Key Points
New function-disrupting mutations in CalDAG-GEFI, p.S381F and p.R113X, were identified in 2 unrelated families of distinct ethnic origin.
Homozygous carriers of these mutations displayed relevant bleeding diathesis and moderate impairment and/or delay in platelet aggregation.
Abstract
In addition to mutations in ITG2B or ITGB3 genes that cause defective αIIbβ3 expression and/or function in Glanzmann’s thrombasthenia patients, platelet dysfunction can be a result of genetic variability in proteins that mediate inside-out activation of αIIbβ3. The RASGRP2 gene is strongly expressed in platelets and neutrophils, where its encoded protein CalDAG-GEFI facilitates the activation of Rap1 and subsequent activation of integrins. We used next-generation sequencing (NGS) and whole-exome sequencing (WES) to identify 2 novel function-disrupting mutations in RASGRP2 that account for bleeding diathesis and platelet dysfunction in 2 unrelated families. By using a panel of 71 genes, we identified a homozygous change (c.1142C>T) in exon 10 of RASGRP2 in a 9-year-old child of Chinese origin (family 1). This variant led to a p.Ser381Phe substitution in the CDC25 catalytic domain of CalDAG-GEFI. In 2 Spanish siblings from family 2, WES identified a nonsense homozygous variation (c.337C>T) (p.Arg113X) in exon 5 of RASGRP2. CalDAG-GEFI expression was markedly reduced in platelets from all patients, and by using a novel in vitro assay, we found that the nucleotide exchange activity was dramatically reduced in CalDAG-GEFI p.Ser381Phe. Platelets from homozygous patients exhibited agonist-specific defects in αIIbβ3 integrin activation and aggregation. In contrast, α- and δ-granule secretion, platelet spreading, and clot retraction were not markedly affected. Integrin activation in the patients’ neutrophils was also impaired. These patients are the first cases of a CalDAG-GEFI deficiency due to homozygous RASGRP2 mutations that are linked to defects in both leukocyte and platelet integrin activation.
Introduction
One of the most severe inherited platelet disorders1,2 is Glanzmann’s thrombasthenia, which is caused by recessive mutations in either ITG2B or ITGB3 leading to defects in αIIbβ3 integrin activation and/or expression, and consequently to severely impaired platelet aggregation in response to all platelet agonists.3,4 Impaired platelet aggregation and bleeding can also be caused by mutations in genes for signaling proteins that are critical to the inside-out activation of αIIbβ3. RASGRP2 was recently identified as one such gene affected in patients with a platelet function defect and a bleeding complication.5 RASGRP2 codes for the protein CalDAG-GEFI, a guanine nucleotide exchange factor for the small GTPase Rap1. In platelets, Rap1 activity is regulated by 2 independent yet synergistic signaling pathways6 : (1) a fast but reversible activation pathway mediated by an increase in the intracellular calcium ion concentration and the activation of CalDAG-GEFI and (2) a slow but sustained pathway that requires signaling by protein kinase C and the adenosine 5′-diphosphate (ADP) receptor P2Y12, the target of antiplatelet drugs such as clopidogrel. Although CalDAG-GEFI mediates guanosine-5′-triphosphate (GTP) loading (ie, the activation of Rap1), signaling by P2Y12 leads to the inhibition of Rasa3, a GTPase-activating protein critical in the inactivation of Rap1.7 In mice, genetic deletion of Rasgrp2 did not affect embryonic development and survival. Adult mice lacking CalDAG-GEFI showed marked protection from experimental thrombosis and a significant impairment in hemostasis.8,9 Furthermore, these mice exhibited a mild defect in the adhesive function of neutrophils, another cell type that expresses significant levels of CalDAG-GEFI.10 The study by Canault et al5 provided the first genetic evidence that (1) CalDAG-GEFI is also a critical regulator of integrin-mediated adhesion in human platelets, and (2) the molecular machinery controlling Rap1 activity in human platelets closely resembles that of murine platelets. No defects in neutrophil adhesion mediated by integrins were observed.
In this study, we report 2 pedigrees characterized by a bleeding tendency and a platelet dysfunction due to impaired αIIbβ3 integrin activation. Neutrophils from homozygous patients also showed a defect in β2 integrin activation in vitro, but no relevant infections were reported. These patients carry novel mutations in RASGRP2, a nonsynonymous change c.1142C>T in family 1, and a nonsense change c.337C>T in family 2. Both mutations led to a marked reduction in CalDAG-GEFI expression. Thus, these patients are the first cases of a CalDAG-GEFI deficiency due to homozygous RASGRP2 mutations that are linked to defects in both leukocyte and platelet integrin activation.
Patients and methods
Patients, blood sampling, and DNA collection
The study involved 3 index cases from 2 unrelated families of Chinese (family 1) and Spanish (family 2) origin. Venous blood was drawn from each patient and a parallel healthy control into 7.5% K3 EDTA tubes (for blood counts and DNA isolation), and into buffered 0.105 M sodium citrate (for platelet and neutrophil function studies) by using a 20-gauge needle. Samples were maintained at room temperature until processing. Genomic DNA from EDTA blood samples was isolated by using a DNeasy blood and tissue kit, following the manufacturer’s protocol (Qiagen, Hilden, Germany). DNA concentration was measured by using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA).
This investigation followed the Helsinki Declaration and was approved by the Ethics Committee of the Hospital Reina Sofía (Murcia, Spain). All patients provided written informed consent.
Next-generation sequencing gene panel
A clinical next-generation sequencing (NGS) panel was designed (Design Studio, Illumina, San Diego, CA) with probes targeting all exons, 3′UTR (untranslated region), and flanking regions of each of the 71 genes depicted in supplemental Table 1 (available on the Blood Web site). The NGS was performed on a MiSeq Instrument, running MiSeq Control Software and according to a Nextera sequencing design (Illumina). This NGS panel was applied in the index case of family 1. In brief, 50 ng of the DNA was sequenced following Illumina’s standardized protocol. DNA libraries were normalized to 4 nM and pooled in equal volumes. We used the 300-cycle reagent kit. Specifically, the sample was sequenced by using paired 150 nt reads, multiplexing 12 dual indexed samples per run. Each run performed in this study contained 24 samples. Minimum coverage per base was 100 reads. High sequence quality was based on a Phred score of >20, quality >20, and read coverage >30 at each position within the reads. Sequence data were mapped to the revised Cambridge Reference Sequence by using an integrated computer software platform from MiSeq (MiSeq Reporter, Illumina).
Whole-exome sequencing
In 2 patients from family 2, the whole-exome sequencing (WES) was performed essentially as described.11 After enrichment of coding regions and intron/exon boundaries with the SureSelect Human All Exon 50 Mb kit (Agilent Technologies), captured libraries were sequenced on the Illumina HiSequation 2500 (Illumina) with 100 bp paired-end reads. Bioinformatic analysis was carried out as in previous studies.11,12
Sanger sequencing
Verification of the 2 mutations in the RASGRP2 gene that were identified in these patients by NGS and WES and their segregation in the pedigrees was achieved by standard Sanger sequencing with specific primers by using an ABI 3730 automated sequencer. The following primer pairs were designed by using the ExonPrimer script (https://ihg.helmholtz-muenchen.de/ihg/ExonPrimer.html):
(a) mutation C.1142 C>T [p.Ser381Phe]
RASGRP2 × 5F: GTCGACCTTTGGCCCTG
RASGRP2 × 5R: AGTGCTCCGGCAAACTGG
(b) mutation C.337 C>T [p.Arg113]
RASGRP2 × 5F: ….. GGTGGTTGTCTCAAGGGTCT
RASGRP2 × 5R: …. CGAGAGAGCAACATGACCCT
Sequences in the polymerase chain reaction products were analyzed by using MutationSurveyor software available from SoftGenetics (http://www.softgenetics.com/mutationSurveyor.php).
Platelet aggregation
Platelet-rich plasma (PRP) was obtained by centrifugation of citrated whole blood at 150g for 10 minutes. Platelet-poor plasma (PPP) was prepared by a further centrifugation step of the same tube at 1000g for 20 minutes. Light transmission aggregometry was performed essentially as described3 by using undiluted PRP in an Aggrecorder II aggregometer (Menarini Diagnostics, Florence, Italy). Time course changes in the maximal percentage of light transmission of PRP over baseline (PPP) were recorded for 300 seconds upon addition of various platelet agonists (1.5 mM arachidonic acid, 25 μM thrombin receptor-activating peptide (such as protease-activated receptor 1 agonist peptide [PAR1p]), 2 and 10 μg/mL collagen, 10 μM ADP, or 100 nM phorbol 12-myristate 13-acetate (PMA).
Platelet 14C-serotonin release
PRP was adjusted with homologous PPP to a final platelet count of 250 × 109/L and labeled with 1 μM 14C-serotonin (5-OH-14C-labeled tryptamine [14C-serotonin]; GE Healthcare, Barcelona, Spain) for 45 minutes at 37°C as described.13 Platelet activation and/or aggregation in the labeled PRP samples was performed as above with selected agonists, and a 1:6 volume of EDTA (0.05 M):formaldehyde (0.633 M) was added to stop reactions. Then, cuvettes were centrifuged (12 000g for 2 minutes) to pellet platelets, and 14C-serotonin released into the supernatant was quantified by a Wallac 1409 liquid scintillation counter (Wallac Oy; AG&G Company, Turku, Finland) and was reported as a percentage of the total radioactivity incorporated after correction for background.13
Clot retraction
Clot retraction assays were performed essentially as described.14 Briefly, PRP samples adjusted with homologous PPP to a final platelet count of 225 × 109/L were mixed in aggregation cuvettes with Tyrodes-2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid buffer containing a small number of red blood cells. Clot formation was induced at room temperature with thrombin (1 U/mL) and CaCl2 (2 mM), a sealed glass pipette was set at the center of the cuvette, and clot retraction was monitored over time.
Platelet flow cytometry
Platelet expression of major platelet membrane glycoproteins (GPs) such as GPIa (integrin α2), GPIbα, GPIIb (αIIb), and GPIIIa (β3) was assessed by flow cytometry in citrated whole blood diluted 1:10 in saline buffer through a direct standard technique with appropriate labeled monoclonal antibodies in a FACScalibur platform (Becton Dickinson, San Jose, CA). For analysis of surface-expressed P-selectin (marker of α-granule release) and binding of fibrinogen (marker for activated αIIbβ3), diluted PRP (∼20 × 109/L platelets) was stimulated under static conditions (30 minutes at room temperature) with the desired agonist concentration in the presence of both anti-CD62-phycoerythrin antibody (BD Biosciences, Madrid, Spain) and fibrinogen-Alexa488 (Thermo Fisher, Madrid, Spain). Samples were then run in a FACSCalibur flow cytometer (Becton Dickinson), and the fluorescence of positively stained cells was analyzed by using CellQuest software.
Platelet spreading
Washed platelets were prepared as described.15 Coverslips were coated overnight in 100 µg/mL fibrinogen (Enzyme Research Laboratory, South Bend, IN) in sterile phosphate-buffered saline (PBS) at 4°C. The coverslips were blocked in denatured sterile filtered bovine serum albumin 5 mg/mL (First Link UK Ltd, Birmingham, United Kingdom) in PBS. Three hundred microliters of washed platelets, adjusted to 20 × 109/L, were added to each well and allowed to spread for 45 minutes at 37°C. Spread platelets were washed in PBS and fixed at room temperature in 4% paraformaldehyde for 5 minutes. Images were taken by using a DM IRE2 Leica inverted microscope with an SP2 confocal system running Leica Confocal Software Version 2.61 Build 1537. Platelet-spreading assays were imaged by using reflectance microscopy with the 488-nm line of an argon-ion laser (457-514 nm) with an HCX Plan Apo Ibd.BL 63× NA 1.4 Olympus objective. To calculate platelet spread area, 10 platelets from 10 different fields of view (100 platelets per sample) were outlined by using the ROI tool in ImageJ (1.48V). The area measurements were then logged. Statistical significance was assessed by one-way analysis of variance.
Immunoblotting
Total lysates from washed human platelets were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis on 4% to 20% gradient gels and transferred to polyvinylidene fluoride membranes (Millipore). Standard western blotting procedures were used. RAP1 (α-RAP1 clone 121; Santa Cruz Biotechnology, Dallas, TX) and CalDAG-GEFI (polyclonal rabbit α-RasGRP2; Thermo Fisher Scientific, Waltham, MA) proteins were detected by using IRDye 800-conjugated goat anti-rabbit secondary antibodies (Li-Cor Biosystems, Lincoln, NE). RASA3 (polyclonal goat α-GAP1-InsP4BP; Santa Cruz Biotechnology) and β-actin (anti-β-actin clone AC-15; Sigma-Aldrich, St. Louis, MO) proteins were detected by using IRDye 680-conjugated donkey anti-goat and goat anti-mouse secondary antibodies (Li-Cor Biosystems), respectively, and visualized with the Odyssey Infrared Imaging System (Li-Cor Biosystems).
CalDAG-GEFI expression and purification
Human CalDAG-GEFI and Rap1B proteins were overexpressed in Escherichia coli and purified by using a combination of immobilized metal affinity and size exclusion chromatography. Purified proteins were homogenous on the basis of sodium dodecyl sulfate polyacrylamide gel electrophoresis. Wild-type and mutant forms of CalDAG-GEFI were expressed as truncations lacking 58 N-terminal residues predicted to be unstructured (CalDAG-GEFI [1-551]), whereas Rap1B was truncated at Cys181, and this residue mutated to serine (Rap1B [C181S]). CalDAG-GEFI harboring S381F was not submitted to size exclusion chromatography because of protein instability.
Nucleotide exchange activity assay
Purified proteins were used to determine the guanine nucleotide exchange factor (GEF) activity toward Rap1B of wild-type and mutant CalDAG-GEFI. Nucleotide exchange on Rap1B was monitored by using Bodipy fluorophore (FL) guanosine 5′-diphosphate (GDP), a fluorescent analog of GDP. The fluorescence of Bodipy FL GDP is quenched in solution and increases upon binding to Rap1B. To determine the GEF activity of CalDAG-GEFI mutants, purified Rap1B (1 μM) was added to exchange buffer (20 mM tris(hydroxymethyl)aminomethane [pH 7.5], 150 mM NaCl, 2 mM MgCl2, 0.08% NP-40, 5% [v/v] glycerol, and 1 mM dithiothreitol) containing 1 μM Bodipy GDP (Thermo Fisher) in the absence or presence of 400 nM wild-type or mutant CalDAG-GEFI. Fluorescence data were collected on a Pherastar microplate reader (BMG LABTECH, Ortenberg, Germany) with λex = 485 nm, λem = 520 nm, and 2-nm slits.
Leukocyte studies
Citrated whole blood samples containing 1 × 106 white blood cells were resuspended in ice-cold hypotonic NaCl solution (0.2%) for 30 seconds, after which hypertonic (1.6%) NaCl solution (v/v) was added to restore isotonicity, and cells were pelleted (350g at 10°C for 5 minutes). The lysis step was repeated once, and white blood cells were resuspended in saline. To analyze surface GP expression, monoclonal antibodies for membrane GPs CD11a and CD11b were used (Becton Dickinson). β2 integrin activation in stimulated neutrophils was analyzed by determining the ability of these cells to bind soluble fibrinogen or the conformation-specific antibody m24 (Abcam, Cambridge, MA). To analyze granule release, cells were stained with an anti-CD11b Mac-1 antibody (Becton Dickinson). Blood leukocytes were incubated with 60 μg/mL Alexa Fluor 488 fibrinogen (Thermo Fisher) and PerCP-anti CD45 (Becton Dickinson), phycoerythrin-anti-Mac-1 and PerCP-anti-CD45, or with 10 μg/mL m24. Saline, PMA, N-formylmethionyl-leucyl-phenylalanine, or manganese chloride (Mn+2) were added during this phase for 30 minutes at room temperature. Cells were washed and resuspended in saline, or tubes containing m24 were further incubated with a PerCP-anti mouse immunoglobulin G1 (IgG1; Santa Cruz Biotechnology) for 30 minutes, washed again, and resuspended in saline.
To analyze β1 integrin expression, leukocytes were incubated with soluble VCAM-1/Fc chimera protein (R&D Systems, Minneapolis, MN), washed, and stained with a fluorescein isothiocyanate–conjugated antibody to human IgG (Sigma-Aldrich, Madrid, Spain). Flow cytometry was conducted immediately after the final wash.
A FACSCalibur flow cytometer (Becton Dickinson) and CellQuest and Paint-A-Gate software were used for analysis. Analysis of leukocyte subpopulations was performed on the basis of a CD45/side scatter (fibrinogen binding, CD11a and Mac-1 expression) or light scatter (m24 and VCAM-1 binding) gating procedure.
Statistical analysis
All functional assays were performed with samples from 3 patients and 3 healthy controls. Because of the limited sample number, statistical comparisons could not be performed.
Results
Patient general information
In this study we have characterized 3 patients from 2 unrelated families. The proband in family 1 (P1-family 1) is a 9-year-old Chinese child suffering from severe epistaxis requiring hospitalization and medical intervention, including nasal packing, tranexamic acid treatment, and transfusion of red blood cells and platelets. In the second family, the index case (P1-family 2) is a 55-year-old Spanish woman who has a lifelong history of spontaneous bruising, petechiae, epistaxis, and gingival and gastrointestinal bleeding. Minor bleeding has been treated mainly with antifibrinolytic drugs. Menorrhagia required oral contraceptives and iron therapy. Her 2 episodes of childbirth, dental extractions, and minor surgical procedures were successfully managed with prophylactic platelet transfusions. Her 46-year-old brother, the second proband in this family (P2-family 2) also suffered from epistaxis and gingival and digestive bleeding in childhood that required platelet transfusion. In adulthood, he has experienced few bleeding episodes, but minor surgical procedures (colonoscopy and dental extractions) required the use of prophylactic desmopressin and antifibrinolytic drugs.
In addition to their bleeding symptoms, these 3 patients displayed, on repeated testing, a normal platelet count and volume and mild anemia (Table 1). Blood smears, leukocyte count and blood differential, markers of liver and kidney function, prothrombin time, activated partial thromboplastin time, fibrinogen, and von Willebrand factor levels were within normal ranges. Our patients did not display overt immune defects or susceptibility to bacterial infections other than P2-family 2 suffering from ulcerative colitis.
Platelet function studies
A clinical suspicion of inherited platelet disorder was established in these patients because of their prolonged bleeding time, extended platelet function assay 100 (PFA-100) closure times (Table 1), and the severely reduced platelet aggregation in response to ADP and low-dose collagen (Figure 1A). The expression level of major adhesive GPs, including GPIb/IX, GPIa, and GPVI, was normal (not shown). Glanzmann’s thrombasthenia was ruled out for the 3 patients because (1) the surface expression of αIIbβ3 was not affected (not shown), and (2) the platelet aggregation was normal or minimally impaired in response to high concentrations of PAR1p, collagen, arachidonic acid, or PMA (Figure 1A). Compared with controls, the lag time (ie, the time required to reach 10% aggregation) was markedly increased in platelets from patients stimulated with 10 μg/mL collagen (0.93 ± 0.27 vs 0.55 ± 0.07 minutes), 1.6 mM arachidonic acid (0.62 ± 0.12 vs 0.24 ± 0.01 minutes), or 10 μM ADP (0.43 ± 0.11 vs 0.18 ± 0.09 minutes), all expressed as mean ± standard deviation (n = 3). No delay in the aggregation response to PMA or PAR1p was observed in the patient’s platelets. Consistent with the delay in aggregation, platelets from the 3 patients exhibited markedly impaired fibrinogen binding upon stimulation with most agonists (ADP, PAR1p, PAR4p, or collagen-related peptide [CRP]) (Figure 1B). In contrast, the patients’ platelets showed only minor defects in agonist-induced α- or δ-granule secretion (supplemental Figure 1A-B), clot retraction (supplemental Figure 2), and spreading on fibrinogen (supplemental Figure 3).
Platelet aggregation and integrin activation is markedly impaired in index cases with lifelong bleeding diathesis from 2 unrelated families. (A) Platelet aggregation in response to the indicated platelet agonists was evaluated in unadjusted PRP from index cases (P) and a healthy and unrelated control (C). (B) Platelets from index cases and healthy and unrelated controls (controls) (combined data from 3 participants) were stimulated under static conditions (30 minutes at room temperature) with agonist (5 and 25 μM ADP [ADP-5 and ADP-25], 25 μM PAR1p, 250 μM PAR4p, 2 and 10 μg/mL collagen-related peptide [CRP-2 and CRP-10], 2 μg/mL convulxin [Cvx], and 100 nM PMA) in the presence of fibrinogen-Alexa 488. The samples were evaluated by flow cytometry and the median fluorescence intensity (MFI) for fibrinogen-A488 binding is shown.
Platelet aggregation and integrin activation is markedly impaired in index cases with lifelong bleeding diathesis from 2 unrelated families. (A) Platelet aggregation in response to the indicated platelet agonists was evaluated in unadjusted PRP from index cases (P) and a healthy and unrelated control (C). (B) Platelets from index cases and healthy and unrelated controls (controls) (combined data from 3 participants) were stimulated under static conditions (30 minutes at room temperature) with agonist (5 and 25 μM ADP [ADP-5 and ADP-25], 25 μM PAR1p, 250 μM PAR4p, 2 and 10 μg/mL collagen-related peptide [CRP-2 and CRP-10], 2 μg/mL convulxin [Cvx], and 100 nM PMA) in the presence of fibrinogen-Alexa 488. The samples were evaluated by flow cytometry and the median fluorescence intensity (MFI) for fibrinogen-A488 binding is shown.
Identification of deleterious mutations in RASGRP2
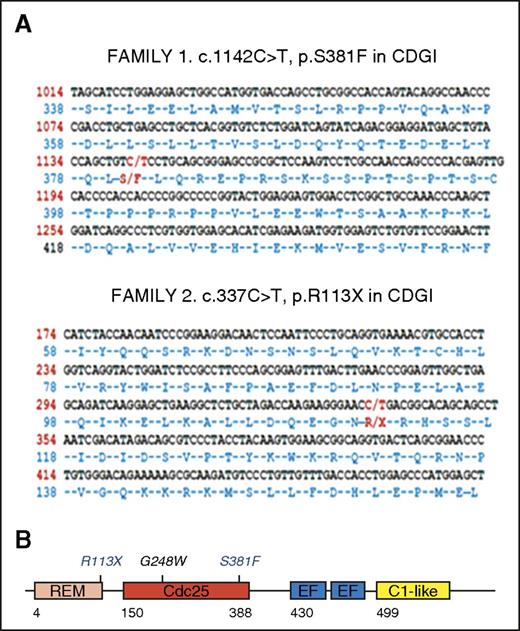
To identify the molecular defects responsible for the bleeding phenotype in these patients, we analyzed DNA from the Chinese boy and the Spanish siblings by using a novel NGS panel that covered 71 genes known to, or suspected to, play a relevant role in inherited platelet disorders (supplemental Table 1) and WES, respectively. As shown in Figure 2A, these molecular analyses identified novel homozygous changes in the RASGRP2 gene in the index cases: (1) a nonsynonymous change c.1142C>T (p.Ser381Phe) in exon 10 in family 1 and (2) a nonsense change c.337C>T (p.Arg113X) in exon 5 in family 2. Sanger sequencing confirmed the presence of these mutations in the probands and their inheritance from the consanguineous parents (Figure 3).16 CalDAG-GEFI, the RASGRP2-encoded protein, contains an N-terminal catalytic region that comprises a Ras exchange motif and a CDC25 domain that provides GEF activity and a C-terminal regulatory region with two Ca2+ binding EF-hand domains and a C1-like domain17 (Figure 2B). The nonsense c.337C>T (p.Arg113X) variant is expected to truncate CalDAG-GEFI synthesis at the level of the Ras exchange motif, whereas the p.Ser381Phe substitution is located at the C-terminal end of the CDC25 catalytic domain, and it is predicted by PolyPhen (http://genetics.bwh.harvard.edu/pph2/) to be probably damaging with a score of 1.0. The p.Ser381Phe variant is reported with an allelic frequency of 8.47 × 10−6 in the Exome Aggregation Consortium database (ExAC; http://exac.broadinstitute.org), and it is unreported in the 1000 Genomes project database (http://browser.1000genomes.org). The p.Arg113X is reported as a somatic mutation in the Catalog of Somatic Mutations in Cancer (COSMIC) project (http://cancer.sanger.ac.uk).
Localization of the novel c.1142C>T (p.S381F) and c.337C>T (p.R113X) mutations within the RASGRP2 sequence and the encoded protein CalDAG-GEFI. DNA from index cases was analyzed by NGS or WES, and novel mutations in RASGRP2 were identified. (A) Localization of the novel c.1142C>T (p.S381F; family 1) and c.337C>T (p.R113X; family 2) mutations within the RASGRP2 sequence. (B) Schematic representation for CalDAG-GEFI showing the different domains: Ras exchange motif (REM), catalytic domain (Cdc25), calcium-binding EF hands (EF), and C1-like domain (unknown function). The positions of the recently reported G248W mutation5 and the novel p.R113X and p.S381F mutations within the REM and Cdc25 domains are shown. CDGI, CalDAG-GEFI.
Localization of the novel c.1142C>T (p.S381F) and c.337C>T (p.R113X) mutations within the RASGRP2 sequence and the encoded protein CalDAG-GEFI. DNA from index cases was analyzed by NGS or WES, and novel mutations in RASGRP2 were identified. (A) Localization of the novel c.1142C>T (p.S381F; family 1) and c.337C>T (p.R113X; family 2) mutations within the RASGRP2 sequence. (B) Schematic representation for CalDAG-GEFI showing the different domains: Ras exchange motif (REM), catalytic domain (Cdc25), calcium-binding EF hands (EF), and C1-like domain (unknown function). The positions of the recently reported G248W mutation5 and the novel p.R113X and p.S381F mutations within the REM and Cdc25 domains are shown. CDGI, CalDAG-GEFI.
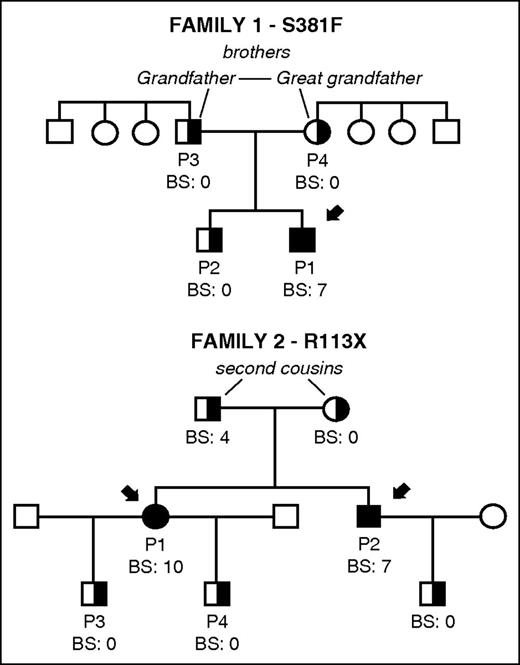
Family pedigrees and bleeding scores in 2 unrelated families carrying novel mutations S381F and R113X in CalDAG-GEFI. The index cases in each family are indicated with black arrows. Bleeding in patients and available family members was evaluated and a bleeding score was assigned by using the International Society on Thrombosis and Haemostasis Bleeding Assessment tool.16 Filled and partially filled black symbols indicate homozygosis and heterozygosis for the corresponding mutation in RASGRP2. Other family members (white symbols) were not available for study.
Family pedigrees and bleeding scores in 2 unrelated families carrying novel mutations S381F and R113X in CalDAG-GEFI. The index cases in each family are indicated with black arrows. Bleeding in patients and available family members was evaluated and a bleeding score was assigned by using the International Society on Thrombosis and Haemostasis Bleeding Assessment tool.16 Filled and partially filled black symbols indicate homozygosis and heterozygosis for the corresponding mutation in RASGRP2. Other family members (white symbols) were not available for study.
Novel RASGRP2 mutations impair CalDAG-GEFI expression and function
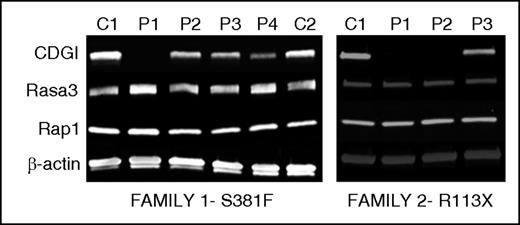
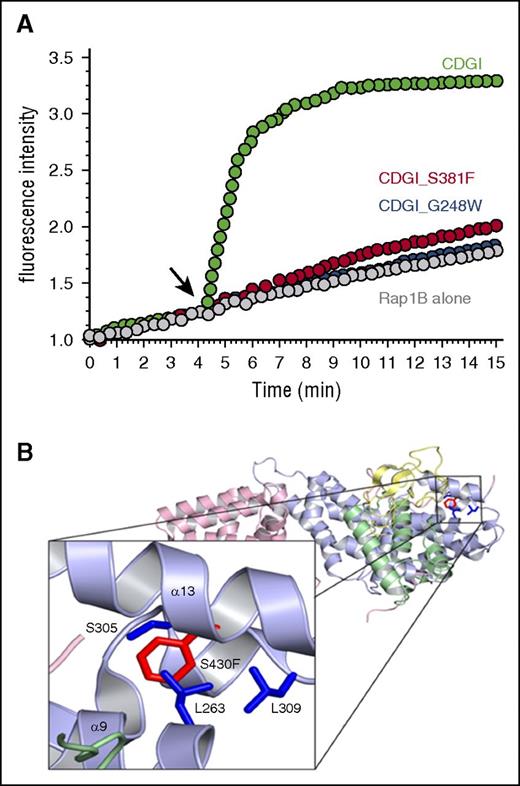
Immunoblotting analysis demonstrated that CalDAG-GEFI expression was markedly reduced in platelets from index cases homozygous for p.Ser381Phe or p.Arg113X and moderately reduced in platelets from available heterozygous relatives. In contrast, expression of Rap1 itself and Rasa3, a main Rap1 GTPase activating protein in platelets,7 was comparable in platelets from affected individuals and healthy volunteers (Figure 4). To evaluate whether the Ser381Phe substitution also impairs the GEF activity of CalDAG-GEFI, we purified the wild-type and mutant protein and subjected them to a novel, fluorescence-based assay to monitor nucleotide exchange on purified Rap1B. In this assay, nucleotide exchange on Rap1B is monitored with the help of Bodipy FL GDP, a fluorescent analog of GDP that is quenched in solution but shows increased fluorescence upon binding to Rap1B. As shown in Figure 5A, loading of Rap1 with Bodipy FL GDP occurred very slowly unless it was catalyzed by the addition of CalDAG-GEFI. Interestingly, large amounts of CalDAG-GEFI (Ser381Phe), but not CalDAG-GEFI (wild-type), precipitated during the purification process, suggesting altered stability of the patient protein. Nonaggregated CalDAG-GEFI (Ser381Phe) protein showed strongly reduced GEF activity when added to Rap1B at the same concentration as CalDAG-GEFI (wild-type). The reduction in the catalytic activity was similar to that observed with purified CalDAG-GEFI bearing a Gly248Trp substitution, the only other known CalDAG-GEFI mutation in humans5 (Figure 5A). Notably, whereas Gly248 is positioned in the interface between the CDC25 domain and Rap1,5 Ser381 localizes to an adjacent region (Figure 5B).18 On the basis of structural modeling, we propose that the p.Ser381Phe substitution clashes with Leu263, Ser305, and Leu309 and thus causes a conformational change in CalDAG-GEFI, which affects both protein stability and nucleotide exchange activity.
Novel mutations R113X and S381F severely affect the expression of CalDAG-GEFI. Immunoblot analysis for CalDAG-GEFI (CDGI), Rasa3, Rap1, and β-actin in platelet lysates from carriers of the indicated mutations in CalDAG-GEFI. Left: p.S381F (P1 homozygous; P2, P3, P4 heterozygous); Right: p.R113X mutation (P1 and P2 homozygous; P3 heterozygous). Protein expression in lysates from healthy and unrelated controls (C) analyzed in parallel is also shown.
Novel mutations R113X and S381F severely affect the expression of CalDAG-GEFI. Immunoblot analysis for CalDAG-GEFI (CDGI), Rasa3, Rap1, and β-actin in platelet lysates from carriers of the indicated mutations in CalDAG-GEFI. Left: p.S381F (P1 homozygous; P2, P3, P4 heterozygous); Right: p.R113X mutation (P1 and P2 homozygous; P3 heterozygous). Protein expression in lysates from healthy and unrelated controls (C) analyzed in parallel is also shown.
The S381F substitution alters CalDAG-GEFI structure and markedly impairs its nucleotide exchange activity. (A) Bodipy fluorescence-based assay to monitor nucleotide exchange on purified Rap1B. Black arrow indicates where wild-type (green) or S381F (red) or G248W (blue) CalDAG-GEFI was added. The increase in fluorescence intensity, a measure of nucleotide exchange, over time is shown. (B) A structural model of the S381F substitution using the mutagenesis feature in pymol. This model was built on the basis of the crystal structure of CalDAG-GEFII18 and represents phenylalanine substituted for serine at position 430, the homologous position of serine 381 of CalDAG-GEFI. On the basis of this model, there are 3 residues that clash with the phenylalanine substitution at S430: leucine 263, serine 305, and leucine 309.
The S381F substitution alters CalDAG-GEFI structure and markedly impairs its nucleotide exchange activity. (A) Bodipy fluorescence-based assay to monitor nucleotide exchange on purified Rap1B. Black arrow indicates where wild-type (green) or S381F (red) or G248W (blue) CalDAG-GEFI was added. The increase in fluorescence intensity, a measure of nucleotide exchange, over time is shown. (B) A structural model of the S381F substitution using the mutagenesis feature in pymol. This model was built on the basis of the crystal structure of CalDAG-GEFII18 and represents phenylalanine substituted for serine at position 430, the homologous position of serine 381 of CalDAG-GEFI. On the basis of this model, there are 3 residues that clash with the phenylalanine substitution at S430: leucine 263, serine 305, and leucine 309.
Neutrophil function studies
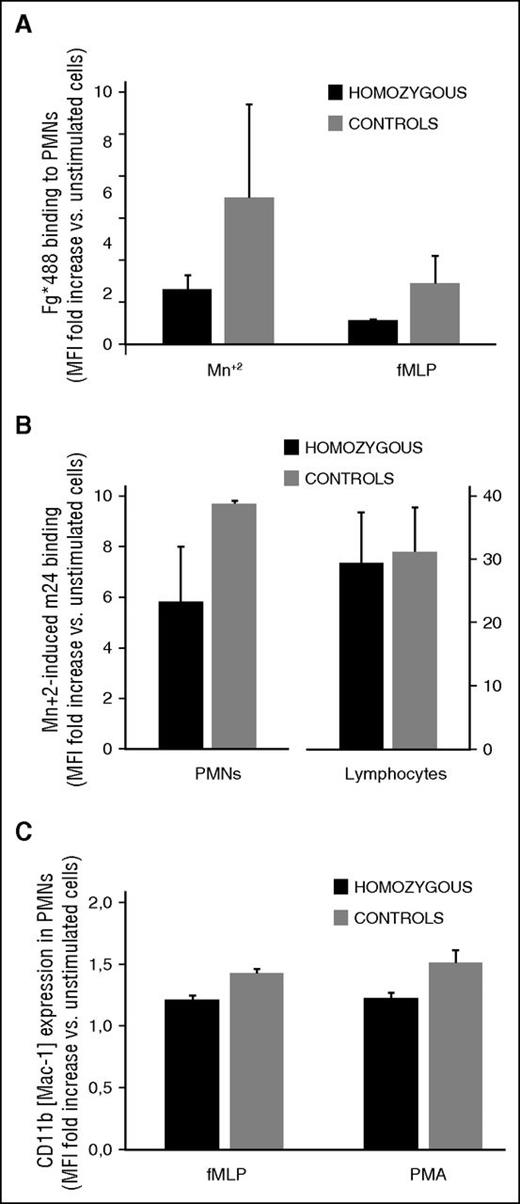
Studies in mutant mice identified a role for CalDAG-GEFI in neutrophil integrin affinity modulation and adhesion.10 We thus investigated whether neutrophil function was also affected in homozygous carriers of the CalDAG-GEFI S381F and R113X mutations (Figure 6). Compared with controls, neutrophils from homozygous patients showed normal β2 integrin and reduced β1 integrin expression (supplemental Table 2). β2 integrin activation was impaired in stimulated neutrophils from homozygous RASGRP2 patients (Figure 6A-B).
Impaired integrin activation in leukocytes from homozygous carriers of the CalDAG-GEFI S381F and R113X mutations. (A,B) β2 integrin activation. Neutrophils were kept resting or were stimulated with the indicated agonists in the presence of (A) Alexa Fluor 488-fibrinogen or (B) m24 antibody to determine the activation state of αMβ2 and αLβ2, respectively. C) Granule secretion. Neutrophils were kept resting or were stimulated with N-formylmethionyl-leucyl-phenylalanine (fMLP; 1 mM) or PMA (100 nM) in the presence of a phycoerythrin-labeled antibody to CD11b (Mac-1). Results are expressed as the mean fold increase, plus standard error, in median fluorescence intensity from data obtained in the 3 homozygous patients (black bars) and 3 healthy controls (gray bars).
Impaired integrin activation in leukocytes from homozygous carriers of the CalDAG-GEFI S381F and R113X mutations. (A,B) β2 integrin activation. Neutrophils were kept resting or were stimulated with the indicated agonists in the presence of (A) Alexa Fluor 488-fibrinogen or (B) m24 antibody to determine the activation state of αMβ2 and αLβ2, respectively. C) Granule secretion. Neutrophils were kept resting or were stimulated with N-formylmethionyl-leucyl-phenylalanine (fMLP; 1 mM) or PMA (100 nM) in the presence of a phycoerythrin-labeled antibody to CD11b (Mac-1). Results are expressed as the mean fold increase, plus standard error, in median fluorescence intensity from data obtained in the 3 homozygous patients (black bars) and 3 healthy controls (gray bars).
Thus, when activated with N-formylmethionyl-leucyl-phenylalanine, which causes inside-out activation, or by agents such as Mn2 that increase integrin affinity by direct and indirect (signaling) effects,19 patient neutrophils displayed a markedly impaired ability to bind fibrinogen (Figure 6A). Consistently, the binding of an antibody that detects the active conformation of lymphocyte function-associated antigen 1 (LFA-1 or m24) was also reduced in Mn+2-activated patient neutrophils when compared with controls (Figure 6B). In contrast, granule secretion was only minimally affected in neutrophils from homozygous patients (Figure 6C).
Discussion
Here we report on 2 families with bleeding diathesis and platelet dysfunction as a result of novel mutations in RASGRP2 that affect the expression and/or function of CalDAG-GEFI. This defect causes a markedly impaired aggregation response to low concentrations of certain agonists, most prominently ADP and collagen, and a delay in aggregation, even when stimulated with high concentrations of most agonists. Our findings, together with those of an earlier study,5 strengthen the molecular heterogeneity of RASGRP2 as a cause of inherited platelet disorder associated with impaired αIIbβ3 integrin activation.
In addition to the defect in platelet αIIbβ3 activation, we observed alterations in the expression or function of β1 and β2 integrins in neutrophils. Neutrophils from homozygous patients exhibited reduced surface expression of β1 integrins, a finding that is in accordance with results reported in Rasgrp2 knockout mice.10 Furthermore, patient neutrophils showed defects in agonist-induced inside-out activation of β2 integrins, confirming that CalDAG-GEFI is critical for the affinity modulation of various integrin subfamilies.10,20 The fact that fibrinogen binding to β2 integrins was also impaired in patient neutrophils stimulated with Mn+2 supports the concept that Rap1 is involved in both inside-out and outside-in signaling, as previously suggested.21 Interestingly, recent studies by Canault et al5 did not reveal any defects in integrin-mediated adhesion of neutrophils from patients with a loss-of-function mutation in RASGRP2. As discussed by the authors, the absence of an adhesion defect in CalDAG-GEFI (pG248W) neutrophils may be explained by the fact these cells express normal levels of mutant protein (ie, this mutation may spare structural domains that facilitate leukocyte adhesive functions). Further studies will be required to test this hypothesis.
Mice lacking CalDAG-GEFI are characterized by a very strong protection from experimental thrombosis, a moderate deficit in hemostasis, and a very mild immunodeficiency.6,8-10 Including the patients described in this article, we are now aware of a total of 6 patients (4 males and 2 females) with 3 distinct mutations in RASGRP2. All of the patients are characterized by impaired platelet aggregation to select agonists and markedly prolonged bleeding times. They all exhibit lifelong bleeding complications, including spontaneous bruising, petechiae, epistaxis, and gingival and gastrointestinal bleeding. Interestingly, these bleeding complications seem to be less severe in adults, such as the 46-year old proband in family 2 who has experienced few bleeding episodes during adulthood. None of the reported patients showed clinical signs of immunodeficiency. Because of the small number of patients, it is not possible to evaluate whether mutations in RASGRP2 also lead to protection from thrombotic complications associated with cardiovascular disease (in humans). This would seem likely, however, given the critical role of CalDAG-GEFI in platelet adhesion at high shear stress conditions and the fact that the Rap1 signaling response in human platelets is very similar to that in murine platelets.
In summary, we here report 2 novel mutations in RASGRP2 that lead to partially impaired integrin activation in platelets and neutrophils. Homozygous patients display a life-long, bleeding tendency, but they do not show signs of immunodeficiency. Only with the availability of increased numbers of patients will it become clear whether they are protected against thrombotic disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was conducted according to the aims of the Project “Functional and Molecular Characterization of Patients with Inherited Platelet Disorders” (approved by the Hemorrhagic Diathesis Working Group of the Spanish Society of Thrombosis and Haemostasis). The authors thank the families for providing samples and our clinical and laboratory colleagues for their help. The authors also thank Itziar Astigarraga, María Soler, Isabel Sanchez Guiu, Javier Corral, Constantino Martínez, José Padilla, and José Miguel Rivera Caravaca for their help and advice in handling clinical samples, platelet studies, molecular screening, and graph design.
Research by the group of J.R., M.L.L., F.F.-M., and V.V. is supported by grants from Instituto de Salud Carlos III and Feder (PI14/01956 and CB15/00055). Research by the group of N.M., S.P.W., S.J.F., and B.J. was conducted under the scope of the United Kingdom Genotyping and Phenotyping of Platelets study group and is supported by the British Heart Foundation (RG/ PG/13/36/30275; RG/09/007). W.B. is supported by National Institutes of Health National Heart, Lung, and Blood Institute grants P01 HL120846 and R01 HL121650.
Authorship
Contribution: M.L.L., S.P.W., W.B., and J.R. designed the research and wrote the paper; V.V., G.I., R.A.-P., and A.R.C. provided patient samples and clinical data; J.R., M.L.L., F.F.-M., and S.J.F. carried out the platelet phenotyping; A.C., D.S.P., J.S., J.R., and W.B. carried out protein analysis and cell modeling; J.M.B., J.R.G.-P., and J.M.H.-R. performed the next-generation sequencing gene panel; N.M. and B.J. performed whole-exome sequencing and Sanger sequencing; and all authors critically reviewed and approved the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.L.L., F.F.-M., V.V., and J.R. is Grupo de investigación CB15/00055 del Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), Instituto de Salud Carlos III (ISCIII), Madrid, Spain.
Correspondence: José Rivera, Centro Regional de Hemodonación-Universidad de Murcia, Ronda de Garay s/n, Murcia 30003, Spain; e-mail: jose.rivera@carm.es; and Wolfgang Bergmeier, University of North Carolina, 120 Mason Farm Rd, Campus Box 7260, Chapel Hill, NC 27599; e-mail: bergmeie@email.unc.edu.

![Figure 1. Platelet aggregation and integrin activation is markedly impaired in index cases with lifelong bleeding diathesis from 2 unrelated families. (A) Platelet aggregation in response to the indicated platelet agonists was evaluated in unadjusted PRP from index cases (P) and a healthy and unrelated control (C). (B) Platelets from index cases and healthy and unrelated controls (controls) (combined data from 3 participants) were stimulated under static conditions (30 minutes at room temperature) with agonist (5 and 25 μM ADP [ADP-5 and ADP-25], 25 μM PAR1p, 250 μM PAR4p, 2 and 10 μg/mL collagen-related peptide [CRP-2 and CRP-10], 2 μg/mL convulxin [Cvx], and 100 nM PMA) in the presence of fibrinogen-Alexa 488. The samples were evaluated by flow cytometry and the median fluorescence intensity (MFI) for fibrinogen-A488 binding is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/9/10.1182_blood-2015-11-683102/4/m_1282f1.jpeg?Expires=1766051794&Signature=4qAFcpibgU1J1L8xQjuWhAqvwehdhgwliTY-oO-Ph7jwwqy2bfjaQIpKHNnIKh9QoCXPFUOQNsMvtzZ9Qmy3obQrNlQptTCoyj3VMXALSCoTqcouKqrqIMHbSInCqNO2ghp~ijlpBc8TV8JTUrKJoRlqpfR-9buIEIZnAgnZPdDpKBZpuNeeOmPtsvELhTEeTNqYPdgn0eRLdtYy5RyXZL2ngeBjkZC2wVhoPVQA4miOBuvf~UD5AAUTZr5hd0l5Ld0QeSxVZP6RiGZHCWlFUj7T5L6iCRqI0bsWZKiNf-6VE1O1PrRrKQ8OJUnINFHMTJnG2W2Sznsc5kWdrLIydw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)