Key Points
CSF1R is expressed on the earliest fetal B-cell progenitors, and CSF1R deficiency impairs fetal B-cell development.
CSF1R+ fetal ProB cells have a B-myeloid gene signature and possess B and myeloid potential.
Abstract
Although it is well established that unique B-cell lineages develop through distinct regulatory mechanisms during embryonic development, much less is understood about the differences between embryonic and adult B-cell progenitor cells, likely to underpin the genetics and biology of infant and childhood PreB acute lymphoblastic leukemia (PreB-ALL), initiated by distinct leukemia-initiating translocations during embryonic development. Herein, we establish that a distinct subset of the earliest CD19+ B-cell progenitors emerging in the E13.5 mouse fetal liver express the colony-stimulating factor-1 receptor (CSF1R), previously thought to be expressed, and play a lineage-restricted role in development of myeloid lineages, and macrophages in particular. These early embryonic CSF1R+CD19+ ProB cells also express multiple other myeloid genes and, in line with this, possess residual myeloid as well as B-cell, but not T-cell lineage potential. Notably, these CSF1R+ myeloid-primed ProB cells are uniquely present in a narrow window of embryonic fetal liver hematopoiesis and do not persist in adult bone marrow. Moreover, analysis of CSF1R-deficient mice establishes a distinct role of CSF1R in fetal B-lymphopoiesis. CSF1R+ myeloid-primed embryonic ProB cells are relevant for infant and childhood PreB-ALLs, which frequently have a bi-phenotypic B-myeloid phenotype, and in which CSF1R-rearrangements have recently been reported.
Introduction
B-lymphoid cells are generated from pluripotent, self-renewing hematopoietic stem cells (HSCs) by a complex differentiation process in the liver during fetal development and in the bone marrow (BM) after birth. Several molecular and functional aspects distinguish fetal from adult B lymphopoiesis, such as interleukin 7 (IL7) dependency1,2 and the ability of fetal but not adult progenitors to produce B1-a cells, critically involving Lin28b, which is exclusively expressed in fetal progenitors.3,4
B-lymphopoiesis in the embryo has been suggested to be closely linked with the myeloid lineage, including the identification of fetal liver (FL) progenitor cells with a combined B-cell and macrophage lineage potential,5 although these progenitors were later shown to also partially possess T-cell potential.6 A rare B220–CD19+ bipotential B–macrophage progenitor has also been implicated in adult BM,7 but the exact identity and developmental origin of such a progenitor remain unclear. Further evidence for a link between fetal lymphoid and myeloid lineage development derived from studies suggesting the existence of bipotential lympho-myeloid precursors in the FL giving rise to either T and myeloid, or B and myeloid cells,8 although each of these proposed fetal B-myeloid and T-myeloid precursors have yet to be prospectively identified and characterized. Moreover, overexpression of myeloid lineage transcription factors or the colony-stimulating factor 1 receptor (CSF1R) has been shown to promote a switch from a B cell to a macrophage lineage fate.9,10
The CSF1R, also known as macrophage colony-stimulating factor receptor (M-CSFR), is a member of the tyrosine kinase receptor family class III, and together with its ligand (CSF1L), has a critical role in regulating the survival, proliferation, and differentiation of myeloid cells, in particular the macrophage lineage, as well as bone formation.11,12 The loss of CSF1R in mice results in a postnatal morbidity and lethality caused by an osteopetrotic phenotype precluding or complicating studies of postnatal hematopoiesis.12 In adult hematopoiesis, CSF1R has been shown to be highly and selectively expressed on myeloid and dendritic cells, but to be undetectable on lymphocytes, including progenitors and mature cells of the B-cell lineage.13,14 Recently early lympho-myeloid progenitor cells in the mouse embryo, possessing combined B, T, and myeloid-lineage potential, but not megakaryocyte-erythroid–lineage potential, were found to coexpress CSF1R and IL7 receptor (IL-7R),15,16 suggesting that CSF1R might not exclusively play a role in myelopoiesis, and a potential role in early embryonic lympho-myeloid development. Thus, although a number of studies have provided support for a strong link between fetal B and myeloid lineage development and the potential existence of a fetal B myeloid–restricted progenitor, such a progenitor has yet to be prospectively identified and characterized. Moreover, the potential expression and role of CSF1R in fetal B-cell or B-myeloid progenitors lacking T-cell potential has yet to be investigated.
A potentially close developmental connection between the B and myeloid lineages during fetal development is highly relevant for infants carrying chromosomal translocations of the mixed lineage leukemia (MLL) gene diagnosed with a B acute lymphoblastic leukemia (B-ALL), which notably can switch to an acute myeloid leukemia (AML) at relapse.17,18 In addition, biphenotypic leukemias with characteristics of the B-lymphoid as well as the myeloid lineage have been reported in childhood and infant B-ALL,19 and the ample evidence of an in utero origin of the translocations frequently seen in infant and childhood B-ALL supports that the target cell for these leukemia-initiating genomic lesions might be a distinct fetal progenitor cell.20 Of particular relevance for our studies, several cases have been reported in which CSF1R is rearranged in childhood PreB B-ALL.21-23
In light of these findings, we investigated the potential expression and role of CSF1R in normal fetal B lymphopoiesis by specifically investigating its expression in the FL CD19+ ProB-cell progenitor compartment. Herein we demonstrate that CSF1R is expressed and involved in regulation of a distinct and developmentally highly restricted early myeloid-primed fetal B-cell progenitor with residual myeloid lineage potential.
Methods
Animals
Wild-type (Wt) C57BL/6 (CD45.1 or CD45.2) (JBmsd) mice were obtained from the Jackson Laboratory. Csf1r−/− embryos and Wt (Csf1r+/+) littermate controls were generated by breeding Csf1r+/− mice (on C57BL/6 background) that were kindly provided by E. Richard Stanley.12 Flk2−/− mice were kindly provided by lhor R. Lemischka24 and were on C57BL/6 background. Csf1r−/−Flk2−/−and Flk2−/− littermate embryos used in experiments were generated by breeding of Csf1r+/−Flk2−/− mice. For timed pregnancies, mice were mated late afternoon and females were checked the following morning for the presence of a vaginal plug designated as embryonic day 0.5 (E0.5). All mice were maintained under specific pathogen-free conditions at Lund University Animal Facility. The Ethical Committee at Lund University approved all the experimental procedures and performed studies.
Dissections and cell preparations
The FL (E13.5, E14.5, and E17.5) and fetal spleen (E17.5) were dissected and mechanically disrupted with a syringe. BM cells were extracted using a mortar. Single-cell suspensions were prepared in phosphate-buffered saline (Thermo Scientific) containing 5% of fetal bovine serum (FBS) (Hyclone) and filtered through a 70-μm cell strainer (BD Biosciences). Cells were counted with the Sysmex (KX-21N) hematology analyzer or manually in a Neubauer chamber with trypan blue.
Flow cytometry and fluorescence-activated cell sorting
Dissected fetal tissues and adult BM cells were treated with purified anti-CD16/32 antibody (Fc-block) and then stained with specific mouse monoclonal antibodies (mAb). mAbs used to stain cell-surface markers are listed in supplemental Table 1, available on the Blood Web site. Fluorescence-minus-one (FMO) controls, isotype controls or Csf1r−/− embryos were used to determine the positive signals (supplemental Figure 1A-B). 7-aminoactinomycinD (7-AAD, Sigma-Aldrich) or TO-PRO-1 iodide (1 mM, Invitrogen) were used to exclude dead cells from the analysis. Samples were analyzed on an LSRII (BD Biosciences) and analysis was performed using FlowJo software (version 9.3; TreeStar). All sorts were performed on a BD FACSAriaIIu (BD Biosciences) with purity reproducibly >94%. Single cells were index-sorted using a single cell depositor. For all the displayed flow cytometry profiles, singlet viable cells were first gated as lineage-negative, and further gating is indicated with arrows.
Cytokine response assay
For cytokine response studies, single cells of indicated cell populations were plated using the single-cell depositor unit on an AriaIIu (Becton Dickinson) directly into Terasaki plates containing X-vivo15 medium (BioWhittaker), supplemented with 1% penicillin/streptomycin (Sigma-Aldrich), 1% l-glutamine (Sigma-Aldrich), 1% 10−2 M 2-β-mercaptoethanol (Sigma-Aldrich), 10% FBS, and 50 ng/mL human CSF1L (PreproTech). Wells were scored, with an inverted microscope, for clonal growth after 7 days of culture.
In vitro evaluation of lineage potentials
For evaluation of lineage potential, 20 cells per well were plated onto ∼80% confluent monolayers of OP9 or OP9-DL1 stroma cells in OPTIMEM (Gibco) medium supplemented with 1% penicillin/streptomycin, 1% 10−2 M 2-β-mercaptoethanol, 10% FBS, and cytokines: 25 ng/mL stem cell factor, 25 ng/mL FLT3 Ligand (FLT3L), and 20 ng/mL IL7 (PreproTech) for OP9 and 25 ng/mL stem cell factor (only first week) and 25 ng/mL FLT3L for OP9-DL1. Cells were analyzed by fluorescence-activated cell sorting (FACS) after 7 days for B-cell and myeloid potential and after 14 to 16 days for T-cell potential. Wells were scored as positive for proliferation when containing >30 viable cells, positive for B-cell potential when >20 cells were Topro–NK1.1–B220+CD19+, positive for myeloid potential when >10 cells were Topro–NK1.1–B220–CD19–Gr1+Mac1+, and positive for T-cell potential when >10 cells were Topro–NK1.1–B220–MAC1–GR1–CD25+THY1.2+, and/or Topro–NK1.1–B220–MAC1–GR1–CD4+CD8a+.
Gene expression profiling
For bulk gene expression analysis, 25 cells were sorted directly into 96-well plates containing: 2.5 μL gene-specific 0.2× TaqMan gene expression assays (Applied Biosystems), 5 μL of CellsDirect 2× Reaction mix (Invitrogen), 1.2 μL SSIII/PlatinumTaq mix, 1.2 μL TE buffer, and 0.1 μL SUPERase-In RNase Inhibitor (Ambion) followed by reverse-transcription polymerase chain reaction (RT-PCR) and preamplification. For single-cell gene expression analysis, single cells were index-sorted directly into 96-well plates containing: 4 μL of lysis buffer containing 0.4% NP40, deoxynucleoside triphosphates, dithiothreitol, and RNase OUT (Invitrogen), and snap frozen. CellsDirect reaction mix containing SSIII/PlatinumTaq and 48 TaqMan assays to a final dilution of 0.05× each were added to the thawed cell lysate for RT-PCR and preamplification. Preamplified products (25 cycles for single cells and 22 cycles for bulk) were diluted fivefold in TE buffer before real-time quantitative PCR (qPCR) analysis was performed on individual samples using the BioMark 48.48 Dynamic Array platform (Fluidigm) according to the manufacturer’s instructions. TaqMan primers used for the analysis are shown in supplemental Table 2. Data were analyzed using the ΔCt method; results were normalized to hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1) expression and presented as mean expression level relative to Hprt1. Gene filter: (1) for each gene, including controls, data with CtCall = FAILED and CtQuality < threshold were removed; (2) for each gene, including controls, CtValues ≥ 32.0 were removed to filter out very low expression genes; (3) for each gene, a difference of <2 cycles between the CtValues compared with the CtValue of the no RT control samples were removed; and (4) samples with a nondetectable level of Hprt1 were removed.
Transplantation assay
Lethally irradiated (900 cGy) 14- to 16-week-old C57BL/6 CD45.1 Wt recipient mice were injected IV with, for competitive transplantation assay, 400 000 unfractionated Csf1r−/− (CD45.2) or Wt Csf1r+/+ littermate (CD45.2) E14.5 FL cells together with 400 000 unfractionated Wt CD45.1 competitor E17.5 FL cells or for noncompetitive transplantation, 5 × 106 unfractionated Csf1r−/− (CD45.2) or Wt Csf1r+/+ littermate (CD45.2) E14.5 FL cells. Seven or 16 weeks after transplantation, donor-derived reconstitution was analyzed in the BM.
Statistics
Prism software (GraphPad Software) was used for all statistical analysis. Statistical significances were determined using an unpaired Mann-Whitney U test. Group comparison was done with a Kruskal-Wallis test and followed by a multiple comparisons using Dunn’s correction. The significance level was set at P < .05.
Results
CSF1R is expressed on ProB cell progenitors in the early fetal liver
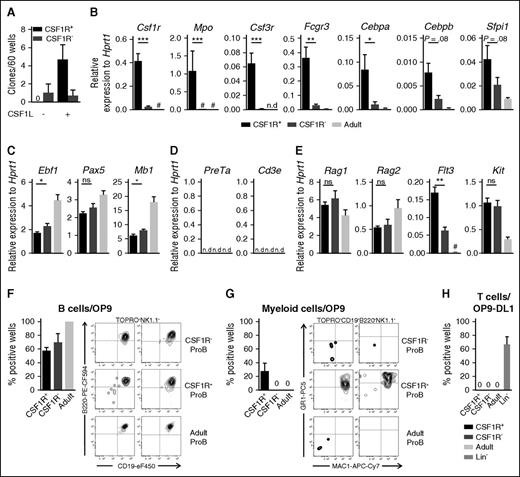
To assess the potential role of CSF1R in regulation of B lymphopoiesis, we investigated whether it is expressed on B-cell progenitors. We analyzed the earliest B cell–committed progenitors, ProB cells (Lin– B220+CD43+CD19+CD24+CD93+), at E13.5, which is the first time when ProB cells can be detected in the FL,16,25,26 and we compared the expression pattern for Csf1r with the adult BM counterpart. Notably, whereas Csf1r was undetectable in adult BM ProB cells, it was reproducibly expressed in E13.5 FL ProB cells (Figure 1A). We next analyzed the surface expression of CSF1R at different stages of ontogeny using flow cytometry. In the E13.5 FL, we found that >20% of ProB cells expressed CSF1R (Figure 1B-C), which decreased to 4.1% at E14.5 (Figure 1B-C). In agreement with the lack of transcriptional expression, we found also cell surface CSF1R expression to be absent in adult BM ProB cells, and in fact already in E17.5 FL ProB cells (Figure 1B-C). We also found no CSF1R expression on the PreB cells (Lin–B220+CD43–CD19+IgM–) in the E17.5 FL and in adult BM (Figure 1B). We confirmed the specificity of the anti-CSF1R staining on ProB cells by analyzing FL ProB cells from littermate CSF1R-deficient (Csf1r−/−) embryos or using isotype control antibodies as a negative control (supplemental Figure 1A-B).
CSF1R is expressed on early B-cell progenitors in the fetal liver but not in adult bone marrow. (A) Mean (± standard error of the mean [SEM]) Csf1r expression levels measured by RT-qPCR in 20 E13.5 FL (n = 7 embryos) and 8- to 10-week-old adult BM (n = 6 mice) ProB (Lin–B220+CD43+CD19+CD24+CD93+) cells (2-3 experiments). Results are presented relative to Hprt1 expression. **P < .01. (B) Representative FACS profiles of CSF1R expression on ProB at E13.5, E14.5, and on ProB and PreB (Lin–B220+CD19+CD43–IgM– cells) at E17.5 FL and in 8- to 10-week-old adult BM (2-5 experiments). (C) Mean percentage (± SEM) of ProB cells expressing CSF1R at E13.5 (n = 26 embryos), E14.5 (n = 21 embryos), E17.5 (n = 12 embryos), and in 8- to 10-week-old adult BM (n = 17 mice). (D) Representative FACS profiles of CSF1R expression on HSCs (Lin–KIT+SCA-1+CD48–CD150+FLT3–) and LMPPs (Lin–KIT+SCA-1+FLT3high) in E13.5 FL and 8- to 10-week-old adult BM (2-5 experiments). (B, D) Numbers represent mean percentage of CSF1R+ cells in the investigated populations. (E) Mean percentage (± SEM) of HSCs and LMPPs expressing CSF1R at E13.5 (n = 21 embryos), E14.5 (n = 13-30 embryos), and 8- to 10-week-old adult BM (n = 6-17 mice). (F) Gene expression analysis of single E13.5 FL CSF1R+ ProB and CSF1R+ FL LMPP cells expressing Csf1r mRNA (2-3 experiments). Results are presented relative to Hprt1 expression. Each column represents a single cell.
CSF1R is expressed on early B-cell progenitors in the fetal liver but not in adult bone marrow. (A) Mean (± standard error of the mean [SEM]) Csf1r expression levels measured by RT-qPCR in 20 E13.5 FL (n = 7 embryos) and 8- to 10-week-old adult BM (n = 6 mice) ProB (Lin–B220+CD43+CD19+CD24+CD93+) cells (2-3 experiments). Results are presented relative to Hprt1 expression. **P < .01. (B) Representative FACS profiles of CSF1R expression on ProB at E13.5, E14.5, and on ProB and PreB (Lin–B220+CD19+CD43–IgM– cells) at E17.5 FL and in 8- to 10-week-old adult BM (2-5 experiments). (C) Mean percentage (± SEM) of ProB cells expressing CSF1R at E13.5 (n = 26 embryos), E14.5 (n = 21 embryos), E17.5 (n = 12 embryos), and in 8- to 10-week-old adult BM (n = 17 mice). (D) Representative FACS profiles of CSF1R expression on HSCs (Lin–KIT+SCA-1+CD48–CD150+FLT3–) and LMPPs (Lin–KIT+SCA-1+FLT3high) in E13.5 FL and 8- to 10-week-old adult BM (2-5 experiments). (B, D) Numbers represent mean percentage of CSF1R+ cells in the investigated populations. (E) Mean percentage (± SEM) of HSCs and LMPPs expressing CSF1R at E13.5 (n = 21 embryos), E14.5 (n = 13-30 embryos), and 8- to 10-week-old adult BM (n = 6-17 mice). (F) Gene expression analysis of single E13.5 FL CSF1R+ ProB and CSF1R+ FL LMPP cells expressing Csf1r mRNA (2-3 experiments). Results are presented relative to Hprt1 expression. Each column represents a single cell.
Next, we assessed whether CSF1R is expressed in the fetal multipotent stem and progenitor cells preceding B-cell lineage commitment. In agreement with previous studies,15,27 23% and 9% of lymphoid-primed multipotent progenitors (LMPPs; Lin–KIThighSCA-1highFLT3high) expressed CSF1R in E13.5 and E14.5 FL, respectively, whereas 6% of LMPPs in adult BM were CSF1R-positive (Figure 1D-E). In contrast, there was almost no detectable CSF1R expression on HSCs (Lin–KIThighSCA-1highCD48–CD150+FLT3–) in the FL or in adult BM (Figure 1D-E).
To unequivocally establish that CSF1R+ E13.5 FL ProB cells (Lin–CD43+B220+CD19+CD24+CD93+) were indeed B-cell progenitors, we sorted E13.5 single CSF1R+ and CSF1R– ProB cells to high purity (supplemental Figure 1C), as well as CSF1R+ LMPPs and performed single-cell multiplex gene expression analysis. The majority of sorted single E13.5 CSF1R+ ProB cells and CSF1R+ LMPPs, which expressed Csf1r mRNA, coexpressed the receptors Il7r, Kit, and Flt3, known to be important for early B lymphopoiesis,1,24,28,29 as well as the early lymphoid genes Rag1 and Rag2, while being devoid of the megakaryocyte and erythroid genes Gata1 and Vwf (Figure 1F). In contrast, B lineage–specific genes Mb1, Pax5, Ebf1, and Vpreb1 (Figure 1F) were highly expressed in CSF1R+ ProB cells but undetectable (Mb1, Pax5, and Ebf1) or low (Vpreb1) in CSF1R+ LMPPs, confirming the definitive B-cell progenitor identity of CSF1R+ ProB cells. Together these data show that CSF1R is expressed on fetal B-cell progenitors, but is restricted to the earliest stages of B-cell development.
CSF1R+ ProB cells possess B and myeloid lineage potential
Because we identified for the first time the expression of the myeloid cytokine receptor CSF1R on B-cell progenitors, we next investigated its potential functional relevance. Using in vitro clonal cytokine responsiveness assays, we established that purified CSF1R+ but not CSF1R– ProB cells proliferated in response to CSF1L alone (Figure 2A). To further characterize CSF1R+ ProB cells and their potential ability to also produce myeloid cells, we next performed quantitative mRNA expression analysis for genes of the myeloid lineage. In addition to Csf1r being highly expressed in CSF1R+ ProB cells, several other myeloid-affiliated genes (Mpo, Csf3r, Fcgr3) were expressed almost exclusively in the FL E13.5 CSF1R+ ProB fraction compared with the E13.5 CSF1R– ProB population and adult BM ProB cells (Figure 2B). Notably, the mRNA expression of the transcription factors Cebpa, Cebpb, and Sfpi1 (encoding PU.1), known regulators of Csf1r expression and myeloid development,30,31 was much higher in CSF1R+ than CSF1R– FL ProB cells, and adult BM ProB cells expressed virtually undetectable levels of Cebpa and Cebpb and lower levels of Sfpi1 (Figure 2B). We also investigated other lineage-affiliated programs and observed a small increase in the expression levels of the early B-cell genes Ebf1 and Mb1 in CSF1R– compared with CSF1R+ FL ProB cells (Figure 2C), whereas early T-cell genes were undetectable in all ProB cell populations (Figure 2D). Although Rag genes were expressed at similar levels, Flt3 was more highly expressed in CSF1R+ than CSF1R– FL ProB cells, and undetectable in adult ProB cells (Figure 2E).
Fetal CSF1R+ ProB cells possess B- and myeloid-lineage potential. (A) Response to CSF1L in single-sorted E13.5 ProB CSF1R+ (black) and CSF1R– (dark gray) cells. Mean percentage (± SEM) of clones (3 experiments, 30-60 cells per population per experiment). (B-E) Mean (± SEM) mRNA expression levels of (B) myeloid genes; (C) B-cell genes; (D) early T-cell genes; (E) early lymphoid genes, as measured by RT-qPCR in CSF1R+ (black) and CSF1R– (dark gray) E13.5 FL (n = 8 litters); and 8- to 10-week-old adult BM (light gray, n = 9 mice) ProB cells (25 cells/replicate, 3 experiments, 2-3 replicates per biological sample). Results are presented relative to Hprt1 expression. #Expressed but at too low a level to be visualized on the applied scale; n.d., Not detectable. Statistical significance was tested between CSF1R+ (black) and CSF1R– (dark gray) ProB population. ***P < .001; **P < .01; *P < .05; ns, not significant. (F-G) Cells generated on OP9 stroma after 7 days from sorted E13.5 FL CSF1R+ and CSF1R– (n = 8 litters), and 8- to 10-week-old adult BM (n = 6 mice) ProB cells (20 cells/replicate, 2-3 experiments, 4-10 replicates per biological sample). Mean percentage (± SEM) of wells with (F) B cell (Topro–NK1.1–B220+CD19+) and (G) myeloid (Topro–NK1.1–B220–CD19–GR1+MAC1+) cells. (H) T-cell generation on OP9–DL1 stroma after 14 to 16 days from sorted E13.5 FL CSF1R+ and CSF1R– (n = 9 litters), and 8- to 10-week-old adult BM (n = 9 mice) ProB cells or E13.5 FL-lineage negative cells (n = 2 litters) (20 cells/replicate, 2 experiments, 3-9 replicates per biological sample). Mean percentage (± SEM) of wells with T (Topro–NK1.1–B220–MAC1–GR1–CD25+THY1.2+ or Topro–NK1.1–B220–MAC1–GR1–CD4+CD8a+) cells.
Fetal CSF1R+ ProB cells possess B- and myeloid-lineage potential. (A) Response to CSF1L in single-sorted E13.5 ProB CSF1R+ (black) and CSF1R– (dark gray) cells. Mean percentage (± SEM) of clones (3 experiments, 30-60 cells per population per experiment). (B-E) Mean (± SEM) mRNA expression levels of (B) myeloid genes; (C) B-cell genes; (D) early T-cell genes; (E) early lymphoid genes, as measured by RT-qPCR in CSF1R+ (black) and CSF1R– (dark gray) E13.5 FL (n = 8 litters); and 8- to 10-week-old adult BM (light gray, n = 9 mice) ProB cells (25 cells/replicate, 3 experiments, 2-3 replicates per biological sample). Results are presented relative to Hprt1 expression. #Expressed but at too low a level to be visualized on the applied scale; n.d., Not detectable. Statistical significance was tested between CSF1R+ (black) and CSF1R– (dark gray) ProB population. ***P < .001; **P < .01; *P < .05; ns, not significant. (F-G) Cells generated on OP9 stroma after 7 days from sorted E13.5 FL CSF1R+ and CSF1R– (n = 8 litters), and 8- to 10-week-old adult BM (n = 6 mice) ProB cells (20 cells/replicate, 2-3 experiments, 4-10 replicates per biological sample). Mean percentage (± SEM) of wells with (F) B cell (Topro–NK1.1–B220+CD19+) and (G) myeloid (Topro–NK1.1–B220–CD19–GR1+MAC1+) cells. (H) T-cell generation on OP9–DL1 stroma after 14 to 16 days from sorted E13.5 FL CSF1R+ and CSF1R– (n = 9 litters), and 8- to 10-week-old adult BM (n = 9 mice) ProB cells or E13.5 FL-lineage negative cells (n = 2 litters) (20 cells/replicate, 2 experiments, 3-9 replicates per biological sample). Mean percentage (± SEM) of wells with T (Topro–NK1.1–B220–MAC1–GR1–CD25+THY1.2+ or Topro–NK1.1–B220–MAC1–GR1–CD4+CD8a+) cells.
The fact that other myeloid genes were expressed in CSF1R+ ProB cells together with B cell–specific genes raised the possibility that these fetal CSF1R+ ProB cells might also possess myeloid lineage potential. We investigated this using the OP9 coculture system.27,32 Whereas a comparable fraction of wells plated with limited numbers of CSF1R+ and CSF1R– E13.5 FL, as well as adult BM ProB cells, gave rise to B cells (Figure 2F), only FL CSF1R+ ProB cells generated MAC1+GR1+ myeloid cells (also negative for NK1.1, B220, and CD19) (Figure 2G). Moreover, CSF1R+ ProB cells lost their CSF1R expression after coculturing on OP9, suggesting their position upstream of CSF1R– ProB cells (supplemental Figure 2A). Importantly, neither CSF1R+ nor CSF1R– FL ProB cells, nor adult ProB cells, had any T-cell potential (Figure 2H; supplemental Figure 2B). These data demonstrate that the earliest FL CD19+ ProB cell progenitors expressing CSF1R uniquely coexpress myeloid and B cell–lineage genes and, in addition to B-lineage potential, possess residual myeloid but not T cell–lineage potential.
Deletion of CSF1R expression impairs specifically fetal B lymphopoiesis
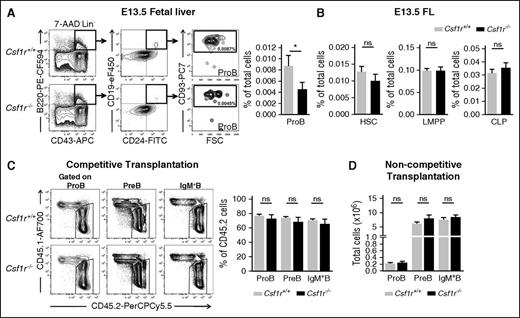
To address whether CSF1R has a nonredundant role in fetal B lymphopoiesis, we investigated the impact of CSF1R loss of function on the emergence of the first B cell–committed progenitors. Because of a high postnatal lethality of Csf1r−/− mice, we used Csf1r−/− and Wt Csf1r+/+ littermate control embryos generated from intercrossing of Csf1r+/− animals.12 Csf1r−/− embryos were present at the expected Mendelian frequency at the time of analysis (E13.5-E17.5). A distinct and highly reproducible, almost 50% reduction in ProB cells was observed in E13.5 Csf1r−/− embryos (Figure 3A). Because CSF1R is also expressed on LMPPs at this time in the development (Figure 1D-E), we also investigated this compartment together with the upstream HSC population. Importantly, no reduction was found in E13.5 Csf1r−/− embryos in the upstream HSCs, LMPPs, or common lymphoid progenitors (CLPs), suggesting a distinct requirement of CSF1R at the ProB cell progenitor stage (Figure 3B; supplemental Figure 3A-B). To investigate whether the role of CSF1R on B-cell progenitors was restricted to embryonic B lymphopoiesis, as suggested by the fetally restricted expression of CSF1R on ProB cells, we transplanted unfractionated FL cells from E14.5 Csf1r−/− and Csf1r+/+ littermate controls, but did not observe any impairment in replenishment of any stages of B-cell progenitors in the BM of recipient mice 7 weeks after a competitive transplantation (Figure 3C) nor at 16 weeks after a noncompetitive transplantation (Figure 3D). Thus, in agreement with the fetal restriction of CSF1R expression on early B-cell progenitors, CSF1R signaling is fully dispensable for adult B-cell progenitors.
CSF1R deficiency impairs the emergence of ProB cells in the fetal liver. (A) (Left) Representative FACS profiles of ProB cells in E13.5 FL of Csf1r+/+ (Wt) and Csf1r−/− littermate embryos (3 experiments). Numbers represent mean percentage of gated populations relative to total FL cells. (Right) Mean percentage (± SEM) of ProB cells in E13.5 FL of Csf1r+/+ (light gray) and Csf1r−/− littermate embryos (black) (n = 12-13 embryos per genotype). (B) Mean percentage (± SEM) HSCs, LMPPs, and CLPs (Lin–KITlowSCA-1lowIL7R+FLT3+) in E13.5 FL of Csf1r+/+ and Csf1r−/− littermate embryos (n = 6-12 embryos per genotype, 2-3 experiments). (C) Lethally irradiated adult Wt recipient (CD45.1) mice were transplanted with 400 000 unfractionated Csf1r−/− (CD45.2) or Csf1r+/+ littermate (CD45.2) E14.5 FL cells together with 400 000 unfractionated Wt CD45.1 competitor E17.5 FL cells. (Left) Representative FACS profile of CD45.2/CD45.1 distribution within ProB, PreB, or IgM+ B cells (Lin–B220+CD19+CD43–IgM+cells) in BM from the different genotypes at 7 weeks post-transplantation. (Right) Mean percentage (± SEM) of CD45.2 cells within the indicated population in mice transplanted with Csf1r+/+ and Csf1r−/− littermate embryos (n = 7 mice per genotype with 4 to 5 different donor embryos per genotype). (D) Lethally irradiated adult Wt recipient (CD45.1) mice were transplanted with 5 × 106 unfractionated Csf1r−/− (CD45.2) or Csf1r+/+ littermate (CD45.2) E14.5 FL cells. Mean number (± SEM) of donor-derived ProB, PreB, and IgM+ B cells in BM reconstituted with Csf1r+/+ or Csf1r−/− littermate embryos at 16 weeks post-transplantation (n = 7 mice per genotype with 4 different donor embryos per genotype). Statistical significance was tested between Csf1r+/+ and Csf1r−/−.*P < .05. ns, not significant.
CSF1R deficiency impairs the emergence of ProB cells in the fetal liver. (A) (Left) Representative FACS profiles of ProB cells in E13.5 FL of Csf1r+/+ (Wt) and Csf1r−/− littermate embryos (3 experiments). Numbers represent mean percentage of gated populations relative to total FL cells. (Right) Mean percentage (± SEM) of ProB cells in E13.5 FL of Csf1r+/+ (light gray) and Csf1r−/− littermate embryos (black) (n = 12-13 embryos per genotype). (B) Mean percentage (± SEM) HSCs, LMPPs, and CLPs (Lin–KITlowSCA-1lowIL7R+FLT3+) in E13.5 FL of Csf1r+/+ and Csf1r−/− littermate embryos (n = 6-12 embryos per genotype, 2-3 experiments). (C) Lethally irradiated adult Wt recipient (CD45.1) mice were transplanted with 400 000 unfractionated Csf1r−/− (CD45.2) or Csf1r+/+ littermate (CD45.2) E14.5 FL cells together with 400 000 unfractionated Wt CD45.1 competitor E17.5 FL cells. (Left) Representative FACS profile of CD45.2/CD45.1 distribution within ProB, PreB, or IgM+ B cells (Lin–B220+CD19+CD43–IgM+cells) in BM from the different genotypes at 7 weeks post-transplantation. (Right) Mean percentage (± SEM) of CD45.2 cells within the indicated population in mice transplanted with Csf1r+/+ and Csf1r−/− littermate embryos (n = 7 mice per genotype with 4 to 5 different donor embryos per genotype). (D) Lethally irradiated adult Wt recipient (CD45.1) mice were transplanted with 5 × 106 unfractionated Csf1r−/− (CD45.2) or Csf1r+/+ littermate (CD45.2) E14.5 FL cells. Mean number (± SEM) of donor-derived ProB, PreB, and IgM+ B cells in BM reconstituted with Csf1r+/+ or Csf1r−/− littermate embryos at 16 weeks post-transplantation (n = 7 mice per genotype with 4 different donor embryos per genotype). Statistical significance was tested between Csf1r+/+ and Csf1r−/−.*P < .05. ns, not significant.
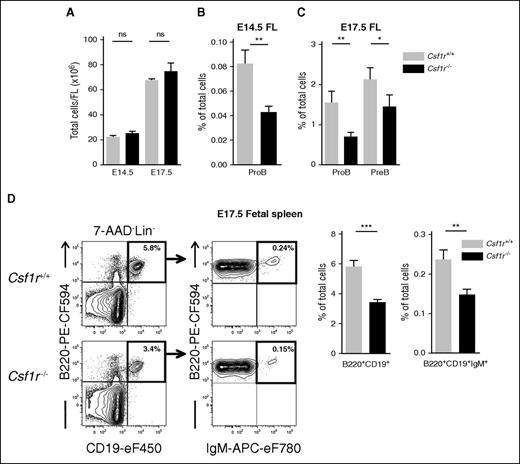
Because CSF1R is involved specifically in embryonic B lymphopoiesis, we next investigated the impact of CSF1R deficiency on later stages of fetal B-cell progenitors. In E14.5 and E17.5 FL, at which time the total FL cellularity remained unaltered in Csf1r−/− embryos (Figure 4A), the frequency of Csf1r−/− ProB cells remained reduced by ∼50%, despite CSF1R expression being downregulated at these later stages of development (Figure 4B-C; supplemental Figure 3C-D). At E17.5, PreB cells can also be detected in the FL, and although they lack CSF1R expression, we found them to be reduced by >30% in the Csf1r−/− FL (Figure 4C; supplemental Figure 3D). B-cell progenitors can also be observed in the spleen already at E14.533 and it has been suggested that they migrate there from the FL through the circulation.6,34 The spleen is a site of fetal B-cell maturation and it was therefore of interest to assess the impact of Csf1r deletion on splenic B cells. At E17.5 we found a 41% decrease in the percentage of total B220+CD19+ cells in Csf1r−/− spleens, and a comparable decrease of mature B220+CD19+IgM+ B cells (Figure 4D).
CSF1R deficiency affects fetal B-cell differentiation. (A) Total FL cells in E14.5 and E17.5 Csf1r+/+ (light gray) and Csf1r−/− littermate embryos (black) (n = 11-16 embryos per genotype). (B-C) Mean percentage (± SEM) of (B) ProB cells in E14.5 FL, and (C) ProB and PreB cells in E17.5 FL of Csf1r+/+ and Csf1r−/− littermate embryos (n = 11-16 embryos per genotype, 3-5 experiments). (D) (Left) Representative FACS profiles of total B (B220+CD19+) and mature B (B220+CD19+IgM+) cells in E17.5 fetal spleen of Csf1r+/+ and Csf1r−/− littermate embryos (4 experiments). Numbers represent mean percentage of gated populations relative to total spleen cells. (Right) Mean percentage (± SEM) of total B cells and mature B cells out of total E17.5 spleen cells in Csf1r+/+ and Csf1r−/− littermate embryos (n = 14-15 embryos per genotype). Statistical significance was tested between Wt (Csf1r+/+) and Csf1r−/−. ***P < .001; **P < .01; *P < .05. ns, not significant.
CSF1R deficiency affects fetal B-cell differentiation. (A) Total FL cells in E14.5 and E17.5 Csf1r+/+ (light gray) and Csf1r−/− littermate embryos (black) (n = 11-16 embryos per genotype). (B-C) Mean percentage (± SEM) of (B) ProB cells in E14.5 FL, and (C) ProB and PreB cells in E17.5 FL of Csf1r+/+ and Csf1r−/− littermate embryos (n = 11-16 embryos per genotype, 3-5 experiments). (D) (Left) Representative FACS profiles of total B (B220+CD19+) and mature B (B220+CD19+IgM+) cells in E17.5 fetal spleen of Csf1r+/+ and Csf1r−/− littermate embryos (4 experiments). Numbers represent mean percentage of gated populations relative to total spleen cells. (Right) Mean percentage (± SEM) of total B cells and mature B cells out of total E17.5 spleen cells in Csf1r+/+ and Csf1r−/− littermate embryos (n = 14-15 embryos per genotype). Statistical significance was tested between Wt (Csf1r+/+) and Csf1r−/−. ***P < .001; **P < .01; *P < .05. ns, not significant.
Collectively, these results show that lack of CSF1R expression has a specific impact on the earliest fetal ProB-cell progenitors, as well as the subsequent stages of fetal B lymphopoiesis, while not affecting the upstream HSC, LMPP, and CLP compartments.
Concomitant loss of FLT3 and CSF1R leads to a severe reduction in early fetal B-cell progenitors
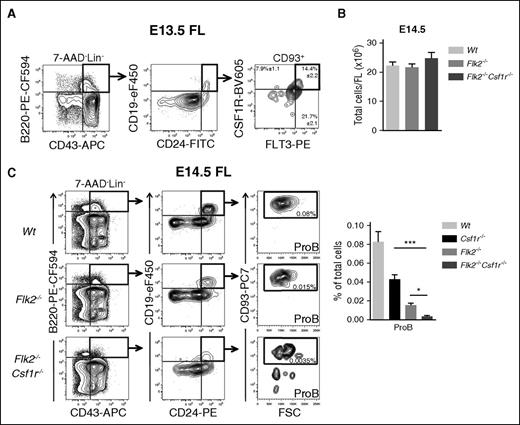
Our single-cell gene expression analysis showed that the majority of E13.5 CSF1R+ ProB also expressed high levels of Flt3 (Figures 1F and 2E), encoding another cytokine tyrosine kinase receptor of the same family as CSF1R, critically involved in B lymphopoiesis.28 In agreement with this, we found a high level of coexpression of CSF1R and FLT3 also at the cell surface of E13.5 FL ProB cells (Figure 5A). Previous studies have demonstrated that Flt3-Ligand–deficient mice have a reduction in B-cell progenitors in the E14.5 FL.35 Here we reproduced this finding in Flt3 receptor–deficient (Flk2−/−) mice, and found that although the FL cellularity was not affected (Figure 5B), the frequency of ProB cells was further and dramatically reduced in the E14.5 FL of Flk2−/−Csf1r−/− double-deficient embryos, when compared with Csf1r−/− and Flk2−/− single-deficient embryos (Figure 5C). This finding supports that CSF1R and FLT3 have complementary and critical roles in regulation of the emergence of fetal B-cell progenitors.
Concomitant loss of FLT3 and CSF1R expression leads to a severe reduction in fetal B-cell progenitors. (A) Representative FACS profiles of CSF1R and FLT3 coexpression on E13.5 FL ProB cells. Numbers represent mean percentage (± SEM) of CSF1R+FLT3+, CSF1R+FLT3–, and CSF1R–FLT3+ ProB cells (n = 16 embryos, 3 experiments). (B) Total FL cells in E14.5 FL of Wt control (light gray), Flk2−/− (gray), and Flk2−/−Csf1r−/− (dark gray) embryos (n = 16-21 embryos per genotype). (C) (Left) Representative FACS profiles of ProB cells in E14.5 FL of Wt, Flk2−/−, and Flk2−/−Csf1r−/− embryos. Numbers represent mean percentage of gated populations relative to total FL cells (4-5 experiments). (Right) Mean percentage (± SEM) of ProB in E14.5 FL of Wt control (light gray), Csf1r−/− (black), Flk2−/− (gray), and Flk2−/−Csf1r−/− (dark gray) embryos (n = 14-21 embryos per genotype). Statistical significance was tested between Csf1r−/− and Csf1r−/−Flk2−/−, and between Flk2−/− and Csf1r−/−Flk2−/−. Because of multiple comparisons, Dunn’s correction of P values was performed (see Methods). ***P < .001; *P < .05.
Concomitant loss of FLT3 and CSF1R expression leads to a severe reduction in fetal B-cell progenitors. (A) Representative FACS profiles of CSF1R and FLT3 coexpression on E13.5 FL ProB cells. Numbers represent mean percentage (± SEM) of CSF1R+FLT3+, CSF1R+FLT3–, and CSF1R–FLT3+ ProB cells (n = 16 embryos, 3 experiments). (B) Total FL cells in E14.5 FL of Wt control (light gray), Flk2−/− (gray), and Flk2−/−Csf1r−/− (dark gray) embryos (n = 16-21 embryos per genotype). (C) (Left) Representative FACS profiles of ProB cells in E14.5 FL of Wt, Flk2−/−, and Flk2−/−Csf1r−/− embryos. Numbers represent mean percentage of gated populations relative to total FL cells (4-5 experiments). (Right) Mean percentage (± SEM) of ProB in E14.5 FL of Wt control (light gray), Csf1r−/− (black), Flk2−/− (gray), and Flk2−/−Csf1r−/− (dark gray) embryos (n = 14-21 embryos per genotype). Statistical significance was tested between Csf1r−/− and Csf1r−/−Flk2−/−, and between Flk2−/− and Csf1r−/−Flk2−/−. Because of multiple comparisons, Dunn’s correction of P values was performed (see Methods). ***P < .001; *P < .05.
Discussion
Herein, we establish that a distinct subset of the earliest CD19+ ProB cells emerging for the first time in the E13.5 mouse FL express CSF1R, previously demonstrated to play an important role during the development of myeloid cells, in particular the macrophage lineage.11,36 Importantly, CSF1R+ ProB cells are unique for fetal hematopoiesis as they, in agreement with previous studies,13 do not persist in adult BM. In fact, our analysis demonstrates that they only persist during a short developmentally restricted window of time, because already at E17.5 the FL exclusively contains CSF1R-CD19+ ProB cells. This finding along with in vitro experiments demonstrating that CSF1R+CD19+ ProB cells give rise to CSF1R–CD19+ ProB cells, suggests that CSF1R+CD19+ ProB cells precede and subsequently give rise to CSF1R–CD19+ ProB cells. Most importantly, our characterization of FL CSF1R+CD19+ ProB cells demonstrates that they possess B-cell, as well as residual myeloid lineage, but not T-lineage potential. In agreement with this, they express B-lymphoid, as well as myeloid genes, including key transcription factors for both lineages, but no genes characteristically and specifically expressed in early T-cell progenitors. In contrast, FL CSF1R–CD19+ ProB cells express much lower levels of myeloid lineage genes, although somewhat higher than in adult ProB cells, and in agreement with this are fully B cell–restricted in their lineage potentials in the same assays.
Previous studies of fetal hematopoiesis had provided a number of findings implicating a close link between B-lymphocyte and granulocyte-macrophage lineage development, including the existence of a fetal B myeloid–restricted progenitor,5,6,8 although such a progenitor had not previously been prospectively identified and characterized. Previous work established that B220+CD43+CD24+ ProB cells from E16 FL and the adult BM counterpart have DJ but not VDJ immunoglobulin heavy-chain gene rearrangement,25 express similar levels of λ5, VpreB, Rag1, and Rag2 gene, whereas TdT and PLRLC genes were absent in the FL but highly expressed in the adult BM.37 Both the adult and E16 FL ProB cells generated IgM+ B cells in vitro and after transplantation into immune-deficient mice but failed to produce T cells.25 However, their myeloid lineage potential was not studied, nor was their potential coexpression of B-cell and myeloid lineage genes.25,37
Recently, we identified early FL immune-restricted LMPPs, coexpressing early lymphoid and myeloid genes, as well as cell surface CSF1R and IL7R, and possessing combined myeloid, B-cell, and T-cell potential.15 As confirmed here, and in contrast to the CSF1R+ fetal CD19+ ProB cells identified here, these FL LMPPs lack detectable transcriptional expression of Cd19 as well as the transcription factors Pax5 and Ebf1, which are critical for B-lineage commitment. In another study, a similar FL immune-restricted LIN–KIT+CSF1R+IL7R+ LMPP was characterized, which also possessed myeloid, B-, and T-lineage potential.16 However, at variance with our strategy for purification of FL LMPPs,15 these LIN–KIT+CSF1R+IL7R+ LMPPs also expressed Pax5 and Ebf1 at the cell population level.16 Therefore, whether these early LIN–KIT+CSF1R+IL7R+ lympho-myeloid progenitor cells (which were sorted as CD19–) also included contaminating fully B cell–restricted progenitors, B myeloid–restricted CD19+ ProB-cell progenitors as identified here, or even earlier CD19–CSF1R+ B cell progenitors, remains to be investigated.
A major role of CSF1R in bone remodeling has complicated the interpretation of previous studies of Csf1r−/− mice postnatally, in particular with regard to distinguishing intrinsic from extrinsic/secondary effects on hematopoiesis.38,39 Our studies of fetal hematopoiesis before establishment of BM hematopoiesis, for which bone modeling and CSF1R is critically involved, allowed us to directly investigate the expression and role of CSF1R in fetal B lymphopoiesis. Upon loss of CSF1R, the earliest CD19+ ProB cells were decreased, demonstrating, for the first time, the importance of this cytokine receptor for early fetal B-cell progenitors. The finding that HSCs and LMPPs were unaffected by CSF1R deficiency supports the distinct requirement for CSF1R in early fetal B-cell progenitors. However, because a subfraction of LMPPs also express CSF1R, it cannot be ruled out that the observed reduction in early B-cell progenitors could reflect a requirement for CSF1R function in the generation of CSF1R+ ProB cells from LMPPs. We can also not rule out that the observed reduction in fetal ProB cells in Csf1r−/− embryos might partially or fully be extrinsically mediated through the loss of macrophages in Csf1r−/− embryos. Additional studies with conditional deletion of Csf1r expression specifically in early B-cell progenitors could more decisively address both of these possibilities.
The sustained impact on B lymphopoiesis at subsequent stages and different time points suggests that the earliest and distinct CSF1R+ fetal ProB-cell compartment is important for subsequent fetal development of CSF1R– B lymphopoiesis. However, in agreement with our data showing that CSF1R is not expressed on B-cell progenitors in the adult, Csf1r−/− FL cells transplanted into adult lethally irradiated recipients, in a competitive as well as noncompetitive setting, showed no impairment in the reconstitution of distinct stages of B-cell progenitors. This further supports a distinct role of CSF1R in fetal B lymphopoiesis.
The established critical role of FLT3 in lymphopoiesis24,40 and the herein identified novel role of CSF1R in fetal B-cell development made us also investigate the impact of concomitant loss of both FLT3 and CSF1R, which resulted in a much more severe impairment in B lymphopoiesis than either alone, suggesting critical and complementary roles of FLT3 and CSF1R signaling in fetal B lymphopoiesis.
Identifying and characterizing distinct fetal B-cell progenitors is of considerable relevance toward establishing the identity of the fetal progenitors targeted by preleukemic and leukemic genetic lesions in infant and childhood PreB-ALL.20 The herein identification of a developmentally restricted CSF1R+CD19+ B-myeloid progenitor is intriguing in light of the frequent bi-phenotypic (B cell–myeloid) nature of childhood and infant PreB-ALL,19 although further mechanistic studies will be required to establish the potential significance of fetal B myeloid–restricted progenitors for childhood and infant PreB-ALL. Of further potential clinical relevance, recent studies have found CSF1R to be rearranged in childhood ALLs,21,23 and in vitro studies using cell lines and human leukemic PreB ALL cells have demonstrated their sensitivity to a CSF1R-specific inhibitor,22 supporting that CSF1R might be a relevant therapeutic target in childhood ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Linda Hansson for expert animal support, Trine Kristiansen for discussions, Ihor R. Lemischka for providing the Flk2−/− mice, and E. Richard Stanley for providing the Csf1r+/− mice.
This work was supported by the Swedish Childhood Cancer Foundation (E.S., S.E.W.J.), the Gunnar Nilssons Foundation (E.S.), the ALF Clinical Research Award from Lund University Hospital (E.S.), the Hemato-Linne (E.S., S.E.W.J.), Stem Therapy (E.S.) programs, and a unit grant (MC_UU_12009/5) from the Medical Research Council UK (S.E.W.J.).
C.B. was supported by a postdoctoral fellow grant from the Swedish Childhood Cancer Foundation. E.S. has an Associate Professor position supported by the Swedish Childhood Cancer Foundation. J.C.A.G. is a recipient of the Alexander Thatte Fellowship.
Authorship
Contribution: A.Z., E.S., and S.E.W.J. designed and conceptualized the overall research and analyzed the data; A.Z. performed the experiments; L.W. provided expertise in the animal work; C.B. contributed to the design, execution, and analysis of experiments; J.C.A.G. provided expert advice on B-cell progenitor analysis; P.S.W. contributed with expert advice and input on gene expression analysis; and A.Z., C.B., E.S., and S.E.W.J. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ewa Sitnicka, Division of Molecular Hematology, Department of Laboratory Medicine, Lund University, BMC, B12, Klinikgatan 26, 221-84 Lund, Sweden; e-mail: ewa.sitnicka@med.lu.se; or Sten Eirik W. Jacobsen, Karolinska Institutet, Hematopoietic Stem Cell Laboratory, Department of Medicine Huddinge, Center for Hematology and Regenerative Medicine (HERM), Karolinska University Hospital, 141-86 Stockholm, Sweden; e-mail: sten.eirik.jacobsen@ki.se.
References
Author notes
S.E.W.J. and E.S. contributed equally to this study.

![Figure 1. CSF1R is expressed on early B-cell progenitors in the fetal liver but not in adult bone marrow. (A) Mean (± standard error of the mean [SEM]) Csf1r expression levels measured by RT-qPCR in 20 E13.5 FL (n = 7 embryos) and 8- to 10-week-old adult BM (n = 6 mice) ProB (Lin–B220+CD43+CD19+CD24+CD93+) cells (2-3 experiments). Results are presented relative to Hprt1 expression. **P < .01. (B) Representative FACS profiles of CSF1R expression on ProB at E13.5, E14.5, and on ProB and PreB (Lin–B220+CD19+CD43–IgM– cells) at E17.5 FL and in 8- to 10-week-old adult BM (2-5 experiments). (C) Mean percentage (± SEM) of ProB cells expressing CSF1R at E13.5 (n = 26 embryos), E14.5 (n = 21 embryos), E17.5 (n = 12 embryos), and in 8- to 10-week-old adult BM (n = 17 mice). (D) Representative FACS profiles of CSF1R expression on HSCs (Lin–KIT+SCA-1+CD48–CD150+FLT3–) and LMPPs (Lin–KIT+SCA-1+FLT3high) in E13.5 FL and 8- to 10-week-old adult BM (2-5 experiments). (B, D) Numbers represent mean percentage of CSF1R+ cells in the investigated populations. (E) Mean percentage (± SEM) of HSCs and LMPPs expressing CSF1R at E13.5 (n = 21 embryos), E14.5 (n = 13-30 embryos), and 8- to 10-week-old adult BM (n = 6-17 mice). (F) Gene expression analysis of single E13.5 FL CSF1R+ ProB and CSF1R+ FL LMPP cells expressing Csf1r mRNA (2-3 experiments). Results are presented relative to Hprt1 expression. Each column represents a single cell.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/2/10.1182_blood-2016-01-693887/4/m_217f1.jpeg?Expires=1769185708&Signature=APyqfN3S2OXbq2IoCL2kes-E6mAe5CuunPbYUF6tF76aT4cAKdZ7wp2juT7Kenw0xk9ic~9oyyWYhNoJLcutKfCwqpxN8l7Dec1vTEhzToVkA-8aISkoNPZedQ8JVqjD638Nqy-Qj4zuYsVjDqNRf7V7H02-nI6cpnhIAF0bxw3OmngT5J8S3o1oPAxcZ6Of5ocTQCXgJxBz8GxQhgRuHSwLJOEPygtHin7xMmowuGuMVYAVjWXbz7ilDXF5NmLy4fxU-3mpZIRKmzs6l5SqBMlM55n8gPs55F9a7eZEpeiNbWlXKXwgHVh83cfTekSt4HLwweL6VphX1U3HO6E1jA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)