To the editor:
Large granular lymphocyte (LGL) leukemia is a group of chronic lymphoproliferative disorders of cytotoxic T or natural killer (NK) cells frequently complicated with cytopenia and autoimmune phenomena.1,2 In the current World Health Organization (WHO) classification, T-LGL leukemia and chronic lymphoproliferative disorder of NK cells (CLPD-NK) are included in this category.3
Recurrent somatic mutations in the Src homology 2 (SH2) domain of the signal transducer and activator of transcription 3 (STAT3) gene have been found in T-LGL leukemia and CLPD-NK,4,5 leading to constitutive activation of STAT3 and dysregulation of genes downstream of STAT3. More recently, mutations outside the SH2 domain have been discovered in T-LGL leukemia.6 Activating mutations in the SH2 domain of the STAT5B gene were also identified in 2% of LGL leukemia patients,7 which further underlines the importance of the JAK/STAT signaling pathway in LGL leukemia.
The majority of T-LGL leukemia cases present with a clonal expansion of the CD8+ LGLs. However, in a small percentage of cases, the tumor cells have a CD4+ phenotype.8-10 Cytomegalovirus-derived stimulation and restricted use of the T-cell receptor (TCR)-Vβ region has been associated with CD4+ T-LGL cases,11 but this rare disease entity still remains poorly described. To further elucidate the pathogenesis of this rare subgroup of T-LGL leukemia, we explored the mutational landscape of CD4+ cases using exome and targeted amplicon sequencing. Patients diagnosed with T-LGL leukemia and CLPD-NK were recruited. The diagnostic criteria were based on the WHO classifications of 2008. Three patient cohorts (described in detail in the supplemental Appendix, available on the Blood Web site) were included in this study.
Exome sequencing was performed on 3 CD4+ T-LGL leukemia patients’ sorted tumor (CD4+ or CD4+CD8+ T cells) and control (CD4−) fractions. The exome was captured with Nimblegen SeqCap EZ Exome Library v2.0, and sequencing was performed with the Illumina HiSeq2000 sequencing platform. Candidate somatic mutations were identified with a bioinformatics pipeline described earlier,4 as well as a novel pipeline described in more detail in the supplemental Appendix. Through exome sequencing, we were able to identify novel somatic missense mutations in the transactivation domain of STAT5B in 2 CD4+ T-LGL leukemia patients. Patient 1 had a Q706L mutation at a variant allele frequency (VAF) of 45% in the CD4+CD8+ tumor fraction. Patient 2 displayed an S715F mutation (VAF, 36%) in the CD4+ fraction (Figure 1A). Only wild-type (WT) STAT5B was observed in the CD4− fractions, confirming that the mutations were somatic. The third patient with CD4+ T-LGL leukemia did not show any mutations in STAT5B or STAT3 genes, but mutations in members of the protein tyrosine phosphatase family (PTPN14, PTPN23) regulating cell proliferation and tumor suppressor MLL2 were observed (supplemental Table 3).
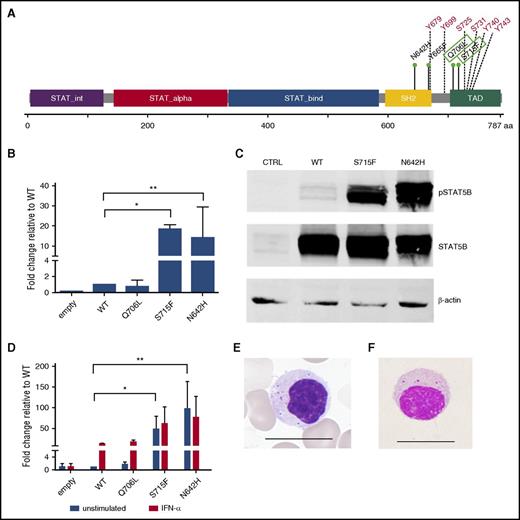
STAT5B mutation characterization. (A) Linear representation of the STAT5B protein structure. Previously known LGL leukemia mutations in STAT5B are marked in the SH2 domain, whereas the novel Q706L and S715F mutations in the transactivation domains are marked with green boxes. Multiple tyrosine and serine phosphorylation sites are marked in red. (B) STAT5B reporter assay results. Mutated STAT5B constructs (pCMV6-XL6 STAT5B) were generated through site-directed mutagenesis followed by transfection and expression of WT and mutated STAT5B (Q706L, S715F, N642H) in HeLa cells together with a STAT5B reporter. Dual-reporter luciferase assay was used to determine activation and phosphorylation of mutated STAT5B. The experiment was repeated 3 times. Columns represent mean of the fold-change activity. Error bars indicate the standard error of the mean (SEM), and the statistical significance was calculated with a 1-way analysis of variance (ANOVA; *P < .05, **P < .001). (C) To investigate the phosphorylation status of the variants, HeLa cells transfected with the abovementioned variants were analyzed by western blot with a phosphoSTAT5 (Tyr694) specific antibody. Protein lysates of the different variants were separated on an sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and transferred to a nitrocellulose membrane. STAT5 protein levels of the different variants were used to normalize for the transfection efficacy. β-Actin was used as a loading control. (D) Transfected HeLa cells were stimulated with 100 ng/mL interferon-α for 6 hours. A dual-reporter luciferase assay was used to determine activation and phosphorylation of mutated STAT5B. The experiment was repeated 2 times. Columns represent mean of the fold-change activity. Error bars indicate the SEM, and the statistical significance was calculated with a 1-way ANOVA (*P < .05, **P < .001). (E) Typical morphology of a representative LGL cell in a STAT5B mutated T-LGL patient. Scale bar, 15 μm. (F) Morphology of lymphocyte expressing CD4, CD56, and TCRαβ in a healthy individual. CD4+CD56+ and TCRαβ-type lymphocytes were sorted by the FACS method and stained with Wright-Giemsa stain. A representative cell is shown. Scale bar, 15 μm.
STAT5B mutation characterization. (A) Linear representation of the STAT5B protein structure. Previously known LGL leukemia mutations in STAT5B are marked in the SH2 domain, whereas the novel Q706L and S715F mutations in the transactivation domains are marked with green boxes. Multiple tyrosine and serine phosphorylation sites are marked in red. (B) STAT5B reporter assay results. Mutated STAT5B constructs (pCMV6-XL6 STAT5B) were generated through site-directed mutagenesis followed by transfection and expression of WT and mutated STAT5B (Q706L, S715F, N642H) in HeLa cells together with a STAT5B reporter. Dual-reporter luciferase assay was used to determine activation and phosphorylation of mutated STAT5B. The experiment was repeated 3 times. Columns represent mean of the fold-change activity. Error bars indicate the standard error of the mean (SEM), and the statistical significance was calculated with a 1-way analysis of variance (ANOVA; *P < .05, **P < .001). (C) To investigate the phosphorylation status of the variants, HeLa cells transfected with the abovementioned variants were analyzed by western blot with a phosphoSTAT5 (Tyr694) specific antibody. Protein lysates of the different variants were separated on an sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and transferred to a nitrocellulose membrane. STAT5 protein levels of the different variants were used to normalize for the transfection efficacy. β-Actin was used as a loading control. (D) Transfected HeLa cells were stimulated with 100 ng/mL interferon-α for 6 hours. A dual-reporter luciferase assay was used to determine activation and phosphorylation of mutated STAT5B. The experiment was repeated 2 times. Columns represent mean of the fold-change activity. Error bars indicate the SEM, and the statistical significance was calculated with a 1-way ANOVA (*P < .05, **P < .001). (E) Typical morphology of a representative LGL cell in a STAT5B mutated T-LGL patient. Scale bar, 15 μm. (F) Morphology of lymphocyte expressing CD4, CD56, and TCRαβ in a healthy individual. CD4+CD56+ and TCRαβ-type lymphocytes were sorted by the FACS method and stained with Wright-Giemsa stain. A representative cell is shown. Scale bar, 15 μm.
To study the functional properties of the novel variants, we generated STAT5B expression vectors for WT, Q706L, and S715F mutations and previously described activating N642H mutation.7 The transcriptional activity of the mutants was studied with luciferase reporter assays with and without interferon-α stimulation, and the phosphorylation status was analyzed by western blotting. In HeLa cells, the mutated STAT5B S715F construct significantly enhanced the transcription of the cotransfected STAT5 reporter (18-fold compared with WT STAT5B) similarly to the N642H mutation (Figure 1B), whereas the Q706L mutation activation was equal to WT. In the western blot analysis, S715F and N642H mutations showed significantly increased phosphorylation compared with WT STAT5B (Figure 1C), whereas no increased phosphorylation was observed with the Q706L mutation. The location of the novel S715F mutation in a serine phosphorylation site is likely to increase the phosphorylation of STAT5B. Stimulation with interferon-α revealed that the Q706L mutation behaved as the WT, whereas stimulation was not able to further increase the transcriptional activity of the S715F and N642H mutants (Figure 1D).
To elucidate whether STAT5B mutations are more prevalent in CD4+ T-LGL leukemia cases, deep amplicon sequencing was used for screening of the SH2 and transactivation domains of STAT5B in CD4+ (n = 8), STAT3-mutated CD8+ (n = 37) and nonmutated CD8+ (n = 58) T-LGL leukemia patients. Targeted STAT5B amplicon sequencing covering exons 14 to 19 was done with an in-house–developed deep amplicon sequencing panel using the Illumina Miseq platform.7 The data were analyzed with a bioinformatics pipeline described previously.12 A variant was called when the variant base frequency was 0.5% of all reads covering a given a position. Additionally, the same regions were screened with Sanger sequencing in Japanese and Chinese LGL leukemia cohorts consisting of CD8+ and CLPD-NK cases (n = 57). None of the patients with CD8+ T-LGL leukemia or CLPD-NK had STAT5B mutations. In contrast, 4 of 8 CD4+ T-LGL leukemia cases had STAT5B mutations. Of the 4 patients with STAT5B mutations, 3 possessed the earlier described N642H mutation and 1 the Y665F mutation. Sanger sequencing–negative patients and healthy controls (n = 50) were also screened with allele-specific PCR for N642H and Y665F mutations, but no additional mutations were found. Altogether, the STAT5B mutation frequency in CD4+ T-LGL leukemia patients in our cohort was 55% (6 of 11 patients). This is significantly higher than in the previous study (2%) of 211 CD8+ T- and NK-cell LGL leukemia cases where STAT5B SH2 domain mutations were initially discovered.7 Most of the STAT5B mutations found in CD4+ T-LGL leukemia have also been seen in various T-cell neoplasms, including γδ hepatosplenic T-cell lymphoma,13 T-cell acute lymphoblastic leukemia,14,15 T-cell prolymphocytic leukemia,16 type II enteropathy-associated T-cell lymphoma,17 and extranodal NK/T-cell lymphoma,18 suggesting that these are shared with other T-cell malignancies. The analyses of STAT5 target genes with chromatin immunoprecipitation sequencing have shown that STAT5B is a key factor in T-cell development, binding to molecules such as DOCK8, SNX9, FOXP3, and IL2RA.19 Together these results suggest that the STAT5B pathway plays a central role in the development of T-cell neoplasms.
In contrast to other more aggressive T-cell malignancies with STAT5B mutations, the disease course in our CD4+ T-LGL leukemia cohort was indolent, and none of the patients with STAT5B mutations needed therapy during the observation time (median follow-up, 4 years). Rheumatoid arthritis (RA) is commonly associated with CD8+ T-LGL leukemia, and especially patients with multiple STAT3 mutations more often have RA.12 In our cohort, none of the 11 cases with CD4+ T-LGL leukemia suffered from RA. Two patients showed neutropenia and 1 patient had anemia (Table 1).
All STAT5B mutated CD4+ T-LGL cases possessed a TCRαβ T-cell phenotype with CD16−CD56+ and CD57+ (Figure 1E). Two cases were CD8−, 2 were weakly positive for CD8, and 2 were clearly positive for CD8 (supplemental Table 4). This is in accordance with the earlier reports8-10 of monoclonal CD4+ T-LGL cells, which have shown expression of TCRαβ, variable levels of CD8, and a typical cytotoxic (granzyme B+, CD56+, CD57+, CD11b+/−) and activated/memory T-cell (CD2+bright, CD7−/+dim, CD11a+bright, CD28−, CD62L−HLA-DR+) phenotype. Interestingly, all 6 patients with STAT5B mutations had large monoclonal TCR-Vβ expansions where the mutations were located, whereas significant proportions of STAT3 mutations in CD8+ T-LGL leukemia and CLPD-NK are detected in small subclones.
Because the CD4+CD56+TCRαβ+ immunophenotypes recognized on STAT5B-mutated T-LGL leukemia cells have been poorly defined, we also investigated whether normal lymphocytes with similar phenotypic features exist in peripheral blood of healthy subjects. Among 27 healthy controls, the median percentage of CD4+CD56+TCRαβ+ T cells in lymphocytes was 0.2, and it varied from less than 0.02% to 6.5% (supplemental Figure 2). Fluorescence-activated cell sorter (FACS)-sorted CD4+CD56+TCRαβ cells possessed LGL morphology with cytoplasmic azurophilic granules (Figure 1F; N = 3). Thus, phenotypically similar cells as observed in CD4+ T-LGL leukemia cases can also be observed in healthy individuals in small quantities. However, deep amplicon sequencing of sorted CD4+CD56+ cells from 5 healthy subjects revealed no mutations in the SH2 or transactivation domains of STAT5B.
In conclusion, activating STAT5B mutations can be found in the majority (55%) of CD4+ T-LGL leukemia cases, whereas among patients with CD8+ T-LGL leukemia or CLPD-NK, these are very rare. STAT5B mutations can be considered as a novel diagnostic marker for this specific disease subtype.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: Personnel at the Hematology Research Unit Helsinki and Institute for Molecular Medicine Finland are acknowledged for their expert clinical and technical assistance.
This work was supported by the European Research Council (M-IMM), Academy of Finland, the Finnish Cancer Institute, the Finnish Cancer Societies, the Signe and Ane Gyllenberg Foundation, Sigrid Juselius Foundation, Instrumentarium Science Foundation, Biocentrum Helsinki, state funding for university-level health research in Finland, Swedish Cultural Foundation, Blood Disease Foundation, the Finnish Cultural Foundation, and Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Work of J.P.M. was supported in part by National Institutes of Health National Cancer Institute grants 2K24HL077522, R01 CA127264A, and R01AI085578. Work of T.P.L. was supported by National Cancer Institute grants RO1 CA098472 and RO1 CA178393. Work of Stefania Bortoluzzi. was supported by Cassa di Risparmio di Padova e Rovigo Foundation (Excellence projects 2011/2012), Italian Ministry of Education, Universities and Research (PRIN 2010/11 2010NYKNS7_002), and University of Padova. A.B. is recipient of a fellowship of the Program in Biosciences of the University of Padova.
Contribution: E.I.A., T.T., S.M., and F.I. designed the study, coordinated the project, analyzed the data, and wrote the paper; E.I.A., T.T., T.K., S.L., K.M., and P.E. performed sequence analysis and validated mutations; E.I.A., V.R.G., and Sabrina Bortoluzzi designed and performed the functional experiments; S.E., Stefania Bortoluzzi, A.C., and A.B. designed and performed the bioinformatics analysis; N.S., T.M., N.F., S.N., N.S., H.S., H.N., Y-L.K., T.P.L., and J.P.M. provided patient samples; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: S.M. has received honoraria and research funding from Novartis, Pfizer, and Bristol-Myers Squibb and research funding from Ariad. F.I. has received research funding from Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Satu Mustjoki, Hematology Research Unit Helsinki, University of Helsinki, Haartmaninkatu 8,00290 Helsinki, Finland; e-mail: satu.mustjoki@helsinki.fi; or Fumihiro Ishida, Department of Biomedical Laboratory Sciences, Shinshu University School of Medicine, 3-1-1, Matsumoto, Nagano 3908621, Japan; e-mail: fumishi@shinshu-u.ac.jp.
References
Author notes
E.I.A. and T.T. contributed equally to this work.
S.M. and F.I. are joint senior authors.