In this issue of Blood, McKerrell et al describe a novel next-generation sequencing (NGS)–based platform for the identification of point mutations, common fusion genes, and copy-number alterations in acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS) from a single genomic DNA sample.1
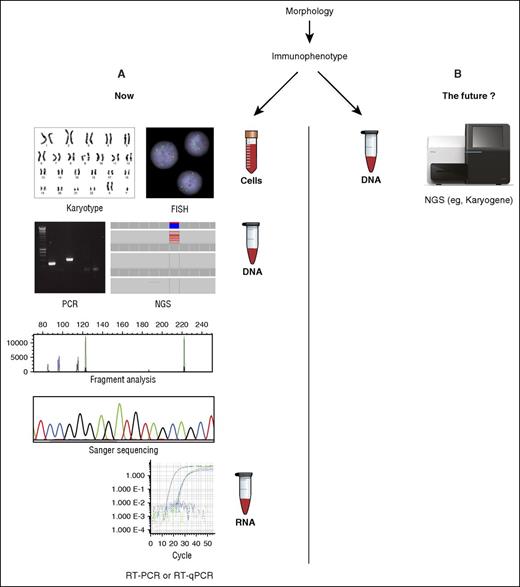
The standard battery of assays required at diagnosis for AML (A) may soon be replaced by a single NGS-based test such as Karyogene (B), described in this issue. FISH, fluorescence in situ hybridization; RT-PCR, reverse transcription PCR.
The standard battery of assays required at diagnosis for AML (A) may soon be replaced by a single NGS-based test such as Karyogene (B), described in this issue. FISH, fluorescence in situ hybridization; RT-PCR, reverse transcription PCR.
Although the diagnosis of MDS is increasingly shifting to panel-based targeted sequencing, the laboratory workup of AML has traditionally involved a variety of different methodologies. It is now clear that AML is driven by a wide range of genomic abnormalities2 and knowledge of these is increasingly being used to personalize therapy and optimize outcomes for patients. Cytogenetic abnormalities provide powerful prognostic information3 and can identify patients suitable for targeted therapies such as all-trans retinoic acid and arsenic trioxide in acute promyelocytic leukemia,4 and gemtuzumab ozogamicin in patients with core-binding factor leukemia.5 Molecular abnormalities provide further prognostic information in those patients lacking a favorable-risk karyotype, also highlighting patients who may benefit from emerging therapies such as small-molecule inhibitors of Fms-like tyrosine kinase 3 (FLT3) and isocitrate dehydrogenase 1/2 (IDH1/2).
In terms of risk stratification, the current European LeukemiaNet guidelines recommend molecular screening for NPM1 mutations and biallelic mutations involving CEBPA, which, in the absence of FLT3–internal tandem duplication (ITD), define further subsets of AML with relatively favorable prognosis.6 Moreover, detection of fusion genes and molecular abnormalities, such as NPM1 exon 12 insertion/deletion (indel) mutations, allows patients to be selected for molecular monitoring using sensitive techniques such as real-time quantitative polymerase chain reaction (qPCR), permitting response-based individualization of treatment.7,8
To deliver optimal therapy and accurately identify patients likely to benefit from allogeneic stem cell transplantation, these genomic abnormalities must therefore be accurately defined at diagnosis; at the present time, this requires a range of different assays to be performed (see figure). Current international guidelines6 stipulate a conventional karyotype, FISH, and molecular testing for a limited number of mutations (NPM1, CEBPA, FLT3-ITD). This has traditionally involved performance of individual DNA- or RNA-based PCR assays and Sanger sequencing. However, many centers now perform NGS panel-based testing for frequently mutated genes to further refine prognosis. This complex array of testing may pose challenges to less specialized laboratories and those with more limited resources.
McKerrell and colleagues now present a potential method to simplify diagnostic testing for AML designated “Karyogene”; this system uses sequence capture and NGS coupled with a range of open-source bioinformatic tools to test for common AML-associated translocations, mutations (in 49 genes recurrently mutated in myeloid malignancy), and copy-number variations and identify regions of copy-neutral loss of heterozygosity from a single sample of genomic DNA. In analysis of a cohort of 112 diagnostic samples (62 AML, 50 MDS), Karyogene could successfully detect the commonest fusion genes including PML-RARA, CBFB-MYH11, RUNX1-RUNX1T1, and KMT2A (mixed-lineage leukemia [MLL]) fusions, which together account for ∼80% of known AML-associated translocations; the authors reported 100% sensitivity and specificity for these abnormalities. Importantly, Karyogene was also able to identify 1 rare and 1 novel KMT2A fusion, neither of which were detected by the standard diagnostic workup.
Important clinically actionable substitutions and indel mutations in NPM1, FLT3, IDH1, IDH2, DNMT3A, and CEBPA were reliably detected, including some CEBPA mutations missed by standard sequencing. Detection of tandem duplications using short-read sequencing is notoriously challenging; however, McKerrell et al describe 2 new publicly available bioinformatic tools: FLT3 Tandem Finder (F-TAFI; to identify FLT3-ITDs) and MLL Tandem Finder (M-TAFI; to detect MLL partial tandem duplications). When used in conjunction with the Karyogene capture panel, these tools were associated with 100% sensitivity and specificity compared with fragment analysis. Karyogene also performed reasonably well in detecting copy-number changes, showing concordance with the karyotype in 93% of cases. In some cases, small but clinically important deletions (eg, involving 17p [leading to TP53 deletion]) not detected by cytogenetics were found.
Although NGS-based platforms such as Karyogene provide an important advance in diagnostic testing for AML, it may be premature for such systems to replace standard cytogenetic testing. Rarer clinically relevant chromosomal rearrangements, such as inv(3)(q21q26)/t(3;3)(q21;q26)/GATA2-EVI1, t(5;11)(q35;p15.5)/NUP98-NSD1, t(6;9)(p23;q34)/DEK-NUP214, t(8;16)(p11;p13)/MYST3-CREBBP, and t(9;22)(q34;q11)/BCR-ABL, were not detectable in the current iteration of the panel. However, there would clearly be scope to expand the panel to address this. Another key limitation to implementation of this approach is the level of bioinformatic expertise, which is likely to be beyond that currently available in many diagnostic laboratories. These challenges are undoubtedly surmountable and the results reported by McKerrell and colleagues provide an important proof of principle, offering a glimpse of how hematologic malignancies might be diagnosed in the future. However, with increasing interest in NGS-based diagnostics, it is critical to ensure that RNA (as well as DNA) is routinely extracted at diagnosis to provide baseline samples to allow molecular monitoring of minimal residual disease, which can provide an important tool to further refine outcome prediction.7,8
Conflict-of-interest disclosure: The authors declare no competing financial interests.