Key Points
The protein disulfide isomerase is involved in VWF dimerization by initiating disulfide bond formation at cysteines 2771 and 2773.
von Willebrand disease-associated mutations in the dimerization domain of von Willebrand factor disturb processing by the protein disulfide isomerase.
Abstract
Multimeric von Willebrand factor (VWF) is essential for primary hemostasis. The biosynthesis of VWF high-molecular-weight multimers requires spatial separation of each step because of varying pH value requirements. VWF is dimerized in the endoplasmic reticulum by formation of disulfide bonds between the C-terminal cysteine knot (CK) domains of 2 monomers. Here, we investigated the basic question of which protein catalyzes the dimerization. We examined the putative interaction of VWF and the protein disulfide isomerase PDIA1, which has previously been used to visualize endoplasmic reticulum localization of VWF. Excitingly, we were able to visualize the PDI–VWF dimer complex by high-resolution stochastic optical reconstruction microscopy and atomic force microscopy. We proved and quantified direct binding of PDIA1 to VWF, using microscale thermophoresis and fluorescence correlation spectroscopy (dissociation constants KD = 236 ± 66 nM and KD = 282 ± 123 nM by microscale thermophoresis and fluorescence correlation spectroscopy, respectively). The similar KD (258 ± 104 nM) measured for PDI interaction with the isolated CK domain and the atomic force microscopy images strongly indicate that PDIA1 binds exclusively to the CK domain, suggesting a key role of PDIA1 in VWF dimerization. On the basis of protein–protein docking and molecular dynamics simulations, combined with fluorescence microscopy studies of VWF CK-domain mutants, we suggest the following mechanism of VWF dimerization: PDI initiates VWF dimerization by forming the first 2 disulfide bonds Cys2771-2773′ and Cys2771′-2773. Subsequently, the third bond, Cys2811-2811′, is formed, presumably to protect the first 2 bonds from reduction, thereby rendering dimerization irreversible. This study deepens our understanding of the mechanism of VWF dimerization and the pathophysiological consequences of its inhibition.
Introduction
von Willebrand factor (VWF) is a multimeric glycoprotein essential for platelet-dependent primary hemostasis. The shear-induced transition between a globular and a stretched conformation of VWF high-molecular-weight multimers (HMWMs) leads to exposure of binding sites for VWF partners, and thus the initiation of platelet adhesion and aggregation. Because smaller multimers experience lower shear forces than larger ones, VWF’s shear stress-activated functions are dependent on multimer size.1,2 Mutations within the VWF gene can lead to the inhibition of the VWF multimerization process, and thereby cause distinct forms of the bleeding disorder von Willebrand disease (VWD). To understand the pathomechanism behind these VWD variants, it is of paramount interest to unravel the details of VWF multimerization, which is established by the formation of intermolecular disulfide bonds.
The individual steps of VWF-HMWM biosynthesis rely on distinct pH conditions, which are realized by spatial separation of the involved processes to different cell organelles. The first multimerization step (dimerization) happens in the endoplasmic reticulum (ER) by disulfide bond formation between the C-terminal cysteine knot (CK) domains. The recently solved crystal structure revealed that 8 of the 11 cysteine residues within the CK domain form intrachain disulfide bonds, whereas 3 (namely, Cys2771, Cys2773, and Cys2811) are used to form the interchain connections for dimerization by formation of the 3 disulfide bonds (Cys2771-2773′, Cys2771′-2773, and Cys2811-2811′),3 as suggested by Katsumi et al.4 Dimerization is strongly disturbed in patients with CK domain mutations.5
The second multimerization step, the formation of N-terminal disulfide bonds between the D′-D3 assemblies of VWF dimers, occurs in the trans-Golgi network and is facilitated by the VWF propeptide that consists of 2 D assemblies that both harbor a CGLC protein disulfide isomerase consensus sequence, which is also present in the D3 assembly. Enabled by the lower pH of the Golgi apparatus, this intrinsic oxidoreductase function is activated by protonation of histidine residues adjacent to these CGLC sequences.6 Mutations within these sequences inhibit multimerization but do not affect dimerization. Therefore, dimerization does not appear to be performed by an intrinsic oxidoreductase activity within VWF.
Because dimerization occurs within the ER at neutral pH, it requires an ER-localized member of the thiol-disulfide oxidoreductase family. The protein disulfide isomerase PDI is one of the few members of this protein family that is known to catalyze disulfide bond formation and reduction, as well as isomerization. It has previously been described that VWF colocalizes with PDI to prove VWF ER localization.7 Here we investigated whether PDIA1 might be the protein that dimerizes VWF.
Materials and methods
Immunofluorescence
Immunofluorescence was performed with transfected HEK293 or nontransfected human umbilical vein endothelial cells (HUVECs), as previously described by Brehm et al.8 Antibodies used were rabbit anti-VWF (DAKO; 1:1000), mouse anti-PDI (abcam, 1:500, PDIA1 specific), goat anti-rabbit AF488 (Invitrogen, 1:5000), and goat anti-mouse AF546 (Invitrogen, 1:5000). Images were captured at room temperature (RT) with a confocal microscope (TCS SP5; Leica, Wetzlar, Germany) or the fluorescence microscope BZ9000 (Keyence). For settings, please refer to the respective figure legends.
Coimmunoprecipitation
Coimmunoprecipitation was performed as described.9 For details, please refer to the supplemental Methods, available on the Blood Web site.
Stochastic optical reconstruction microscopy
Imaging was performed in Ibidi treat 8-well µ-slides (Ibidi), using an imaging buffer containing 100 mM β-mercaptoethylamine in phosphate-buffered saline to facilitate sufficient blinking of the fluorophores.10 Stochastic optical reconstruction microscopy (STORM) data sets were acquired on a Nikon N-STORM microscope equipped with an Apo TIRF 100× oil immersion objective with a numerical aperture of 1.49 (Nikon GmbH, Düsseldorf, Germany), a back-illuminated electron-multiplying charge-coupled device (EMCCD) camera (iXon+ DU-897; Andor Technology Plc., Belfast, Northern Ireland), and a quadband filter composed of a quadline beamsplitter (zt405/488/561/640rpc TIRF; Chroma Technology Corporation, Bellows Falls, VT) and a quadline emission filter (brightline HC 446, 523, 600, 677; Semrock Inc., Rochester, NY) with an inclined illumination scheme11 to achieve a good signal-to-noise ratio. For excitation of Alexa Fluor 647, a 647-nm continuous-wave fiber laser (2RU-VFL-P-300-647; MPB Communications Inc., Montréal, Canada), and for Alexa Fluor 488 the 488-nm line of an argon gas laser (35-IMA-840-019; Melles Griot GmbH, Bensheim, Germany), was used. For multicolor imaging, the lasers were switched on and off alternately, controlled by an acousto-optic tunable filter. The integration time of the EMCCD camera was set to 16 ms per frame, with an EM gain of 300. Super-resolution images were reconstructed from a series of 12 500 images per channel, using the N-STORM analysis module v. 3.3.1.15801 of NIS Elements AR v. 4.20 (Laboratory imaging s.r.o., Prague, Czech Republic), with overlapping peak detection enabled.
Atomic force microscopy
Atomic force microscopy (AFM) imaging was performed in tapping mode in air, using an MFP-3D AFM (Asylum Research, Santa Barbara, CA) and silicon tips (AC160TS-R3; Olympus, Japan), with resonance frequencies of approximately 300 kHz and a nominal spring constant of 26 N/m. Images of 1 μm2 were recorded with a resolution of 1024 × 1024 pixels and analyzed using SPIP software (Image Metrology, Denmark). Substrate preparation was performed as described.12 Protein samples containing purified, recombinant VWF dimers (50 nM), recombinant human PDIA1 (100 nM) (Genway Biotech Inc., San Diego, CA), or a mixture of both proteins were incubated at pH 7.4 for 45 min at RT in buffer (20 mM HEPES, 150 mM NaCl, 20 mM EDTA) and quickly diluted 10-fold before sample deposition on the functionalized mica substrates. For details on AFM, protein expression, purification, and VWF:Ag ELISA, please refer to supplemental Material.
Sample preparation for binding studies
Human PDIA1 (Genway Biotech. Inc, San Diego, CA) was labeled with Alexa Fluor 647 carboxylic acid, succinimidyl ester (Life Technologies GmbH, Darmstadt, Germany). Measurements were conducted in 5 mM Tris buffer containing 500 mM NaCl and 2.5 mM CaCl2 at 21°C. A fixed concentration of labeled PDI (microscale thermophoresis [MST], 25 nM; fluorescence correlation spectroscopy [FCS], 3 nM) was titrated against varying concentrations of recombinant wild-type (wt) VWF, p.Cys2771Arg, and the isolated CK domain. Samples were incubated for 30 min at RT before measurement.
MST
For MST experiments, a Monolith NT.115 system (NanoTemper Technologies, Munich, Germany) was used. Fifteen percent infrared laser and 20% light-emitting diode power were applied for generating local temperature gradients and illumination, respectively. Laser on and off times were set at 60 and 10 s, respectively. Cold fluorescence was averaged over a time period of 5 s before the temperature jump, and the warm fluorescence signal was averaged over 30 s, starting 5 s after the temperature jump. About 5-µL sample volumes were filled into standard treated capillaries (NanoTemper Technologies) for measurements. For the theoretical background and MST data analysis,13,14 please refer to supplemental Material.
FCS
FCS experiments were performed on an Axiovert 200 microscope with a ConfoCor2 unit (Carl Zeiss, Jena, Germany) equipped with a 40× (NA = 1.2) water immersion apochromat objective (Carl Zeiss, Jena). For sample illuminations, a 633 HeNe laser was used. Samples were measured in 8-well LabTekI chamber slides (Nunc, Rochester, NY) 3 times for 10 × 180 s. For details on FCS data analysis and theoretical background, refer to supplemental Material.
PDI inhibitor studies
HUVECs in 8-well chamber slides (Ibidi) were incubated with 80 nM phorbol 12-myristate 13-acetate (PMA) for 25 min at 37°C/5% CO2 to induce VWF release from Weibel Palade bodies (WPBs).15 Cells were washed 5-fold with phosphate-buffered saline, and either medium or medium containing 2 µM 16F16 (Sigma-Aldrich, Taufkirchen, Germany) or 100 nM phenylarsine oxide (PAO, Merck Chemicals, Darmstadt, Germany) was added. After indicated times, VWF was detected by immunofluorescence. To visualize all WPBs within the cells, merged Z-stack images were recorded using the quick-full-focus function of the BZ9000 Keyence fluorescence microscope. In parallel, the same experiment was performed with HUVECs in 6-well plates, and after 4 and 8 h, cell lysates were prepared and investigated by standard Western Blot techniques (for details, refer to the supplemental Material).
Protein–protein docking
Protein–protein docking was employed to study interactions between the VWF CK dimer and the catalytic region of PDI (the A domain). Structures of the CK dimer, in its reduced form, were extracted from molecular dynamics (MD) simulations (for details, see supplemental Material). CK dimer conformations, sampled during the MD simulations, were clustered based on their root mean square deviation (clustering cutoff of 0.15 nm), and 5 representative structures were selected. Five structures of the oxidized PDI A domain were picked from the nuclear magnetic resonance spectroscopy ensemble of conformations (Protein Data Bank ID: 1MEK16 ) by following the identical clustering approach as for CK, but with a cutoff of 0.1 nm. To account for protein-backbone and side chain flexibility, each of the CK dimer structures was docked to each of the PDI A domain structures, resulting in 25 different docking runs. To analyze effects of the p.Cys2771Arg mutation, the same procedure was repeated using a p.Cys2771Arg-containing CK dimer. To investigate the role of the CK C-termini, additional structural models were generated by considering a truncated CK dimer, both in its wt and its mutated form, with the last 5 C-terminal amino acids of each monomer deleted. Docking calculations were carried out with PatchDock,17 a rigid-body docking algorithm, which predicts structures of 2 proteins in complex based on their shape complementarity. Amino acids potentially participating in the binding were specified in the parameter file: the catalytic residue Cys36 of PDI and all the existing cysteines involved in interchain disulfide bonds of CK. All obtained conformations were further refined with FireDock, using suggested parameters for the full refinement.18
Results
Physiological relevance of PDI–VWF interaction
Colocalization of VWF and PDI in the ER has been observed previously,7,19 but this putative interaction and its physiological relevance have not been described. Because the ER is the site of VWF dimerization, we hypothesized that PDI might be the protein that catalyzes the formation of the necessary interdimer disulfide bonds. This reaction would require interaction of PDI with 2 VWF monomers. To visualize these complexes, we performed high-resolution STORM of immunofluorescently labeled endogenous PDIA1 and VWF in HUVEC. We found PDI bound centrally within VWF clusters with a size of ∼200 nm; the expected size of VWF dimers20 (Figure 1; regions of interest 1-4; for antibody specificity, refer to supplemental Figure 1). The average localization precision of the STORM images is about 14 and 13 nm for Alexa Fluor 488 and 647, respectively (supplemental Figure 2). Coimmunoprecipitation of endogenous PDI and VWF from HUVEC lysates provides additional evidence for PDIA1–VWF interaction (Figure 2A). These findings support the hypothesis that PDI is involved in VWF dimerization.
Interaction of PDI and VWF shown by 2-color STORM super-resolution microscopy. Endogenous VWF and PDI in HUVECs were detected by indirect immunofluorescence employing rabbit anti-VWF plus goat anti-rabbit Alexa Fluor 488 (shown in green) and mouse anti-PDI plus goat anti-mouse Alexa Fluor 647 (shown in red), respectively. Colocalization is shown in yellow. Locations of magnified ROIs in the cell (left) are indicated by white squares and numbers. STORM data sets were acquired on a Nikon N-STORM microscope with an Apo TIRF 100× oil immersion objective (NA 1.49), a back-illuminated EMCCD camera, and a quadband filter composed of a quadline beam splitter and a quadline emission filter. For excitation of Alexa Fluor 647 and 488, a 647-nm continuous wave fiber laser, and the 488-nm line of an argon gas laser were used. For multicolor imaging, the lasers were switched on and off alternately, controlled by an acousto-optic tunable filter. The integration time of the EMCCD camera was set to 16 ms per frame, with an EM gain of 300. Super-resolution images were reconstructed from a series of 12 500 images per channel, using the N-STORM analysis module v. 3.3.1.15801 of NIS Elements AR v. 4.20 with overlapping peak detection enabled. Scale bar in the left cell image represents 5 µm; in magnified images, 100 nm.
Interaction of PDI and VWF shown by 2-color STORM super-resolution microscopy. Endogenous VWF and PDI in HUVECs were detected by indirect immunofluorescence employing rabbit anti-VWF plus goat anti-rabbit Alexa Fluor 488 (shown in green) and mouse anti-PDI plus goat anti-mouse Alexa Fluor 647 (shown in red), respectively. Colocalization is shown in yellow. Locations of magnified ROIs in the cell (left) are indicated by white squares and numbers. STORM data sets were acquired on a Nikon N-STORM microscope with an Apo TIRF 100× oil immersion objective (NA 1.49), a back-illuminated EMCCD camera, and a quadband filter composed of a quadline beam splitter and a quadline emission filter. For excitation of Alexa Fluor 647 and 488, a 647-nm continuous wave fiber laser, and the 488-nm line of an argon gas laser were used. For multicolor imaging, the lasers were switched on and off alternately, controlled by an acousto-optic tunable filter. The integration time of the EMCCD camera was set to 16 ms per frame, with an EM gain of 300. Super-resolution images were reconstructed from a series of 12 500 images per channel, using the N-STORM analysis module v. 3.3.1.15801 of NIS Elements AR v. 4.20 with overlapping peak detection enabled. Scale bar in the left cell image represents 5 µm; in magnified images, 100 nm.
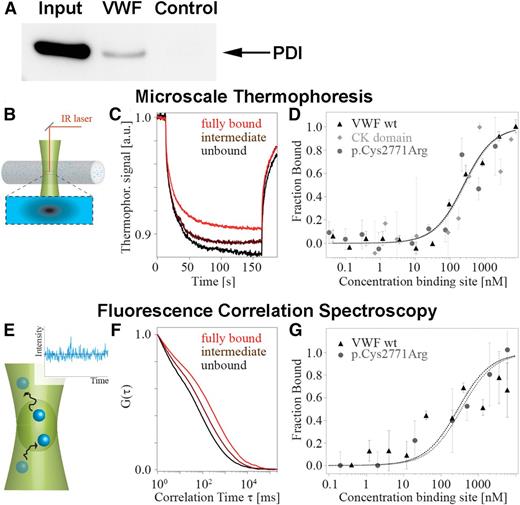
PDI binds VWF. (A) Coimmunoprecipitation of endogenous PDI and VWF from HUVEC lysates. Immunoprecipitation was performed using either rabbit anti-VWF antibody or control rabbit IgG bound to protein G-agarose. Coimmunoprecipitating proteins were separated by sodium dodecyl sulfate gel electrophoresis, and PDI was detected using mouse anti-PDI. Lanes: input, cell lysate of untreated HUVECs; VWF, immunoprecipitate from HUVEC lysate employing anti-VWF antibody; control, immunoprecipitate from HUVEC lysate employing rabbit control IgG. (B-D) MST analyzes the movement of molecules in a temperature gradient (B). (C) Binding of PDI to VWF was detected as a decrease in thermophoretic depletion. (D) PDI binds to wtVWF (black triangles), the isolated CK domain (gray squares), and p.Cys2771Arg (gray circles) with a very similar affinity (KD = 240 ± 100 nM). (E-G) FCS detects the diffusive dynamics of molecules (E). (F) Binding results in prolonged diffusion times visualized by a shift of the autocorrelation curve G(τ). (G) In agreement with the MST measurements, we detect similar affinities for wtVWF (black triangles) and mutant p.Cys2771Arg (gray circles) (KD = 300 ± 100 nM).
PDI binds VWF. (A) Coimmunoprecipitation of endogenous PDI and VWF from HUVEC lysates. Immunoprecipitation was performed using either rabbit anti-VWF antibody or control rabbit IgG bound to protein G-agarose. Coimmunoprecipitating proteins were separated by sodium dodecyl sulfate gel electrophoresis, and PDI was detected using mouse anti-PDI. Lanes: input, cell lysate of untreated HUVECs; VWF, immunoprecipitate from HUVEC lysate employing anti-VWF antibody; control, immunoprecipitate from HUVEC lysate employing rabbit control IgG. (B-D) MST analyzes the movement of molecules in a temperature gradient (B). (C) Binding of PDI to VWF was detected as a decrease in thermophoretic depletion. (D) PDI binds to wtVWF (black triangles), the isolated CK domain (gray squares), and p.Cys2771Arg (gray circles) with a very similar affinity (KD = 240 ± 100 nM). (E-G) FCS detects the diffusive dynamics of molecules (E). (F) Binding results in prolonged diffusion times visualized by a shift of the autocorrelation curve G(τ). (G) In agreement with the MST measurements, we detect similar affinities for wtVWF (black triangles) and mutant p.Cys2771Arg (gray circles) (KD = 300 ± 100 nM).
VWF and PDI are protein-binding partners
Because colocalization and coimmunoprecipitation per se do not prove direct interaction of 2 proteins, we performed extensive binding studies employing MST and FCS. These 2 methods detect complex formation by the change in movement of molecules in a temperature gradient (MST) or while diffusing through a confocal volume (FCS) after addition of a protein-binding partner. We measured binding of recombinant wtVWF to recombinant fluorescently labeled PDIA1 (Figure 2B-G) and demonstrated a direct interaction. To quantify this interaction, the dissociation constants were determined. The values measured by MST and FCS (KD = 236 ± 66 nM and KD = 282 ± 123 nM, respectively) indicate specific binding of intermediate strength, which would be expected for reversible protein interaction.
PDI binds VWF exclusively within its CK domain
VWF is rich in cysteine residues that lock the tertiary structure of the VWF A1, A3, and C domains by formation of intramolecular disulfide bonds. In the C-terminal CK domain, cysteines do not only form stabilizing intramolecular disulfide bonds but also link VWF monomers through intermolecular disulfide bonds.3 To clarify PDI’s role in VWF processing, we further performed MST of PDIA1 and the isolated CK domain (Figure 2D). PDI binding to the CK domain exhibits a dissociation constant (KD of 258 ± 104 nM) similar to the KD of PDI binding to full-length VWF. These data indicate that PDI is not generally involved in VWF folding (which would require binding to all VWF domains); they rather point to exclusive binding of PDI to the CK domain of VWF. To confirm this hypothesis, we imaged adsorbed VWF dimers with PDIA1 in AFM tapping mode (Figure 3). As expected by the KD value for VWF–PDI binding, PDI was not bound to all VWF dimers (supplemental Figure 3). To distinguish VWF dimers without (Figure 3A) and with (Figure 3B) PDI bound to the CK domain, we quantified the apparent height of the CK domain by tracing each dimer individually along its contour (Figure 3C and supplemental Figure 3). A distribution of the maximum height yielded 2 distinct peaks (Figure 3D). The first peak, at 1.2 nm, coincides with the only peak for PDI-free VWF (supplemental Figure 4A), and the second peak, at 1.9 nm, matches the height expected for a complex of the CK domain and PDI (free mean apparent height of PDI is 0.6 nm; supplemental Figure 4B). Importantly, we did not observe binding of PDI to any VWF domain other than CK.
Direct visualization of PDI bound to the VWF CK domain by AFM imaging. AFM images (A-B) and respective height traces (C) of individual VWF dimers co-adsorbed with PDI. PDI-free VWF dimers (A) and PDI-complexed VWF dimers (B) show a clear difference in the apparent height of the CK domain (indicated by arrows). Heights were analyzed by tracing each dimer individually along its contour (for more details, please refer to supplemental Figures 3 and 4). Scale bar represents 20 nm; range of color scale is 2.2 nm. (D) The distribution of the maximum apparent height of n = 246 traced CK domains reveals 2 distinct peaks at approximately 1.2 and 1.9 nm, corresponding to PDI-free and PDI-complexed VWF, respectively.
Direct visualization of PDI bound to the VWF CK domain by AFM imaging. AFM images (A-B) and respective height traces (C) of individual VWF dimers co-adsorbed with PDI. PDI-free VWF dimers (A) and PDI-complexed VWF dimers (B) show a clear difference in the apparent height of the CK domain (indicated by arrows). Heights were analyzed by tracing each dimer individually along its contour (for more details, please refer to supplemental Figures 3 and 4). Scale bar represents 20 nm; range of color scale is 2.2 nm. (D) The distribution of the maximum apparent height of n = 246 traced CK domains reveals 2 distinct peaks at approximately 1.2 and 1.9 nm, corresponding to PDI-free and PDI-complexed VWF, respectively.
PDI activity is required for VWF dimerization
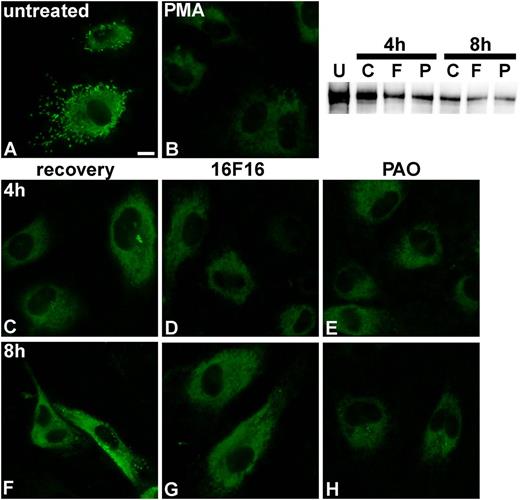
To provide more evidence that it is exclusively the activity of PDI that is required for VWF dimerization, we investigated the effects of PDI inhibition on VWF processing. We stimulated HUVECs (Figure 4A) with PMA to deplete the cells of VWF stored in WPBs (Figure 4B), as previously described.15 Then we observed the recovery of WPB formation in the absence and presence of the PDI inhibitors 16F1621 and PAO.22 As described previously,23 the first small WPB-like structures were observed after 4 h (Figure 4C), and full recovery of cigar-shaped WPBs was reached after 8 h in the absence of PDI inhibitors (Figure 4F). In contrast, WPB formation was completely abolished in cells treated with 16F16 (Figure 4D,G) and strongly reduced in the PAO-treated cells (Figure 4E,H), indicating that VWF dimerization was inhibited, leading to loss of VWF multimers, and thus WPBs. The incomplete inhibition by PAO can most likely be explained by the concentration below the 50% inhibition/inhibitory concentration value (12 µM)24 we had to use because the cells did not survive concentrations of PAO higher than 500 nM. Immunofluorescence after 2 h of recovery (Figure 4B) shows residual VWF within the cells, which most likely consists of monomers and N-terminal dimers. Western blot analysis of these HUVEC lysates showed that the VWF dimer content in the inhibitor-treated cells (Figure 4, lanes F and P) is significantly lower than in the cells that were allowed to recover from PMA treatment without inhibitors (lane C) or untreated cells (lane U), indicating inhibition of the formation of new dimers after PMA treatment by PDI inhibitors. Because 16F16 is specific for PDIA1 and PDIA3,21 these data reduce possible protein candidates for catalysis of VWF dimerization to these 2 isoforms.
Influence of PDI inhibitors on VWF processing. HUVECs (A) were incubated with 80 nM PMA for 25 min at 37°C/5% CO2 to induce VWF release from WPBs (B). Cells were washed 5-fold with phosphate-buffered saline, and either medium (C,F) or medium containing 2 µM 16F16 (D,G) or 100 nM PAO (E,H) was added. After 4 h (C-E) and 8 h (F-H), VWF was detected by immunofluorescence. Images that combine all planes of the cells were imaged, using the quick-full-focus function of the BZ9000 fluorescence microscope (Keyence) equipped with a CFI Plan Apo λ 60× H oil objective (Nikon). Representative images of 1 of 5 independent experiments are shown. Scale bar represents 10 µm. The western blot shows VWF dimers in HUVEC lysates before PMA treatment (U) and after 4 and 8 hours of recovery without PDI inhibitors (C) or in the presence of 16F16 (F) or PAO (P).
Influence of PDI inhibitors on VWF processing. HUVECs (A) were incubated with 80 nM PMA for 25 min at 37°C/5% CO2 to induce VWF release from WPBs (B). Cells were washed 5-fold with phosphate-buffered saline, and either medium (C,F) or medium containing 2 µM 16F16 (D,G) or 100 nM PAO (E,H) was added. After 4 h (C-E) and 8 h (F-H), VWF was detected by immunofluorescence. Images that combine all planes of the cells were imaged, using the quick-full-focus function of the BZ9000 fluorescence microscope (Keyence) equipped with a CFI Plan Apo λ 60× H oil objective (Nikon). Representative images of 1 of 5 independent experiments are shown. Scale bar represents 10 µm. The western blot shows VWF dimers in HUVEC lysates before PMA treatment (U) and after 4 and 8 hours of recovery without PDI inhibitors (C) or in the presence of 16F16 (F) or PAO (P).
Effect of CK domain mutations on PDI–VWF interaction
In patients with VWD type 2A/IID carrying the heterozygous mutation p.Cys2771Arg, the formation of odd-numbered multimers resulting from N-terminal disulfide bonding to wt multimers was observed.8,25 Homozygous expression yields N-terminally connected dimers only, showing a complete inhibition of the C-terminal dimerization. To investigate the mechanism of this inhibitory effect on dimerization, we performed colocalization and binding studies with the recombinant mutant. PDIA1 showed equal colocalization with mutant p.Cys2771Arg (Figure 5B; for quantification, see supplemental Table 1) compared with wtVWF (Figure 5A). The dissociation constants of p.Cys2771Arg binding to PDI, determined by both MST and FCS, exhibited values comparable to those of wtVWF–PDI binding (KD = 235 ± 92 nM [MST] and KD = 350 ± 75 nM [FCS]; Figure 2).
Colocalization of VWF and PDI. wtVWF (A) or VWF mutants p.Cys2771Arg (B) and p.Cys2773Alafs*76 (C) were transiently expressed in HEK293 cells. Forty-eight hours after transfection, cells were fixed and VWF proteins and PDI were detected by indirect immunofluorescence employing rabbit anti-VWF and mouse anti-PDI antibodies; secondary antibodies were goat anti-rabbitAF488 and goat anti-mouseAF546. Images were recorded with a confocal microscope (TCS SP5, Leica), using an HC PL APO CS2 63.0 × 1.40 oil ultraviolet objective and the following settings: image size of 512 × 512 pixels, laser power of the 543 and 488 lasers was set to 9% and 20%, respectively. Colocalization appears yellow in the overlay images; for colocalization analysis, please refer to supplemental Table 1. Scale bars represent 10 µm.
Colocalization of VWF and PDI. wtVWF (A) or VWF mutants p.Cys2771Arg (B) and p.Cys2773Alafs*76 (C) were transiently expressed in HEK293 cells. Forty-eight hours after transfection, cells were fixed and VWF proteins and PDI were detected by indirect immunofluorescence employing rabbit anti-VWF and mouse anti-PDI antibodies; secondary antibodies were goat anti-rabbitAF488 and goat anti-mouseAF546. Images were recorded with a confocal microscope (TCS SP5, Leica), using an HC PL APO CS2 63.0 × 1.40 oil ultraviolet objective and the following settings: image size of 512 × 512 pixels, laser power of the 543 and 488 lasers was set to 9% and 20%, respectively. Colocalization appears yellow in the overlay images; for colocalization analysis, please refer to supplemental Table 1. Scale bars represent 10 µm.
We further investigated colocalization of PDI and the VWF frameshift mutant p.Cys2773Alafs*76. This mutant, previously identified in patients with VWD, exhibits a single base pair deletion (8566delC) that leads to an altered amino acid sequence C-terminally of residue 2772.26 PDI shows normal colocalization with mutant p.Cys2773Alafs*76 (Figure 5C; for quantification, see supplemental Table 1) compared with wtVWF. These data indicate that an initial association between PDI and VWF occurs N-terminally of residue 2772 before the formation of disulfide bonds.
Mechanism of VWF dimerization by PDI
To predict potential binding modes of PDI to VWF and elucidate the order of disulfide bond formation, we first performed protein–protein docking of the catalytic A domain of PDI to the wtVWF CK domain dimer. For all the resulting conformations of the complex, the minimal distance between the catalytic site in the PDI A-domain and cysteines 2771, 2773, and 2811 of the CK dimer was between 0.6 and 1.2 nm (Figure 6A), which is too large to induce disulfide bond formation. Thus, these conformations do not correspond to any relevant catalytic state. This is a result of the presence of the CK C-termini, which obscured the accessibility of the PDI catalytic motif to cysteines Cys2771 and Cys2773 over the entire course of the simulations (Figure 6B). Although the bond linking the 2 C-termini (Cys2811-Cys2811′), was removed in the simulations, the separation between these cysteines was only observed to partially increase in the simulations (supplemental Figure 5A). We thus speculate that the conformational rearrangement of the C-termini, which gives accessibility to the catalytic cysteines Cys2771 and Cys2773, occurs on timescales beyond our simulations. Nevertheless, the lack of this bond increased the flexibility of the C-termini (supplemental Figure 5B). This result emphasizes the role of this bond stabilizing the C-termini in a conformation obstructing the catalytic cysteines. The docking procedure was repeated, this time after removal of the CK C-termini, mimicking open flexible C-termini in the absence of bond Cys2811-2811′. Now, the PDI catalytic motif was found closer to the CK cysteines 2771 and 2773, at distances between 0.35 and 0.7 nm (Figure 6C). These predicted structures of the complex likely represent realistic catalytic conformations (Figure 6C). Similar results were obtained when docking was performed with the mutant p.Cys2771Arg CK dimer: the PDI catalytic site approached the remaining Cys2773 only upon removal of the CK C-termini. The distances are comparable to those observed in calculations with the wtCK dimer (Figure 6A), suggesting binding in a catalytically inactive state. Taken together, our docking results suggest that PDI initiates VWF dimerization by catalyzing the formation of the Cys2771-2773′ and Cys2771′-2773 disulfide bonds. This reaction must occur before closing of the Cys2811-2811′ disulfide bond. The Cys2811-2811′ link thus most likely stabilizes the termini in a conformation shielding the other 2 disulfide bonds and renders them inaccessible to the enzyme and the environment. The stabilizing role of this link is supported by the observation of higher structural diversion and fluctuations of the reduced C-terminal β-hairpin even within the microsecond timescale of our simulations (supplemental Figure 5B).
Implications of the VWF CK-PDI A-domain complex from protein–protein docking. (A) Minimum distance, d, between the PDI A domain catalytic site and the CK dimer cysteines involved in dimerization (sulfur–sulfur distances are shown). PDI was docked to either the wt or the hypothetical p.Cys2771Arg CK dimer. The complete dimer (green) or a truncated CK dimer (blue), lacking the C termini, was considered in each case (n = 25 for each set). (B) Typical conformation of the catalytic A domain of PDI (magenta) in complex with the wtCK dimer (monomers: green and cyan), recovered from docking. The cysteine side chains of CK and the catalytic site of PDI (CS) are shown in stick representation. CK termini prevent the CS from accessing the CK cysteines. (C) Structure of the typical docking conformation of PDI–wtCK complex (same colors and representations as in B) when the CK C-terminal amino acids were removed to mimic opening of C-termini. The PDI CS approaches the indicated CK cysteines. (D) Proposed mechanism of CK dimerization mediated by PDI.
Implications of the VWF CK-PDI A-domain complex from protein–protein docking. (A) Minimum distance, d, between the PDI A domain catalytic site and the CK dimer cysteines involved in dimerization (sulfur–sulfur distances are shown). PDI was docked to either the wt or the hypothetical p.Cys2771Arg CK dimer. The complete dimer (green) or a truncated CK dimer (blue), lacking the C termini, was considered in each case (n = 25 for each set). (B) Typical conformation of the catalytic A domain of PDI (magenta) in complex with the wtCK dimer (monomers: green and cyan), recovered from docking. The cysteine side chains of CK and the catalytic site of PDI (CS) are shown in stick representation. CK termini prevent the CS from accessing the CK cysteines. (C) Structure of the typical docking conformation of PDI–wtCK complex (same colors and representations as in B) when the CK C-terminal amino acids were removed to mimic opening of C-termini. The PDI CS approaches the indicated CK cysteines. (D) Proposed mechanism of CK dimerization mediated by PDI.
Importance of disulfide bond Cys2811-2811′
The existence of bond Cys2811-2811′ in VWF dimers has been described by Zhou et al.,3 but no patients with VWD carrying a Cys2811 mutation have been identified. Therefore, no clinical evidence is available to explain the importance of disulfide bond Cys2811-2811′. To confirm our hypothesis that this bond is necessary to stabilize dimerization, we cloned mutant p.Cys2811Ala and recombinantly expressed it in HEK293 cells. The intracellular localization of this mutant (Figure 7B; supplemental Video 2) was comparable to that of wtVWF (Figure 7A; supplemental Video 1), but multimer analysis revealed residual monomers, odd-numbered multimers (Figures 7G; supplemental Figure 6), and a mild loss of HMWMs (Figure 7G). These data support the hypothesis that bond Cys2811-2811′ is critical to protect bonds Cys2771-2773′ and Cys2771′-2773 from reduction.
Intracellular localization and multimer pattern of mutant p.Cys2811Ala. wtVWF (A) and VWF mutant p.Cys2811Ala (D) were transiently expressed in HEK293 cells. Forty-eight hours after transfection, cells were fixed and VWF proteins (A,D) and PDI (B,E) were detected by indirect immunofluorescence employing rabbit anti-VWF and mouse anti-PDI antibodies, respectively. Z-stacks were recorded with a confocal microscope using an HC PL APO CS2 63.0 × 1.40 oil ultraviolet objective and the following settings: image size of 512 × 512 pixels, laser power of the 543 and 488 lasers was set to 9% and 20%, respectively. Overlays are shown in (C) and (F). Three-dimensional reconstruction was performed using the LAS software (Leica). Scale bars represent 10 µm. For movies of the rotating complete 3-dimensional reconstruction, please refer to supplemental Videos 1 and 2. (G) Multimer analysis of recombinant wtVWF and mutant p.Cys2811Ala was performed by sodium dodecyl sulfate–agarose gel electrophoresis and immunoblotting onto a nitrocellulose membrane with luminescent visualization. The figure is composed of one gel. The black line indicates deleted lanes with mutants not relevant for this study. Additional bands in mutant p.Cys2811Ala resulting from odd-numbered multimers are indicated by black arrows. For resolution of the monomer and dimer band, please refer to supplemental Figure 6.
Intracellular localization and multimer pattern of mutant p.Cys2811Ala. wtVWF (A) and VWF mutant p.Cys2811Ala (D) were transiently expressed in HEK293 cells. Forty-eight hours after transfection, cells were fixed and VWF proteins (A,D) and PDI (B,E) were detected by indirect immunofluorescence employing rabbit anti-VWF and mouse anti-PDI antibodies, respectively. Z-stacks were recorded with a confocal microscope using an HC PL APO CS2 63.0 × 1.40 oil ultraviolet objective and the following settings: image size of 512 × 512 pixels, laser power of the 543 and 488 lasers was set to 9% and 20%, respectively. Overlays are shown in (C) and (F). Three-dimensional reconstruction was performed using the LAS software (Leica). Scale bars represent 10 µm. For movies of the rotating complete 3-dimensional reconstruction, please refer to supplemental Videos 1 and 2. (G) Multimer analysis of recombinant wtVWF and mutant p.Cys2811Ala was performed by sodium dodecyl sulfate–agarose gel electrophoresis and immunoblotting onto a nitrocellulose membrane with luminescent visualization. The figure is composed of one gel. The black line indicates deleted lanes with mutants not relevant for this study. Additional bands in mutant p.Cys2811Ala resulting from odd-numbered multimers are indicated by black arrows. For resolution of the monomer and dimer band, please refer to supplemental Figure 6.
Discussion
Employing different high-resolution methods, we found independent evidence that PDIA1 directly binds to VWF with a strong affinity. Moreover, we identified the CK domain as the only PDIA1 binding domain within VWF. To show that VWF is dimerized by PDIA1 without the compensatory effect of other thiol isomerases, we could not use siRNA because PDIA1 protein levels can only be reduced to 20%, leading to overall reduction of VWF levels without an obvious effect on dimerization27-29 (supplemental Figures 8 and 9). Nonetheless, we were able to rule out effects of PDIA2 because HUVECs do not express this PDI isoform (supplemental Figure 10). By employing the PDIA1- and PDIA3-specific inhibitor 16F16,21 we were able to reduce the candidates to these 2 PDI isoforms. It has previously been reported that PDIA3 exhibits increased binding to misfolded VWF mutants with N-terminal mutations.30 These data indicate that PDIA3 is able to bind to N-terminal regions of VWF, which speaks against an involvement of PDIA3 in dimerization, but for a role in protein repair of VWF mutants. Thus, our data provide strong evidence that VWF dimerization is performed mainly or even exclusively by PDIA1.
The combined data of our docking and MD simulations, as well as the colocalization studies of VWD-associated VWF mutants, suggest the following mechanism of VWF dimerization (Figure 6D): An initial, probably electrostatic, association between PDI and VWF occurs N-terminally of residue 2772. The protein–protein docking calculations showed that the catalytic site of PDI can only interact with Cys2771 and Cys2773 as long as the C-termini are not obstructing them; a condition that is met when the disulfide bond Cys2811-Cys2811′ is not yet formed. Hence, the latter bond has to be formed last. The docking results further showed no preference of PDI for either Cys2771 or Cys2773 in wtCK, suggesting PDI does not distinguish between these 2 cysteines. Therefore, oxidized PDI initially establishes a disulfide bond between its catalytic residue Cys36 and either Cys2771 or Cys2773 in one VWF monomer. Then PDI catalyzes the formation of the first disulfide bond, which can either be Cys2771-2773′ or Cys2771′-2773 (step i, Figure 6D). In the next step, a new oxidized PDI molecule binds (II) and catalyzes (III) the formation of the second disulfide bond. The CK dimer is now stabilized by the 2 formed disulfide bonds, and the CK C-termini assemble into a β-sheet conformation (IV), which brings Cys2811 and Cys2811′ in proximity, enhancing their propensity to form the last disulfide bond Cys2811-2811′ (V). Our simulation data suggest that formation of the latter bond is not necessarily catalyzed by PDI. It could also be a spontaneous process or be catalyzed by a different enzyme or even a small molecule (eg, glutathione). We hypothesize that this far C-terminal disulfide bond acts as a protective cover, rendering oxidation of Cys2771 and Cys2773 by PDI (and thereby VWF dimerization) irreversible by sterically occluding these 2 essential disulfides. Indeed, we found odd-numbered multimers and residual monomers in the supernatant of cells expressing mutant p.Cys2811Ala, indicating partial reopening of VWF dimers by reduction of the first 2 bonds (Figure 7G; supplemental Figure 6). The normal intracellular localization and mild loss of HMWMs of this mutant suggest that mutation of Cys2811 would lead to no or only very mild bleeding symptoms. Together with the low incident of CK domain mutations and an effect only expected for homozygous expression, these data might explain why this mutation has never been identified in individuals with bleeding symptoms.
Our data can further explain the pathomechanism of VWD subtypes induced by CK domain mutations: For mutant p.Cys2771Arg, we found normal binding to PDI and complete abolishment of C-terminal dimerization.5,8 Although MD simulations suggest small distances between the 2 Cys2773 residues within a hypothetical p.Cys2771Arg dimer (supplemental Figure 7), this bond cannot be formed as proven by multimer analysis.5,8 Probably, conformational changes prevent the formation of an alternative Cys2773-2773′ bond, causing the dimerization defect in p.Cys2771Arg. This hypothesis is supported by our previous observation that conformational changes in the Cys2771Arg monomer lead to an extended conformation that impedes the formation of an alternative Cys2773-Cys2773′ bond.8
For mutant p.Cys2773Arg, it has previously been described that this mutation leads to odd-numbered multimers in heterozygous patients.31 However, homozygous expression reduces only formation of HMWMs, showing merely a partial dimerization defect. The intracellular localization and colocalization with PDI were found to be normal.31 Therefore, PDI seems to be able to at least use Cys2771 to form residual dimers by the formation of an alternative Cys2771-2771′ bond.
Summarizing, our data showed, for the first time to our knowledge, direct interaction of VWF and PDIA1. We suggest a mechanism for VWF dimerization and found indication that the pathomechanism of CK domain mutation-derived VWD phenotypes can be explained by inhibition of PDI-catalyzed disulfide bond formation. Further, we used high-resolution methods such as STORM, MST, FCS, and AFM that proved to be valuable tools to investigate protein–protein interactions. These methods are not yet commonly used in medical sciences or diagnostics but could become increasingly important in these fields because they allow the investigation of protein–protein interaction with higher resolution, sensitivity, and specificity than currently preferred methods.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Nikon Center of Excellence for Integrative Microscopy at the Heinrich-Pette-Institute for providing the Nikon N-STORM microscope and the University Medical Center Hamburg-Eppendorf (UKE) Microscopy Imaging Facility for technical support and providing the Leica SP5 microscope. We thank Sarah Vollmers for technical assistance and Florian Langer for helpful discussions. W.V. thanks Research Foundation-Flanders (FWO) for a postdoctoral fellowship and a travel grant for a long stay abroad. This study was supported by research funding from the German Research Foundation to the Research Group FOR1543: “Shear flow regulation of hemostasis - bridging the gap between nanomechanics and clinical presentation” (J.O.R., S.L., R.S., M.A.B., T.O., G.K., C.B., C.A.-S., F.G., J.P.M.).
Authorship
Contribution: S.L. performed MST and FCS experiments and wrote the manuscript. S.L. and J.O.R. analyzed the MST and FCS data. D.E. performed and analyzed STORM experiments. A.L., W.V., and J.P.M. performed and analyzed atomic force microscopy experiments. G.K. performed immunofluorescence, and K.R. performed the inhibitor experiments. T.O. cloned VWF mutants. R.S. designed VWF mutants and critically reviewed the manuscript. K.K. performed the docking calculations and MD simulations. K.K., C.B., C.A.-S., and F.G. designed and analyzed the docking and MD experiments. K.K., C.B., C.A.-S., and F.G. wrote the manuscript. S.S. and U.B. performed multimer analysis. M.A.B. designed this study, performed localization studies, and wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Maria A. Brehm, Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Martinistrasse 52, 20246 Hamburg, Germany; e-mail: m.brehm@uke.de.