific to coding regions. Boxplots show density of Ribo-seq and RNA-seq reads at UTRs or introns relative to that of the associated CDS. (E) Ribosome and RNA profiling of the locus encoding the β-globin–major chain (Hbb-b1). Tracks display Ribo-seq and RNA-seq signal as density of mapped strand-specific reads. The gene model with CDS (blue) and UTR (green) regions is shown at the bottom. Kb, kilobases; RNase I, ribonuclease I.
Global translation profiling during red blood cell development. (A) Workflow for parallel ribosome and RNA profiling during erythropoiesis. (B) Ribosome footprints (RFPs) delineate known CDSs. Metagene plot shows the rise and fall in 28-nt RFP density (reads per million mapped reads, RPM) near starts and ends of annotated CDSs, respectively. The 12-nt and 15-nt offsets from starts and ends reflect distances from RFP 5′ termini to the ribosome P- and A-site codons at translation initiation and termination, respectively (see inset). (C) Subcodon resolution of ribosome footprints. Note that 3-nt codon periodicity relative to the known CDS is seen for 28-nt RFPs but not RNA-seq reads. (D) Ribosome footprints are highly specific to coding regions. Boxplots show density of Ribo-seq and RNA-seq reads at UTRs or introns relative to that of the associated CDS. (E) Ribosome and RNA profiling of the locus encoding the β-globin–major chain (Hbb-b1). Tracks display Ribo-seq and RNA-seq signal as density of mapped strand-specific reads. The gene model with CDS (blue) and UTR (green) regions is shown at the bottom. Kb, kilobases; RNase I, ribonuclease I.
Empirical assessment of noncoding RNA translatability
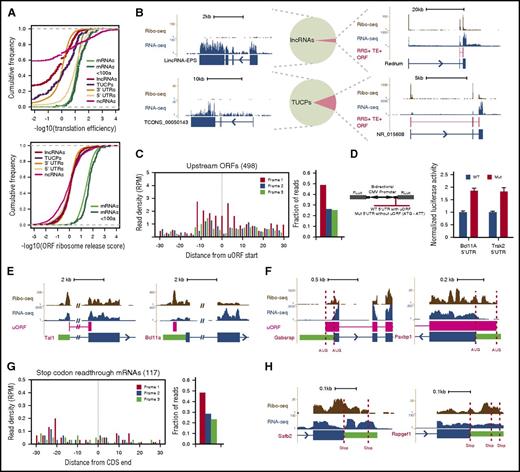
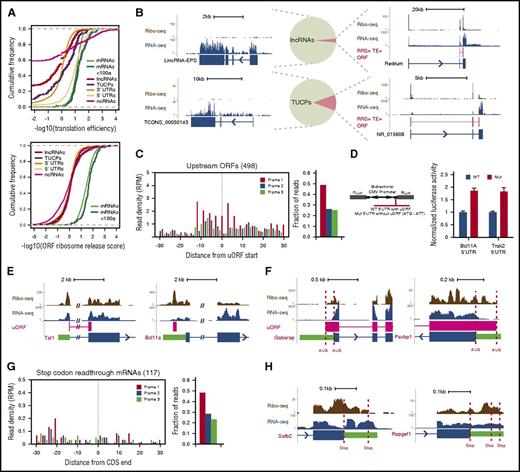
Pervasive transcription of mammalian genomes generates myriad >200 nt RNAs that must be discerned by ribosomes from bona fide mRNAs. We previously defined the landscape of long noncoding RNAs (lncRNAs) in erythroid cells, differentiating them from mRNAs and from uncharacterized transcripts with unclear coding potential (TUCPs) based on their predicted coding capacity.10 To empirically assess the translatability of these RNAs from our data, we looked for known features of translation and benchmarked them against control sets of 5′UTRs, classic noncoding RNAs (ncRNAs), and 3′UTRs. The enrichment of Ribo-seq over RNA-seq reads (translation efficiency [TE]) unambiguously separated lncRNAs, TUCPs, and 3′UTRs from mRNAs, with a median TE difference of ∼6- to 26-fold (P < 10−15 to 10−3, Student t test; Figure 2A, top), but not classic ncRNAs or 5′UTRs, as reported before.15 Conversely, measuring the expected ribosome release after encountering a stop codon by using the ribosome release score (RRS)15 robustly distinguished CDSs of any size from all other RNA classes (∼22- to 30-fold median RRS difference, all P < 10−15, Student t test; Figure 2A, bottom, and supplemental Figure 3A-C). These metrics indicate that our erythroid lncRNAs and TUCPs are predominantly noncoding. Indeed, Ribo-seq reads within lncRNAs and TUCPs are not specific to predicted ORFs, confer poor predicted ORF coverage (median, 0%), and show no codon periodicity or predicted ORF delineation (supplemental Figure 3D-H) as illustrated by lincRNA-EPS (Figure 2B, upper left), which lacks coding capacity.16 Still, a few RNAs resembled micropeptide-encoding mRNAs in their ORF features and conservation, suggesting that some may encode small peptides.
Evidence for RNA translatability, regulatory uORFs, and alternatively translated mRNAs. (A) (Top) The translation efficiency (TE) of lncRNAs and TUCPs distinguishes their translatability from that of 5′UTRs or CDSs of any size. (Bottom) The ribosome release score (RRS) across all possible ORFs separates lncRNAs and TUCPs from translated CDSs of any size. (B) Most erythroid lncRNAs and TUCPs show no evidence of translation. An empirical criterion for productive translation (supplemental Methods) classifies most lncRNAs and TUCPS, such as lincRNA-EPS and TCONS_00050143 (left panels), as noncoding. Others, such as Redrum and NR_015608 (right panels), are predicted to possess translated ORFs (depicted in red). (C) Translation of 5′UTR initiating ORFs. RFPs delineate the starts of high-confidence uORFs and exhibit 3-nt codon periodicity, evidencing their translation. (D) Validation of regulatory uORFs. Transient reporter assays (left, reporter design) evidence uORF-mediated translational control of the Bcl11A and Trak2 mRNAs (right). (E) Examples of uORFs in the Tal1 (left) and Bcl11a (right) mRNAs. (F) Examples of N-terminally extended proteins. Dashed lines mark annotated and upstream start codons. (G) Stop-codon-readthrough mRNA translation. RFPs continuing past known CDSs maintain 3-nt codon periodicity, evidencing readthrough translation. (H) Examples of C-terminally extended proteins, SAFB2 and RAPGEF1 (Rap guanine nucleotide exchange factor 1). Dashed lines mark annotated and next in-frame stop codons. CMV, cytomegalovirus; Fluc, firefly luciferase; Gabarap, GABA(A) receptor–associated protein; Mut, mutant allele; Rluc, Renilla luciferase; WT, wild-type allele.
Evidence for RNA translatability, regulatory uORFs, and alternatively translated mRNAs. (A) (Top) The translation efficiency (TE) of lncRNAs and TUCPs distinguishes their translatability from that of 5′UTRs or CDSs of any size. (Bottom) The ribosome release score (RRS) across all possible ORFs separates lncRNAs and TUCPs from translated CDSs of any size. (B) Most erythroid lncRNAs and TUCPs show no evidence of translation. An empirical criterion for productive translation (supplemental Methods) classifies most lncRNAs and TUCPS, such as lincRNA-EPS and TCONS_00050143 (left panels), as noncoding. Others, such as Redrum and NR_015608 (right panels), are predicted to possess translated ORFs (depicted in red). (C) Translation of 5′UTR initiating ORFs. RFPs delineate the starts of high-confidence uORFs and exhibit 3-nt codon periodicity, evidencing their translation. (D) Validation of regulatory uORFs. Transient reporter assays (left, reporter design) evidence uORF-mediated translational control of the Bcl11A and Trak2 mRNAs (right). (E) Examples of uORFs in the Tal1 (left) and Bcl11a (right) mRNAs. (F) Examples of N-terminally extended proteins. Dashed lines mark annotated and upstream start codons. (G) Stop-codon-readthrough mRNA translation. RFPs continuing past known CDSs maintain 3-nt codon periodicity, evidencing readthrough translation. (H) Examples of C-terminally extended proteins, SAFB2 and RAPGEF1 (Rap guanine nucleotide exchange factor 1). Dashed lines mark annotated and next in-frame stop codons. CMV, cytomegalovirus; Fluc, firefly luciferase; Gabarap, GABA(A) receptor–associated protein; Mut, mutant allele; Rluc, Renilla luciferase; WT, wild-type allele.
To predict micropeptide-encoding RNAs with high confidence, we developed an empirical translatability criterion based on the RRS and TE metrics (supplemental Methods). This approach correctly classified 99% of classic ncRNAs as noncoding and reclassified 23 lncRNAs (3%) and 59 TUCPs (11%) as potentially protein-encoding (Figure 2B; supplemental Figure 3I-K; supplemental Table 2). Most (>85%) of the newly predicted mRNAs encode peptides containing fewer than 50 amino acids (supplemental Figure 3L), including the mouse erythroid lncRNA Redrum (lincRNA-EC2) (Figure 2B), which previously was found to be required for red cell maturation.10,17
Widespread translation of regulatory uORFs and alternative protein isoforms
The enrichment of RFPs at mRNA 5′UTRs in comparison with that at 3′UTRs (Figures 1D and 2A) suggests frequent translation upstream of known CDSs. This may reflect the presence of upstream ORFs (uORFs) or protein N-terminal extensions. To probe these possibilities, we looked for ORFs initiating within 5′UTRs and engaged with translating ribosomes (supplemental Methods). This approach yielded 498 high-confidence, 5′UTR initiating ORFs showing clear RFP codon periodicity and translation start site delineation (Figure 2C; supplemental Table 3) that clearly differ from those of untranslated 5′UTR ORFs (more than twofold greater median RFP density, P < 10−15, Kolmogorov-Smirnov test; supplemental Figure 3M). Most (n = 455) of these were out-of-frame with the downstream CDS and thus reflected uORFs. Notably, uORF-containing mRNAs encode proteins enriched for erythroid-important roles, including the key erythroid regulator TAL1 and the effector of adult hemoglobin switching BCL11A (Figure 2E; supplemental Figures 3N and 4A). Translation of the Tal1 uORF, which overlaps the downstream CDS, fosters synthesis of truncated TAL1 isoforms that favor erythroid lineage choice.18 uORFs can also capture ribosomes to lessen CDS decoding instead of favoring downstream reinitiation. Using dual-luciferase reporters, we validated uORF-mediated translation attenuation of the Bcl11A and Trak2 mRNAs, which are transcriptionally repressed and transcriptionally induced with differentiation, respectively (Figure 2D). Thus, uORFs are common among developmentally regulated genes important for red cell biology.
We also identified 43 5′UTR initiating ORFs in-frame with the known CDS, reflecting N-terminal extensions (Figure 2F; supplemental Figure 4B; supplemental Table 3). These comprise broadly expressed genes and include an alternative start site for PAXBP1, which is conserved and adds 51 amino acids to the known protein, and a 45-aa extension of the GABA(A) receptor–associated protein. Ribosomes can also bypass stop codons in a programmed manner, yielding C-terminal extensions. We looked for mRNAs productively translated past their stop codons, and found 117. These amass substantial 3′UTR RFPs with codon periodicity (Figure 2G; supplemental Table 3) and clearly differ from mRNAs with untranslated 3′UTRs (>10-fold greater median RFP density, P < 10−15, Kolmogorov-Smirnov test; supplemental Figure 3M). The proteins with C terminus extension are enriched for roles in transcriptional regulation and cell homeostasis (supplemental Figure 3O), and include proteins translated until the next in-frame stop codon, such as SAFB2, and proteins extended through several in-frame stop codons, such as regulatory subunits of protein phosphatase-1 (Figure 2H; supplemental Figure 4C). Alternative translation initiation and termination thus reflect additional strategies for proteome diversification in erythroid cells.
Widespread and dynamic translation efficiency control during erythropoiesis
Comparing mRNA ribosome densities during erythropoiesis reveals a marked transition from progenitors to hemoglobinizing erythroblasts (Figure 1E; supplemental Figure 2A), suggesting translation specialization during terminal differentiation. For example, although β-globin mRNA is induced after 24 hours of differentiation, robust translation is seen only after 33 hours (Figure 1E).12 To investigate this further, we calculated TE for all mRNAs at each differentiation stage. TEs varied across five orders of magnitude and were uncorrelated with CDS length or mRNA abundance (supplemental Figure 5A). We found 762 mRNAs whose TE is dynamically regulated (empirical P < .05) during differentiation (Figure 3A; supplemental Figure 5D; supplemental Table 4). Clustering these by TE pattern revealed groups that are jointly translationally up- or downregulated in a stage-specific manner. There were two major groups. Cluster A comprised 123 proteins enriched for roles in cell differentiation and proliferation whose synthesis is elevated in erythroid progenitors (Figure 3A-B, top). Conversely, Cluster D comprised 339 proteins enriched for cell maintenance and specialized functions whose synthesis is upregulated in late differentiating and enucleating cells (Figure 3A-B, bottom; supplemental Text 1). To validate these patterns, we fractionated polysomes from cells collected at 0, 24, and 48 hours of differentiation (supplemental Figure 5F) and quantified mRNAs across the sucrose gradient by quantitative polymerase chain reaction (qPCR). All Cluster A mRNAs tested became depleted in polyribosomes with differentiation, verifying translational repression (Figure 3C, top), and all Cluster D mRNAs became enriched, confirming upregulation (Figure 3C, bottom).
Widespread and dynamic translation efficiency control during erythropoiesis. (A) Translation efficiency of 762 genes differentially regulated (empirical P < .05) during erythropoiesis. Heatmap displays mean row-centered log2 TE values at 0, 24, 33, and 48 hours of ex vivo culture. Letters A through G at the left designate clusters; example genes are listed at the right. (B) Top gene pathways (P < .05, Fisher’s exact test) identified among genes that are translationally downregulated (Cluster A, top panel) or upregulated (Cluster D, bottom panel) during terminal differentiation. (C) Validation of translational repression (top) and activation (bottom) of mRNAs from Clusters A and D during differentiation. (D) Upstream ORFs consistently suppress translation through the stages of erythropoiesis. (E) Translation efficiency of 20 uORF and downstream CDS pairs differentially regulated (empirical P < .05) during erythropoiesis. Heatmaps display mean row-centered log2 TE values. (F-G) Ribosome and RNA profiling of Tal1 (F) and Ergic2 (G). Tal1 is translationally induced with differentiation and has a concordant uORF translation pattern, whereas Ergic2 is repressed and shows an opposite uORF pattern. Gene models with CDS (blue), UTR (green), and uORF (pink) regions are shown at the bottom.
Widespread and dynamic translation efficiency control during erythropoiesis. (A) Translation efficiency of 762 genes differentially regulated (empirical P < .05) during erythropoiesis. Heatmap displays mean row-centered log2 TE values at 0, 24, 33, and 48 hours of ex vivo culture. Letters A through G at the left designate clusters; example genes are listed at the right. (B) Top gene pathways (P < .05, Fisher’s exact test) identified among genes that are translationally downregulated (Cluster A, top panel) or upregulated (Cluster D, bottom panel) during terminal differentiation. (C) Validation of translational repression (top) and activation (bottom) of mRNAs from Clusters A and D during differentiation. (D) Upstream ORFs consistently suppress translation through the stages of erythropoiesis. (E) Translation efficiency of 20 uORF and downstream CDS pairs differentially regulated (empirical P < .05) during erythropoiesis. Heatmaps display mean row-centered log2 TE values. (F-G) Ribosome and RNA profiling of Tal1 (F) and Ergic2 (G). Tal1 is translationally induced with differentiation and has a concordant uORF translation pattern, whereas Ergic2 is repressed and shows an opposite uORF pattern. Gene models with CDS (blue), UTR (green), and uORF (pink) regions are shown at the bottom.
Translational upregulation in Cluster D reflects ongoing protein synthesis despite strong (∼60%) mRNA loss after 48 hours of differentiation (supplemental Figure 5C,E). Intriguingly, upregulated Cluster D mRNAs include translation initiation factors (2B, 4H, 4G, and 5A) and the initiation cofactor RBM42, suggesting upregulation of initiation complexes. Indeed, globally higher TE is the most evident change in translational control between progenitors and late differentiating and enucleating cells (supplemental Figure 5B). Examining TE for all initiation factors, regardless of statistical significance, reveals that they are indeed jointly upregulated with terminal differentiation (supplemental Figure 5G). These data indicate that synthesis of translation initiation machinery is enhanced in maturing red blood cells, suggesting a strategy to sustain ongoing protein synthesis without new mRNA production (supplemental Text 1).
Our data also let us probe the contribution of uORFs to translational dynamics. Comparing patterns of TE for uORFs and their CDSs during differentiation revealed that they typically changed concordantly. uORFs were consistently associated with CDS translation dampening at each differentiation stage (P < 10−7 to 10−4; Student t test; Figure 3D), but dynamics of TE of uORFs tend to follow those of the CDS (Pearson’s r = 0.33-0.51, P < 10−14 to 10−5, Fisher’s exact test; supplemental Figure 5H), as illustrated by the Tal1 uORF (Figure 3F). Still, we found 20 uORFs whose TE pattern significantly differs (empirical P < .05) from that of the downstream CDS (Figure 3E; supplemental Table 5), including cases where uORF TE changes are anticorrelated with TE changes of the CDS (Figure 3E,G).
Global discovery of erythroid translation regulators
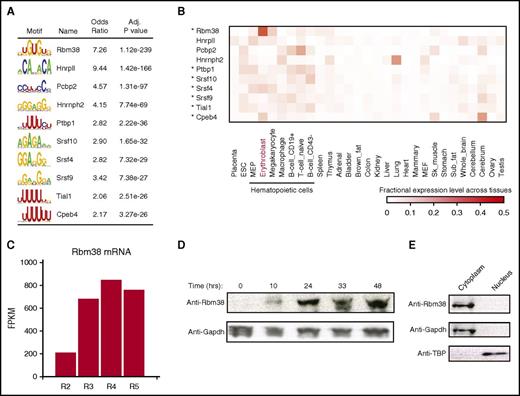
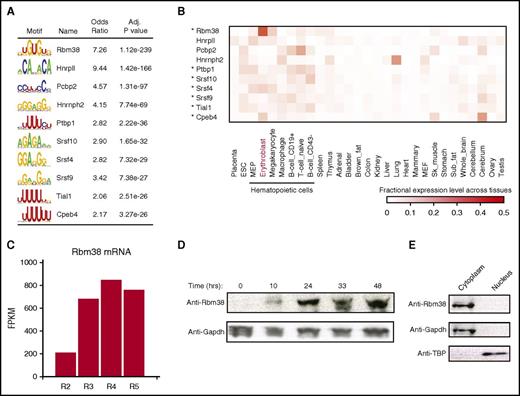
Translation of individual mRNAs can be tuned by UTR cis-elements and trans-acting factors such as RNA-binding proteins (RBPs). To find regulators of mRNAs with dynamic TE during differentiation, we searched their UTRs for known RBP motifs. The most enriched motifs (P < 10−238 to 10−26, Fisher’s exact test; Figure 4A; supplemental Figure 6A; supplemental Table 6), include cytoplasmic polyadenylation element binding protein 4 (CPEB4), a sequence-specific translation repressor in erythroid cells,8 the RBM38 and RBM4B RNA recognition motif family members, and several serine- and arginine-rich factors implicated in splicing but that may also regulate translation.19 Notably, most of these (9 of 15 RBPs) are specifically enriched among hematopoietic cells (P < .05, Kolmogorov-Smirnov test; Figure 4B; supplemental Figure 6B), thus providing a ranked list of candidate translational regulators that may contribute to hematopoietic cell development or function.
Global discovery of erythroid translation regulators. (A) RNA-binding protein motifs enriched within the 3′UTRs of genes translationally regulated during differentiation. The 10 most enriched motifs of erythroid-expressed RBPs are shown along with enrichment statistics (see supplemental Methods). (B) Relative abundance of RBPs from (A) (rows) across 29 primary mouse cell and tissue types profiled by RNA-seq for the ENCODE consortium (columns). Heatmap displays, for each RBP in each cell or tissue type, its fractional expression level out of the total expression across all cell and tissue types examined. Asterisks mark hematopoietic cell–enriched (P < .05, Kolmogorov-Smirnov test) RBPs. (C) Rbm38 is highly induced during terminal erythroid differentiation. Primary differentiating erythroid cells were sorted (R2-R5 fractions) based on cell surface markers,46 and mRNA expression was quantified as fragments per kilobase of exon per million mapped fragments (FPKM). (D) RBM38 is highly induced during terminal erythropoiesis in ex vivo culture. Primary erythroid precursors were differentiated in culture, and protein levels were examined by western blot analysis at the indicated time points. (E) RBM38 is cytoplasmic. Primary erythroblasts were fractionated into cytoplasmic and nuclear components, and protein levels were measured by western blot. Adj., adjusted; Anti-TBP, antibody to TATA box–binding protein. IgG, immunoglobulin G; RNaseA, ribonuclease A; Sk, skeletal.
Global discovery of erythroid translation regulators. (A) RNA-binding protein motifs enriched within the 3′UTRs of genes translationally regulated during differentiation. The 10 most enriched motifs of erythroid-expressed RBPs are shown along with enrichment statistics (see supplemental Methods). (B) Relative abundance of RBPs from (A) (rows) across 29 primary mouse cell and tissue types profiled by RNA-seq for the ENCODE consortium (columns). Heatmap displays, for each RBP in each cell or tissue type, its fractional expression level out of the total expression across all cell and tissue types examined. Asterisks mark hematopoietic cell–enriched (P < .05, Kolmogorov-Smirnov test) RBPs. (C) Rbm38 is highly induced during terminal erythroid differentiation. Primary differentiating erythroid cells were sorted (R2-R5 fractions) based on cell surface markers,46 and mRNA expression was quantified as fragments per kilobase of exon per million mapped fragments (FPKM). (D) RBM38 is highly induced during terminal erythropoiesis in ex vivo culture. Primary erythroid precursors were differentiated in culture, and protein levels were examined by western blot analysis at the indicated time points. (E) RBM38 is cytoplasmic. Primary erythroblasts were fractionated into cytoplasmic and nuclear components, and protein levels were measured by western blot. Adj., adjusted; Anti-TBP, antibody to TATA box–binding protein. IgG, immunoglobulin G; RNaseA, ribonuclease A; Sk, skeletal.
The top-ranked motif was that of RBM38 (RNPC1) (P < 10−238 in 3′UTRs, Fisher’s exact test), which is also the most erythroid-specific of the RBPs, followed by Cpeb4 (Figure 4B; supplemental Figure 6C). RNA-seq reveals that Rbm38 is highly induced in late-differentiating erythroblasts (Figure 4C).20 Indeed, RBM38 is robustly upregulated during ex vivo terminal differentiation (Figure 4D). Further, GATA1 and TAL1 bind to an Rbm38-proximal erythroid-specific enhancer in vivo, and progressive binding of inducible GATA1 at this site results in progressive Rbm38 induction21 (supplemental Figure 6D-F), implying activation by the GATA1–TAL1 complex. Thus, RBM38 is an erythroid-specific RBP potently induced during terminal differentiation by GATA1 and TAL1.
RBM38 interacts with elF4G and promotes target mRNA translation
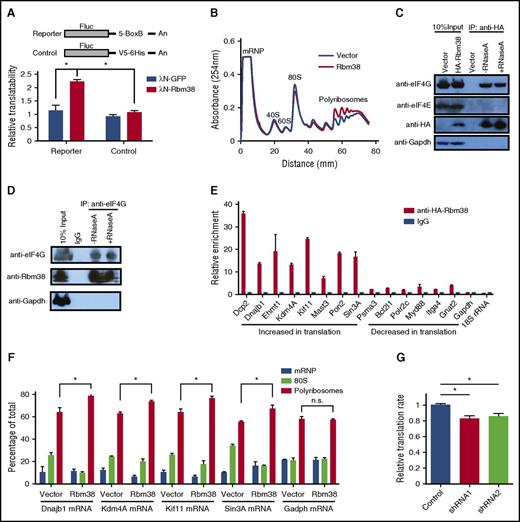
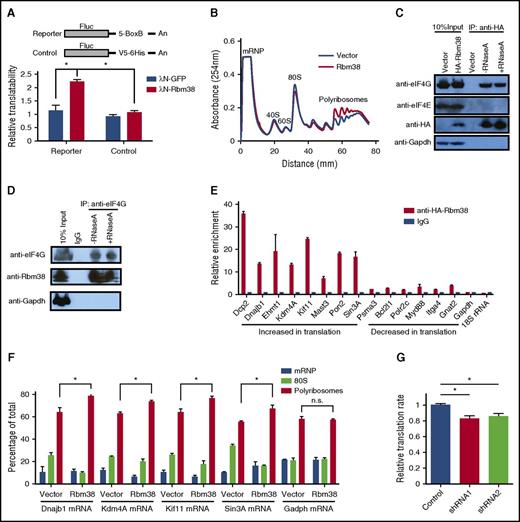
RBM38 has been linked to both mRNA translation and splicing.22-24 In erythroid cells, previous studies have implicated RBM38 in splicing.22,25 The significant enrichment of its binding motif in the UTRs of differentially translated mRNAs (Figure 4A; supplemental Figure 6A), however, suggests that it may also regulate translation in erythroid cells. Fractionation of primary erythroblasts followed by immunoblotting reveals that RBM38 is cytoplasmic, like GAPDH and unlike the TATA box–binding protein (Figure 4E), suggesting that functions in cytoplasmic mRNA translation are indeed plausible. To test this hypothesis, we used tethering assays in human erythroleukemia K562 cells (Figure 5A), which are transfectable erythroid-like cells. We tethered a bacteriophage λN polypeptide–RBM38 fusion protein to the 3′UTR of a luciferase reporter lacking introns, thus enabling specific dissection of translational control without confounding effects from splicing. We found significant (approximately twofold) translation upregulation when RBM38 but not green fluorescent protein was tethered to the reporter (Figure 5A), indicating that RBM38 can promote translation. We thus tested whether RBM38 overexpression can enhance translation in nondifferentiating mouse erythroleukemia (MEL) cells, which enabled specific dissection of the effects of RBM38 on translation without confounding effects from differentiation dynamics. Polysome analysis revealed that RBM38 overexpression indeed favors formation of polyribosomes over 80S monosomes (Figure 5B).
RBM38 interacts with eIF4G and promotes target mRNA translation. (A) RBM38 can stimulate translation. 3′UTR tethering reporter assays (top, reporter design) provide evidence of the capacity of RBM38 to significantly enhance translation of an intronless reporter. (B) RBM38 overexpression promotes polyribosome formation, as evidenced by polysome profiling. (C-D) Specific and RNA-independent interaction between RBM38 and eIF4G. HA-tagged RBM38 was immunoprecipitated from MEL cells (C), and endogenous eIF4G was immunoprecipitated from primary fetal liver erythroblasts (D). The samples were resolved by SDS-PAGE followed by western blotting for the indicated proteins. (E) Validation of RBM38-mediated translational activation of target mRNAs. Selected mRNAs translationally activated or repressed with differentiation were tested for specific immunoprecipitation by an antibody against HA-RBM38. (F) RBM38 overexpression promotes translation of endogenous mRNA targets. The distribution of target or control mRNAs across a polysome gradient was determined in both the presence and the absence of RBM38 expression. (G) Global translation is compromised in RBM38-depleted erythroblasts. Erythroblasts transduced with different shRNA-expressing retroviruses were pulse-labeled with a methionine analog (HPG) and then subjected to a Click-iT assay to determine its rate of incorporation into polypeptides in TER119+ CD71+ erythroblasts (n = 3). mRNP, messenger ribonucleoprotein. *P < .05, Student t test.
RBM38 interacts with eIF4G and promotes target mRNA translation. (A) RBM38 can stimulate translation. 3′UTR tethering reporter assays (top, reporter design) provide evidence of the capacity of RBM38 to significantly enhance translation of an intronless reporter. (B) RBM38 overexpression promotes polyribosome formation, as evidenced by polysome profiling. (C-D) Specific and RNA-independent interaction between RBM38 and eIF4G. HA-tagged RBM38 was immunoprecipitated from MEL cells (C), and endogenous eIF4G was immunoprecipitated from primary fetal liver erythroblasts (D). The samples were resolved by SDS-PAGE followed by western blotting for the indicated proteins. (E) Validation of RBM38-mediated translational activation of target mRNAs. Selected mRNAs translationally activated or repressed with differentiation were tested for specific immunoprecipitation by an antibody against HA-RBM38. (F) RBM38 overexpression promotes translation of endogenous mRNA targets. The distribution of target or control mRNAs across a polysome gradient was determined in both the presence and the absence of RBM38 expression. (G) Global translation is compromised in RBM38-depleted erythroblasts. Erythroblasts transduced with different shRNA-expressing retroviruses were pulse-labeled with a methionine analog (HPG) and then subjected to a Click-iT assay to determine its rate of incorporation into polypeptides in TER119+ CD71+ erythroblasts (n = 3). mRNP, messenger ribonucleoprotein. *P < .05, Student t test.
To study how RBM38 promotes translation, we probed its association with translation initiation factors. Immunoprecipitation of HA-tagged RBM38 in MEL cells revealed a specific and RNA-independent interaction with eIF4G but not eIF4E (Figure 5C). We further confirmed this interaction in the reverse experiment in primary erythroblasts, by immunoprecipitation of endogenous eIF4G followed by immunodetection of RBM38 (Figure 5D). Since eIF4G is a general translation initiation factor that acts as a scaffold recruiting several initiation factors to form the eIF4F complex,26 the RBM38-eIF4G interaction in MEL cells and primary erythroblasts suggests that RBM38 may functionally partner with the eIF4F complex to promote translation of its mRNA substrates. Thus, in addition to its roles in splicing, RBM38 can control translation in erythroid cells, an observation consistent with previous findings in cancer cell lines.2
We then looked to validate endogenous RBM38 targets among mRNAs with dynamic TE during late erythropoiesis. From 191 mRNAs with 3′UTR RBM38 binding sites (supplemental Table 7), we focused on translationally down- or upregulated ones from Clusters A and D, respectively. Given a lack of immunoprecipitation-grade antibodies for endogenous RBM38, we tested 14 such mRNAs by RNA immunoprecipitation of HA-tagged RBM38 in MEL cells, followed by qPCR. We found that all 14 mRNAs specifically bind RBM28 (Figure 5E), unlike Gapdh- or 18S-encoded ribosomal RNA, which are abundant cytoplasmic RNAs without RBM38 binding sites. Notably, translationally upregulated mRNAs from Cluster D show more than fivefold stronger interaction with RBM38 than repressed mRNAs from Cluster A, suggesting that RBM38 selectively promotes their translation. To test this directly, we measured the distribution of these mRNAs across polysome fractions both in the presence and in the absence of exogenous RBM38 (Figure 5F). All mRNAs tested, but not Gapdh, showed higher polyribosome association when RBM38 was expressed, demonstrating specific RBM38-mediated translational activation. Thus, RBM38 is an erythroid-specific factor that promotes translation of select mRNAs with decreasing mRNA levels in late-differentiating and enucleating erythroblasts.
RBM38 is required for terminal erythropoiesis
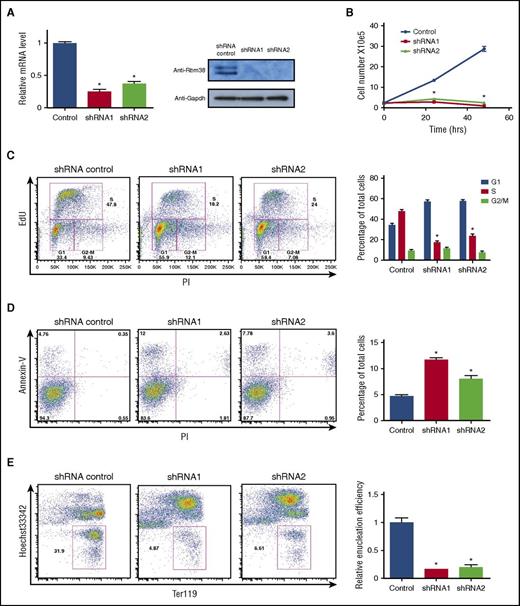
Genetic studies suggest important roles for RBM38 in erythropoiesis. Genome-wide association studies link a single nucleotide polymorphism near Rbm38 to red blood cell volume variation.27 Moreover, mice deficient for RBM38 develop anemia and other pathological conditions.28 To specifically test whether RBM38 is required for terminal erythropoiesis, we separately transduced distinct Rbm38-targeting short hairpin RNAs (shRNAs) into erythroid precursors and cultured them in erythropoietin-containing media to induce terminal proliferation and differentiation. Robust and specific RBM38 knockdown inhibited terminal cell proliferation (Figure 6A-B), blocked cell cycle progression (Figure 6C), induced apoptosis (Figure 6D), and dramatically impaired generation of enucleated reticulocytes (Figure 6E). These data suggest that the in vivo phenotypes of RBM38 deficiency result in part from a critical role in supporting late erythroid development, consistent with previous findings in human erythroid cells.22,25
RBM38 is required for terminal erythropoiesis. (A) RBM38 knockdown in ex vivo–differentiated erythroblasts. (B) RBM38 inhibition blocks proliferation during terminal erythroid differentiation. Growth curves show the number of live ex vivo–TER119+ CD71+ erythroblasts differentiated erythroid cells treated with Rbm38-targeting or non-targeting shRNAs. (C) RBM38-depleted cells accumulate in the G1 cell cycle phase. Plots show the results of DNA staining with propidium iodide (PI) and with a thymidine analog (EdU) in shRNA-transduced cells. The proportion of cells at each cell cycle phase is shown in the bar graph at the right. (D) Elevated apoptosis of RBM38-depleted cells after 24 hours of ex vivo differentiation. Plots display DNA content (PI staining) versus apoptotic status (annexin-V staining) of shRNA-transduced cells. Apoptotic cell fractions are shown in the bar graph at the right. (E) RBM38 inhibition impairs red cell enucleation. Plots display the level of the differentiation surface marker TER119 (fluorescent immunolabeling) versus DNA content (Hoechst staining) of shRNA-transduced live cells after 48 hours of ex vivo culture. Gates mark enucleated reticulocytes. Relative enucleation efficiencies are shown in the bar graph at the right. *P < .05, Student t test.
RBM38 is required for terminal erythropoiesis. (A) RBM38 knockdown in ex vivo–differentiated erythroblasts. (B) RBM38 inhibition blocks proliferation during terminal erythroid differentiation. Growth curves show the number of live ex vivo–TER119+ CD71+ erythroblasts differentiated erythroid cells treated with Rbm38-targeting or non-targeting shRNAs. (C) RBM38-depleted cells accumulate in the G1 cell cycle phase. Plots show the results of DNA staining with propidium iodide (PI) and with a thymidine analog (EdU) in shRNA-transduced cells. The proportion of cells at each cell cycle phase is shown in the bar graph at the right. (D) Elevated apoptosis of RBM38-depleted cells after 24 hours of ex vivo differentiation. Plots display DNA content (PI staining) versus apoptotic status (annexin-V staining) of shRNA-transduced cells. Apoptotic cell fractions are shown in the bar graph at the right. (E) RBM38 inhibition impairs red cell enucleation. Plots display the level of the differentiation surface marker TER119 (fluorescent immunolabeling) versus DNA content (Hoechst staining) of shRNA-transduced live cells after 48 hours of ex vivo culture. Gates mark enucleated reticulocytes. Relative enucleation efficiencies are shown in the bar graph at the right. *P < .05, Student t test.
To directly test whether RBM38-mediated translational control is functionally relevant to terminal erythroid differentiation, we probed the global translation rate in RBM38-depleted erythroblasts. We pulse-labeled cells with a methionine analog and measured its rate of incorporation into polypeptides, focusing on the TER119+ CD71+ subpopulation (supplemental Figure 7) to only compare cells at comparable differentiation stages. RBM38 knockdown by two distinct shRNAs (Figure 6A) reduced the global translational rate by ∼15% to 20% (Figure 5G), consistent with the notion that RBM38 regulates only a subset of mRNAs in erythroblasts. Thus, translation is compromised upon depletion of endogenous RBM38 in primary erythroblasts, arguing that RBM38-mediated translational control functionally contributes to terminal erythropoiesis.
Discussion
We have used ribosome profiling to define and quantify the translational landscape during erythropoiesis, uncovering pervasive and dynamic translational control of protein synthesis. Our study reveals many strategies by which such control is achieved. One of them is widespread and dynamic translation of upstream ORFs. Although thousands of uORFs have been described in mammals, few have been functionally characterized.29 Using reporters, we demonstrate uORF-mediated suppression of downstream translation, including for BCL11A, a key developmental regulator of hemoglobin switching. uORFs are indeed common among developmentally regulated genes important for erythroid-specific processes, supporting the notion that uORFs represent an additional strategy to tightly control synthesis of key cell fate-specifying factors.30,31
Another strategy is programmed alternative translation initiation and termination. We find many novel N-terminal protein extensions, some of which are conserved in humans and may be exploited for regulated synthesis of functionally distinct protein isoforms, as with TAL1.18 We provide only a lower bound for these events, however, as we focused our analysis on canonical AUG-initiation. We also find frequent C-terminal extensions caused by stop codon readthrough, a recognized strategy for natural phenotypic diversity in yeast and fruit flies.32,33 Natural readthrough evidence in higher eukaryotes, however, has been limited to a few examples.34-41 We report several new cases of readthrough events that appear to be regulated, as indicated by ribosome release at specific stop codons after the bypass of several in-frame ones. Our findings thus add to a growing body of evidence suggesting that natural alternative decoding is common among mammals.2
A third strategy is establishment of lineage-specific translational programs by lineage-specific RBPs. These programs involve stage-specific translational up- and downregulation of select mRNAs, highlighting mRNA-specific decoding control as an additional layer enforcing proteome remodeling during erythropoiesis. Importantly, the UTRs of differentially translated mRNAs enrich for recognition sites of hematopoietic cell-specific RBPs, including the erythroid translation repressor CPEB4.8 Our findings thus paint a model where mRNA-specific decoding control arises from the cooperative or competitive action of multiple tissue-intrinsic factors, forming a basis for understanding cell type--specific translational regulation.
A striking aspect of translational control during erythropoiesis is enhanced decoding of select mRNAs depleted in late differentiating and enucleating cells. Specialized translation effectors include ribosomal proteins, translation initiation factors, RBPs, and regulatory RNAs.42,43 Intriguingly, we found coordinate upregulation of initiation factors with terminal differentiation. Much of this initiation machinery must support hemoglobin synthesis, but a fraction likely supports decoding of remaining messages. However, some mRNAs are also translationally repressed with terminal erythropoiesis, such that additional factors must control traffic of initiation complexes toward or away from specific mRNAs. We find that RBM38 can act as such a factor, as it selectively promotes translation of mRNAs exhausted in late differentiating and enucleating cells with decreasing transcriptional activity.
RBM38 is linked to red cell biology by both human and mouse genetics. Although it has been implicated in splicing during late erythropoiesis,22,25 several lines of evidence indicate that it also serves as a translation activator in erythroid cells. First, RBM38 is cytoplasmic in primary erythroblasts and can enhance translation of an intronless reporter. Additionally, RBM38 interacts with the general translation factor eIF4G in an RNA-independent manner. We speculate that the interaction of RMB38 with eIF4G, and its binding to the 3′ ends of its targets, serves to promote their circularization,44 thereby enhancing their translation. Finally, overexpression of RBM38 promotes polyribosome formation, while RBM38-depleted erythroblasts have translational defects. The modest but significant effects of RBM38 on global translation likely reflect the fact that it only fine-tunes translation of a subset of mRNAs in maturing red blood cells. Combined with previous findings, these data cast RBM38 as a bifunctional protein influencing both mRNA splicing and translation, a pattern consistent with the observation that splicing factors can also tune translation.45 An outstanding question is thus whether the splicing and translation roles of RBM38 are dependent on the cell differentiation state. We suspect that in the final stages of erythropoiesis, when cells condense and extrude their nuclei, RBM38 principally regulates cytoplasmic mRNA translation.
Interestingly, while RBM38 interacts with both translationally induced and repressed mRNAs, it binds induced ones much more efficiently. What confers such selectivity? Since both activated and silenced mRNAs have similar numbers of RBM38 target sites in their UTRs, we speculate that productive RBM38 interactions are determined by neighboring RBPs that favor or antagonize its binding. Elucidating how RBPs converge at the same mRNA to determine its ultimate translation fate will thus be essential to delineate translational regulatory networks during mammalian cell differentiation.
Combined with previous findings on CPEB4, our studies begin to dissect such translational networks during erythropoiesis. In developing red cells, tight translational control may be crucial to lessen the dependence of protein synthesis on active mRNA production, which is gradually suppressed and eventually lost as the cells condense and extrude their nuclei. Coordinating these processes involves concomitant translational silencing of expendable mRNAs by CPEB4 and enhanced decoding of irreplaceable ones, promoted by RBM38. We expect that similar translational regulatory networks will be found to orchestrate proteome remodeling during the formation of other functionally specialized cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Harvey F. Lodish for his generous support during the initial stage of this project, and Bingbing Yuan for assistance with bioinformatics analyses.
This work was supported by start-up funding from Mayo Foundation for Medical Education and Research; a grant from the National Institutes of Health, National Heart, Lung, and Blood Institute (R00HL118157) (W.H.); and by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant 2R01DK068348-07 and National Institutes of Health, National Heart, Lung, and Blood Institute grant 2P01HL032262-25 (Harvey F. Lodish).
Authorship
Contribution: J.R.A.-D. and W.H. designed the research and designed, wrote, and revised the manuscript; X.Z. and W.H. performed and interpreted experiments; and J.R.A.-D. performed and interpreted bioinformatics analyses.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wenqian Hu, Department of Biochemistry and Molecular Biology, Mayo Clinic, 200 First St SW, GU16-01A, Rochester, MN 55905; e-mail: hu.wenqian@mayo.edu.