In this issue of Blood, Alvarez-Dominguez et al reveal the strategies used to achieve precise but highly dynamic translational control of protein synthesis during red blood cell development.1
Parallel RNA and ribosome profiling allow comprehensive analysis of translational regulatory dynamics during terminal murine hematopoiesis.
Parallel RNA and ribosome profiling allow comprehensive analysis of translational regulatory dynamics during terminal murine hematopoiesis.
Immature erythroid cells (reticulocytes) lack nuclei and are unable to transcribe messenger RNA (mRNA). Nevertheless, circulating reticulocytes are highly translationally active, synthesizing up to a third of their final hemoglobin content in the period of 48 to 72 hours before they lose ribosomes, organelles, and mRNA as they transition to mature erythrocytes. Although it has been known for decades that the translation of α- and β-globin mRNAs is tightly regulated to maintain equimolar ratios of the encoded α- and β-globin subunits of hemoglobin,2 the mechanisms of translational control during erythropoiesis remain poorly understood. New insights into this process may help shed light on the pathogenesis of the human ribosomopathies caused by mutations in ribosomal proteins or assembly factors.3,4
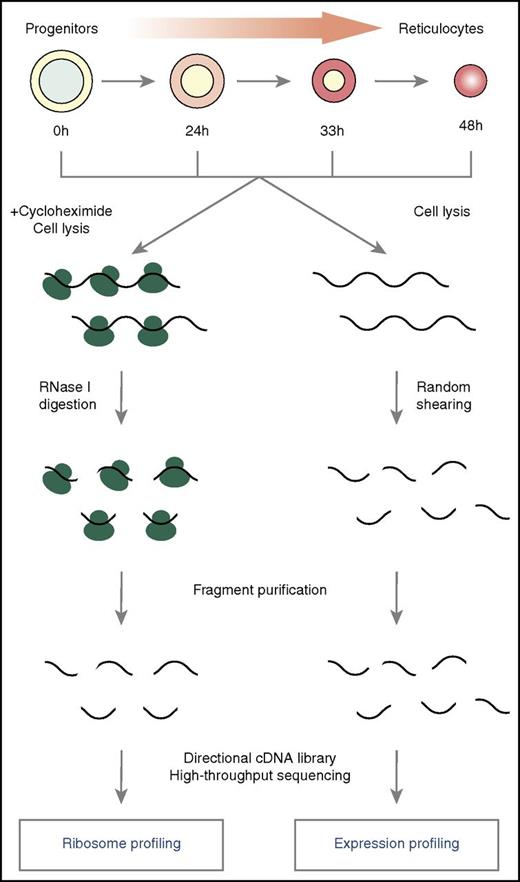
Alvarez-Dominguez et al have comprehensively characterized translation during mouse fetal liver erythroid differentiation using parallel deep sequencing of total RNA (RNA profiling [RNA-seq]) and ribosome-protected fragments (ribosome profiling [Ribo-seq]) (see figure).5 Three key strategies involved in translational regulation during erythropoiesis emerge. First, there is widespread translation of upstream open reading frames (uORFs), with 455 examples identified. The uORFs are cis-regulatory elements that lie in the 5′ untranslated region (UTR) of specific mRNAs, out of frame with the downstream coding sequence, whose functions still remain poorly characterized in mammals. The mRNAs carrying uORFs are enriched for factors with key roles in erythroid maturation, including the basic-helix-loop-helix transcriptional regulator Tal1 and the globin-switching effector Bcl11a.6 Translation of the Tal1 uORF promotes synthesis of a truncated Tal1 protein isoform that favors erythroid lineage choice.7 The demonstration that uORFs are common among the developmentally regulated genes in erythropoiesis supports the idea that uORFs may help tightly regulate the synthesis of key proteins that specify cell fate.
Second, Alvarez-Dominguez et al have identified many examples of programmed alternate translation initiation and termination, revealing that alternate decoding is common in mammalian erythropoiesis. They uncovered 43 novel N-terminal extensions and 117 C-terminal protein extensions caused by stop codon read-through, a strategy known to promote phenotypic diversity in flies and yeast. The authors propose several examples of regulated stop-codon read-through, but the functional significance of these findings requires further experimental work.
A third strategy revealed by Alvarez-Dominguez et al is the establishment of lineage-specific translational programs through engagement of lineage-specific RNA-binding proteins to modulate the translational efficiency (enrichment of Ribo-seq over RNA-seq) of mRNAs at different stages of erythroid maturation. Alvarez-Dominguez et al revealed widespread, dynamic variation in the translational efficiency of specific mRNAs across an astonishing 5 orders of magnitude during different stages of erythroid differentiation. For example, although β-globin mRNA is induced at 24 hours of differentiation, robust translation is only observed at 33 hours. Interestingly, the authors found evidence for coordinate upregulation of translation initiation factors during terminal erythroid differentiation, principally but not exclusively, to support globin synthesis. Taken together, these data suggest a model in which mRNA-specific decoding is positively or negatively regulated during erythropoiesis through cooperative interactions between multiple tissue-specific RNA-binding proteins.
Alvarez-Dominguez et al set out to identify key regulators of mRNAs that exhibited dynamic translational efficiency during erythroid differentiation by searching the UTRs of these mRNAs for specific RNA-binding protein motifs. They uncovered a critical role for RBM38 (previously implicated in splicing8 ) as an erythroid-specific translational activator. The authors found that RBM38 is cytoplasmic in primary erythroblasts, enhances translation of an intronless reporter, and interacts with the general translation factor eIF4G in an RNA-independent manner. Although RBM38 overexpression promotes the formation of polysomes, RBM38-depleted erythroblasts have translational defects and fail to undergo terminal erythropoiesis. Consistent with these data, genetic studies in mice support a role for RBM38 in erythropoiesis.9 It remains unknown how the 2 distinct roles of RBM38 in splicing and translation might be regulated and how RBM38 might specifically discriminate translationally induced mRNAs. Identification of the cytoplasmic polyadenylation element-binding protein 4 (CPEB4) as a sequence-specific translational repressor in erythroid cells10 suggests a model in which erythroid differentiation is controlled by the interplay between the translational silencing of expendable mRNAs by CPEB4 and enhanced decoding of essential mRNAs by RBM38.
A further fascinating tier of translational control has recently been uncovered in human reticulocytes that involves the dynamic regulation of a ribosome rescue pathway.10 Using ribosome footprinting, Mills et al11 revealed dramatic accumulation of posttermination ribosomes on the 3′UTRs of mRNAs in anucleate reticulocytes due to the natural loss of a key ribosome recycling factor (ABCE1) during terminal erythroid differentiation, thus limiting the availability of ribosomes within the actively translating pool. Transient activation of ribosome recycling through the induction of ribosome rescue factors (PELO and HBSL1) provides a mechanism to support ongoing translation of critical mRNAs during terminal differentiation by promoting the recycling of ribosomes into the active cellular pool, thereby maintaining hemoglobin synthesis as ABCE1 levels fall.
Taken together, these interesting new studies begin to uncover multiple tiers of translational regulation and dynamic ribosome recycling that act together to remodel the proteome and maintain ribosome homeostasis during erythropoiesis. Precisely how RNA-binding proteins cooperate to determine the translational fate of specific mRNAs is an exciting area for future investigation. By touching on generic issues of translational control during the dynamic process of differentiation, the significance of the current work extends beyond the field of red blood cell development. Finally, these findings may guide future new therapeutic strategies for the treatment of human ribosomopathies.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal