Key Points
Circular RNAs are hugely enriched in platelets compared with nucleated cell types.
Lack of enrichment in megakaryocte progenitors implicates degradation of platelet linear RNAs.
Abstract
In platelets, splicing and translation occur in the absence of a nucleus. However, the integrity and stability of mRNAs derived from megakaryocyte progenitor cells remain poorly quantified on a transcriptome-wide level. As circular RNAs (circRNAs) are resistant to degradation by exonucleases, their abundance relative to linear RNAs can be used as a surrogate marker for mRNA stability in the absence of transcription. Here we show that circRNAs are enriched in human platelets 17- to 188-fold relative to nucleated tissues and 14- to 26-fold relative to samples digested with RNAse R to selectively remove linear RNA. We compare RNAseq read depths inside and outside circRNAs to provide in silico evidence of transcript circularity, show that exons within circRNAs are enriched on average 12.7 times in platelets relative to nucleated tissues and identify 3162 genes significantly enriched for circRNAs, including some where all RNAseq reads appear to be derived from circular molecules. We also confirm that this is a feature of other anucleate cells through transcriptome sequencing of mature erythrocytes, demonstrate that circRNAs are not enriched in cultured megakaryocytes, and demonstrate that linear RNAs decay more rapidly than circRNAs in platelet preparations. Collectively, these results suggest that circulating platelets have lost >90% of their progenitor mRNAs and that translation in platelets occurs against the backdrop of a highly degraded transcriptome. Finally, we find that transcripts previously classified as products of reverse transcriptase template switching are both enriched in platelets and resistant to decay, countering the recent suggestion that up to 50% of rearranged RNAs are artifacts.
Introduction
Platelets are small, anucleate, circulating blood cells derived from polyploid megakaryocyte progenitors1 and are vital for functions such as hemostasis, wound healing, and angiogenesis.2 Unlike anucleate erythrocytes, which have a lifespan of ∼4 months and have greatly reduced ribosome numbers,3 platelets are short lived (∼8-11 days4 ) and translationally competent,5 and a number of genes have been shown to undergo cytoplasmic splicing on platelet activation.6,7 Consistent with these observations, recent RNAseq analyses have defined a complex platelet transcriptome.8-11 However, some transcripts believed to contribute to platelet function are present at very low levels in RNAseq data,11 and attempts to integrate transcriptome and proteome data have given conflicting results.12-15 In the absence of nuclear transcription, RNAs within platelets are assumed to degrade over time. Although the abundance of individual mRNAs has been shown to fall in platelets at ambient temperatures,16 the stability and integrity of mRNAs within circulating platelets is poorly characterized at the transcriptome level. However, this is highly relevant to functional inferences drawn from RNAseq data.9,11,15,17,18
Transcripts with rearranged exon order are now known to be common in eukaryotic organisms and are characterized by back-splice exon junctions inconsistent with the underlying genomic DNA.19-24 Evidence for linear and polyadenylated structures has been presented,20,25,26 but it is now clear that most are circular (circRNAs) and cytoplasmic.21-23 Recent experiments have established that intronic repeats flanking rearranged exons can promote circularization and that circRNAs primarily use canonical splice sites27-30 and can be regulated by RNA binding proteins.29,31 Although 2 noncoding circRNAs act as miRNA sponges,22,26 and circRNAs with retained introns (exon-intron or EIcircRNAs) can enhance transcription of their parental genes through interaction with U1 snRNP and RNA PolII,32 the vast majority of circRNAs consist of exons from protein encoding genes and have no known function.22,33
circRNAs are resistant to attack by exonucleases involved in the regulation of mRNA,34 and resistance to one, RNAse R, has been used to define them.23,35 Furthermore, blocking of transcription can increase circRNA levels relative to linear.23 This differential stability raises the possibility that abundance of circRNAs relative to linear forms could provide insight into the platelet transcriptome. Here, we analyze the circRNA population of platelets and compare it to populations in nucleated cells, anucleate erythrocytes, and cells where linear RNA has been digested with RNAse R. We show that circRNAs are highly enriched in both platelets and erythrocytes relative to nucleated tissues and confirm this experimentally using quantitative polymerase chain reaction (qPCR). We identify >3000 genes where circRNAs are significantly enriched relative to nucleated cell types, including some genes where only circRNAs producing exons are represented within the RNAseq data. We also use qPCR to establish that circRNAs are not enriched in cultured magakaryocytes and that, over time, the levels of linear RNAs fall relative to circRNAs in platelet preparations. Our results suggest that >90% of mRNAs have been lost from circulating platelets and have potentially important implications for both platelet biology and our understanding of circRNAs.
Materials and methods
Datasets used
In silico analyses were performed on RNAseq data generated during this study and on 36 publicly available datasets: 24 ribosomal RNA depleted total RNA samples sequenced on the Illumina platform,11,23,27,36,37 2 polyA+ selected samples sequenced on the Ilumina platform,11,38 and 10 nonribosomal RNA depleted total RNA samples sequenced on the SOLiD platform.15 For details, see supplemental Methods and supplemental Table S1 available on the Blood Web site.
Erythrocyte RNAseq
Ribosome-depleted RNAseq of total RNA from red blood cells (RBCs) was performed by AROS Applied Biotechnology (Aarhus, Denmark) using the TruSeq RiboZero Stranded mRNA LT kit (Illumina). The library was sequenced using a paired-end 100-bp protocol according to the manufacturer’s recommendations (supplemental Methods).
circRNA identification
Identification of back-splice exon junctions was performed using PTESFinder Pipeline v. 1 (http://sourceforge.net/projects/ptesfinder-v1/). This identifies RNAseq reads that map in inverted order but the same orientation to the same RefSeq entry, generates sequence models of back-splice junctions, and remaps all reads to the models. It then removes reads that do not use known exon junctions or that map equally well or better to the genomic or transcriptome reference (see supplemental Methods and supplemental Figure S1 for details).
Biological samples: origin and preparation
Tissue RNAs were obtained from Biochain (Amsbio, UK). RBCs were obtained from the Scottish National Blood Transfusion Service in accordance with the terms of the standard donor consent form and information and with approval of the Scottish National Blood Transfusion Service Sample Governance Committee. Cord blood was obtained after informed consent under a protocol approved by the National Research Ethics Service (Cambridgeshire 4 Research Ethics Committee ref. no. 07/MRES/44). Whole blood samples were collected from healthy volunteers with approval from Newcastle University’s Faculty of Medical Sciences Ethics Committee. Platelets and platelet-rich plasma (PRP) were isolated according to a protocol that uses a platelet inhibitory cocktail.39 Isolation of peripheral blood mononuclear cells (PBMCs) and RBCs was performed using Histopaque-1.077 density gradients (Sigma-Aldrich). Megakaryoctes were cultured and isolated according to the protocol of Tijssen et al.40 For further details, see supplemental Methods.
RNA isolation and cDNA generation
RNA was isolated using Trizol (Lifetechnologies) and treated with DNase I (Promega) according to manufacturer’s instructions. RNA quantitation was performed using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific), and quality was assessed using the Agilent 2100 Bioanalyser (Agilent Technologies). cDNA was synthesized using high-capacity cDNA kits (Applied Biosystem), random hexamers, and Moloney murine leukemia virus (MMLV) unless stated otherwise.
Real-time PCR
Real-time PCR was performed using Taqman mastermix (Life Technologies) as described previously.20 Primers and assay efficiencies are given in supplemental Table S2. Transcript expression was normalized using the ∆CT method relative to the geometric mean of 4 housekeeping genes (GAPDH, PPIA, TUBB, and GUSB) analyzed using TaqMan gene expression assays (Applied Biosystems). Circular transcripts were also normalized against the linear transcript from the same gene where appropriate. The Wilcoxon rank sum test was used to compare levels of circRNA vs linear RNA across multiple structures (analysis of paired –ΔCt values) and to compare linear vs circRNA expression ratios between conditions or sample types (analysis of –ΔΔCt values across all structures).
Results
Platelets are highly enriched for circRNAs
We first identified 3 publicly available RNAseq datasets from human platelets,9,11,15 one of which had a sequence read length long enough for the identification of back-splice junctions within circRNAs and included samples processed to retain unpolyadenylated RNAs (Array express ID E-MTAB-1846; 100-bp Illumina ribosome-depleted total RNAseq from 3 individuals, and polyA+ RNAseq from 1 individual11 ). Analysis of the 3 total RNAseq samples using a back-splice exon junction discovery pipeline (PTESFinder; see “Materials and methods”) identified 33 829 distinct structures consistent with circRNAs (supplemental Figure S1; supplemental Table S3), ∼4 to 5 times more than reported for fibroblasts23 and ∼15 times more than reported for leukocytes and HEK293 cells22 in libraries of comparable size. Analysis with independent software previously used for identification of both genic and nongenic circRNAs22 also identified an excess in platelets relative to these datasets (supplemental Figure S1). Furthermore, ∼63% of the 33 829 structures have previously been identified within RNAseq data generated following RNAse R digestion (to selectively remove linear transcripts23,27 ), consistent with them being circRNAs. Despite the large number identified, the majority are not platelet specific, as 24 632 (∼73%) were also present in ≥1 of 15 ENCODE (Encyclopedia of DNA Elements) RNAseq datasets previously mined by Salzman et al41 (supplemental Figure S1).
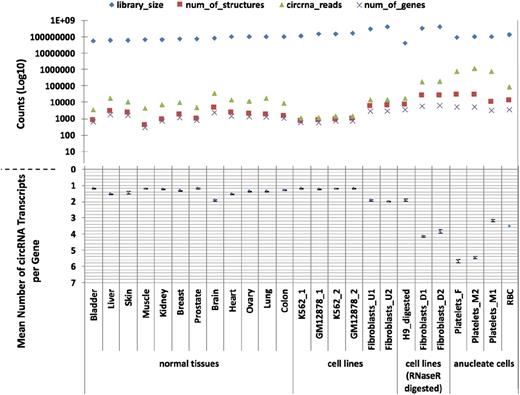
Having identified an apparent abundance of circRNAs in platelets, we then used PTESFinder to identify circRNAs within other publicly available datasets generated using ribosome depleted total RNAseq and sequenced using the Illumina platform. These comprised 12 human tissues,37 ENCODE cell lines,42 and cell lines treated with RNAse R to selectively remove linear RNAs.23,27 To enable comparison with a distinct anucleate cell type, we also generated >120 million 100-bp paired-end reads from total RNA extracted from leukocyte depleted RBCs (GEO record no. GSE69192; see “Materials and methods”). Analyses of these data, summarized in Figure 1 and presented in supplemental Table S3, show that 17 to 188 times more circRNA (back-splice) reads are present in platelets than in human nucleated tissues (7579-11 138 circRNA junction reads per million reads vs 59-447), and 14 to 26 times more than in samples treated with RNAse R (427-463 junction reads per million reads). In addition, the mean numbers of circRNA structures per circRNA producing gene are larger in anucleate and RNAse R digested samples than in others, with 2 of the 3 platelet samples averaging >5 structures per gene compared with 1 to 2 for nucleated tissues and cell lines.
CircRNAs in human tissues. The number of circRNAs (structure counts), junction reads (back-splice read counts), circRNA producing genes (gene counts), mean number of circRNA transcripts per circRNA producing genes, and RNAseq library sizes (read counts) are shown. Samples are presented as 4 groups: nucleated tissues, nucleated cell lines, nucleated cell lines digested with RNAse R, and anucleated cell types. The number of back-splice reads is significantly higher in anucleate samples (platelets and RBCs) than all others (P = 2.7 × 104, Wilcoxon rank sum test; figures adjusted for library size). All data were processed using PTESFinder, and all were publicly available with the exception of the data from RBCs (see “Materials and methods” and supplemental Table S1 for details of datasets, and supplemental Table S3 for details of structures).
CircRNAs in human tissues. The number of circRNAs (structure counts), junction reads (back-splice read counts), circRNA producing genes (gene counts), mean number of circRNA transcripts per circRNA producing genes, and RNAseq library sizes (read counts) are shown. Samples are presented as 4 groups: nucleated tissues, nucleated cell lines, nucleated cell lines digested with RNAse R, and anucleated cell types. The number of back-splice reads is significantly higher in anucleate samples (platelets and RBCs) than all others (P = 2.7 × 104, Wilcoxon rank sum test; figures adjusted for library size). All data were processed using PTESFinder, and all were publicly available with the exception of the data from RBCs (see “Materials and methods” and supplemental Table S1 for details of datasets, and supplemental Table S3 for details of structures).
We also analyzed the platelet polyA+ sample from E-MTAB-1846 and identified 841 circRNA structures from 453 genes. However, their nucleotide composition was found to be significantly enriched for A residues relative to structures present only in platelet total RNA (χ2 = 12 317; P < 2.2e−16; supplemental Table S4). As this suggests these structures are nonpolyadenylated circRNA transcripts rich in A nucleotides, they were not analyzed further.
More than 600 platelet genes generate more than 20 distinct circRNAs
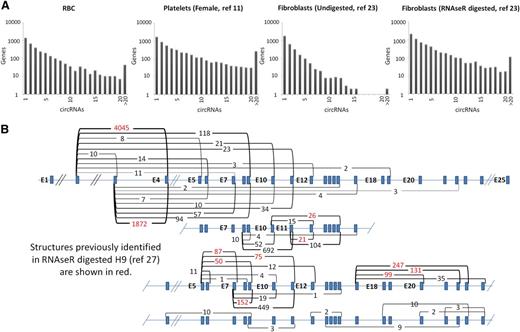
To illustrate the variation in circRNA distributions, the numbers of circRNA structures identified per gene are displayed for 4 samples in Figure 2A. Strikingly, both platelets and fibroblasts digested with RNAse R (which increases circRNA levels by degrading linear RNAs) have large numbers of genes generating >15 distinct circRNA structures. For example, the platelet sample has >600 genes each with >20 different circRNAs. Furthermore, as shown in Table 1, individual structures in anucleate samples are supported by much larger back-splice junction read counts than in nucleated tissues (P = 2.7 × 10−4 anucleate vs nucleated counts, Wicoxon rank sum test). All 3 platelet samples have >1000 structures supported by >100 reads, in sharp contrast to undigested nucleated samples. As an example of the diversity of structures and read support within platelets, the 49 circRNA structures identified within the XPO1 gene are shown in Figure 2B, with supporting read counts ranging from 1 (E8-E5 and E16-E7) to 4045 (E4-E2). It is noteworthy that all 21 internal exons of this gene are encompassed by ≥1 circRNA (illustrating that most exons within some genes can contribute to circRNAs). Only 11 of these structures were identified previously in RNAse R digested H9 RNA.27
circRNA structures per gene. (A) Histograms showing circRNA numbers per gene established using PTESFinder for RBCs (this study), platelets,11 fibroblasts, and fibroblasts digested with RNAse R.23 (B) Schematic diagram showing XPO1 intron/exon organization with read counts and inferred structure of all circRNAs identified within the 3 platelet samples. Inferred structures assume that all internal Refseq exons are present within the structure defined by each back-splice. Read counts for the 11 structures identified previously within RNAse R digested H9 ES cells27 are highlighted in red.
circRNA structures per gene. (A) Histograms showing circRNA numbers per gene established using PTESFinder for RBCs (this study), platelets,11 fibroblasts, and fibroblasts digested with RNAse R.23 (B) Schematic diagram showing XPO1 intron/exon organization with read counts and inferred structure of all circRNAs identified within the 3 platelet samples. Inferred structures assume that all internal Refseq exons are present within the structure defined by each back-splice. Read counts for the 11 structures identified previously within RNAse R digested H9 ES cells27 are highlighted in red.
circRNA read support for structures and their frequency in all samples
| circRNA reads . | Bladder . | Liver . | Skin . | Muscle . | Kidney . | Breast . | Prostate . | Brain . | Heart . | Ovary . | Lung . | Colon . | K562_1 . | GM12878_1 . | K562_2 . | GM12878_2 . | Fibroblasts_U1 . | Fibroblasts_U2 . | H9_D . | Fibroblasts_D1 . | Fibroblasts_D2 . | Platelet_F . | Platelet_M2 . | Platelet_M1 . | RBCs . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| >10 | 55 | 308 | 164 | 136 | 196 | 186 | 71 | 670 | 235 | 263 | 521 | 190 | 3 | 9 | 11 | 6 | 125 | 141 | 227 | 2797 | 3052 | 7281 | 9605 | 8459 | 1272 |
| >100 | 0 | 11 | 3 | 2 | 2 | 3 | 1 | 19 | 5 | 3 | 6 | 5 | 0 | 0 | 0 | 0 | 4 | 2 | 2 | 200 | 224 | 1197 | 1686 | 1325 | 96 |
| >1000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 5 | 83 | 147 | 95 | 2 |
| K/M | 66 | 295 | 170 | 69 | 104 | 126 | 59 | 447 | 148 | 125 | 172 | 84 | 9 | 7 | 9 | 9 | 45 | 36 | 427 | 546 | 464 | 8469 | 11 138 | 7579 | 597 |
| circRNA reads . | Bladder . | Liver . | Skin . | Muscle . | Kidney . | Breast . | Prostate . | Brain . | Heart . | Ovary . | Lung . | Colon . | K562_1 . | GM12878_1 . | K562_2 . | GM12878_2 . | Fibroblasts_U1 . | Fibroblasts_U2 . | H9_D . | Fibroblasts_D1 . | Fibroblasts_D2 . | Platelet_F . | Platelet_M2 . | Platelet_M1 . | RBCs . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| >10 | 55 | 308 | 164 | 136 | 196 | 186 | 71 | 670 | 235 | 263 | 521 | 190 | 3 | 9 | 11 | 6 | 125 | 141 | 227 | 2797 | 3052 | 7281 | 9605 | 8459 | 1272 |
| >100 | 0 | 11 | 3 | 2 | 2 | 3 | 1 | 19 | 5 | 3 | 6 | 5 | 0 | 0 | 0 | 0 | 4 | 2 | 2 | 200 | 224 | 1197 | 1686 | 1325 | 96 |
| >1000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 5 | 83 | 147 | 95 | 2 |
| K/M | 66 | 295 | 170 | 69 | 104 | 126 | 59 | 447 | 148 | 125 | 172 | 84 | 9 | 7 | 9 | 9 | 45 | 36 | 427 | 546 | 464 | 8469 | 11 138 | 7579 | 597 |
For each sample, the number of distinct circRNA structures supported by >10, 100, and 1000 circRNA (back-splice) reads are shown, together with the number of circRNA reads per million reads (K/M).
D, digested with RNAse R; U, undigested, matched sample.
Real-time PCR confirmation of high circRNA transcript levels in anucleate cells
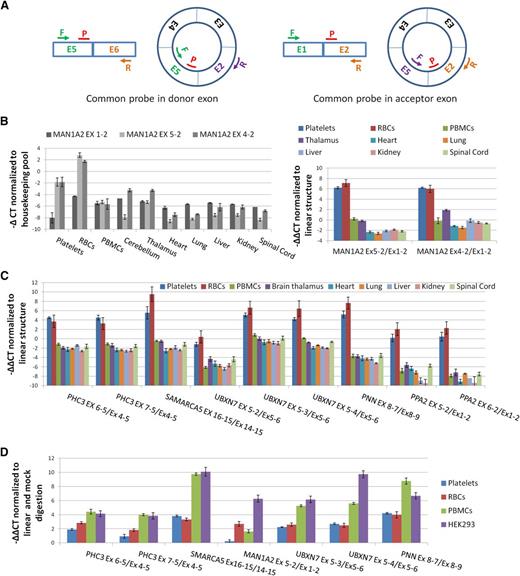
To experimentally confirm enrichment of circRNAs in both anucleate cell types, we used qPCR to assess relative levels of linear and circRNAs. We first assayed the CD45 antigen (PTPRC), which is highly expressed in PBMCs, and performed cell counts to confirm that PBMCs were being efficiently removed during platelet isolation (supplemental Figure S2; supplemental Methods). As a precaution against reverse transcriptase (RT)-specific artifacts,43 we also checked that all circRNAs being analyzed could be amplified using templates generated with both avian myeloblastosis virus (AMV) and MMLV RTs (supplemental Figure S3). We then analyzed the abundance of 2 known circRNA structures from MAN1A2 (E4-E2 and E5-E222 ) relative to the canonical MAN1A2 transcript, 2 structured from PHC3 (a gene expressed in platelets for which assays were already available20 ), and an additional 7 abundant circRNAs with their associated linear RNAs (supplemental Table S2). For each, paired assays with a common probe were used (Figure 3A).
Confirmation of circRNA abundance and resistance to RNAse R. (A) Schema of qPCR assays using an E5-E2 circRNA as an example. All assays use a common reporter probe and use either an exon downstream of the circRNA to assay linear expression (probe in donor exon) or an exon upstream (probe in acceptor exon). (B) (Left) Expression levels (−∆CT values) of linear (Ex1-2) and circular (Ex5-2 and Ex4-2) MAN1A2 transcripts relative to housekeeping pool. (Right) Expression levels of circRNAs relative to linear RNAs from the same loci normalized to housekeeping pool (−∆∆CT values). (C) Expression of 9 circRNAs relative to linear forms from the same loci, normalized to housekeeping pool (−∆∆CT values). (D) Change in CT values of circRNAs relative to linear forms from the same loci in RNAse R digested RNAs, normalized to mock digested RNAs (−∆∆CT values). Templates, circRNAs, and linear forms assayed are indicated.
Confirmation of circRNA abundance and resistance to RNAse R. (A) Schema of qPCR assays using an E5-E2 circRNA as an example. All assays use a common reporter probe and use either an exon downstream of the circRNA to assay linear expression (probe in donor exon) or an exon upstream (probe in acceptor exon). (B) (Left) Expression levels (−∆CT values) of linear (Ex1-2) and circular (Ex5-2 and Ex4-2) MAN1A2 transcripts relative to housekeeping pool. (Right) Expression levels of circRNAs relative to linear RNAs from the same loci normalized to housekeeping pool (−∆∆CT values). (C) Expression of 9 circRNAs relative to linear forms from the same loci, normalized to housekeeping pool (−∆∆CT values). (D) Change in CT values of circRNAs relative to linear forms from the same loci in RNAse R digested RNAs, normalized to mock digested RNAs (−∆∆CT values). Templates, circRNAs, and linear forms assayed are indicated.
Both of the MAN1A2 circRNA junctions are much more abundant in platelets and RBCs than the linear mRNA (Figure 3B). However, they are less abundant than the linear transcripts in virtually all nucleated tissues analyzed. This is consistent with the RNAseq data and previous analyses of nucleated tissues.20,22 The other 9 circRNAs are also expressed at higher levels in platelets and RBCs than their corresponding linear structures (Figure 3C), with circRNAs from SMARCA5, UBXN7, and PNN registering ∼6 to 10 cycles before their corresponding linear RNAs, suggesting they are ∼50- to 1000-fold more abundant. In contrast, all are expressed in nucleated cells at an equivalent or lower level than their linear counterparts. Collectively, the difference in relative expression levels of linear and circRNAs between anucleate and nucleated tissues is highly significant (P = 2.7 × 10−12, Wilcoxon rank sum test).
Finally, the impact of RNAse R digestion on expression was also assessed (Figure 3D). Digestion increases the levels of all back-splice junctions relative to the canonical linear junctions in each cell type analyzed (P = 7.8 × 10−3, Wilcoxon paired rank sum test; mock digest vs RNAse R digested), although the MAN1A2 E5-E2 back-splice shows only a marginal increase in the platelet sample. This confirms that these structures include circRNAs. However, the impact of RNAse R digestion is more pronounced in PBMCs and in HEK293 cells than in either anucleate sample (P < .02, Wilcoxon paired rank sum test for all pairwise nucleated vs anucleated comparisons; Figure 3D).
For some genes, only reads from circRNA producing exons are detected in platelets
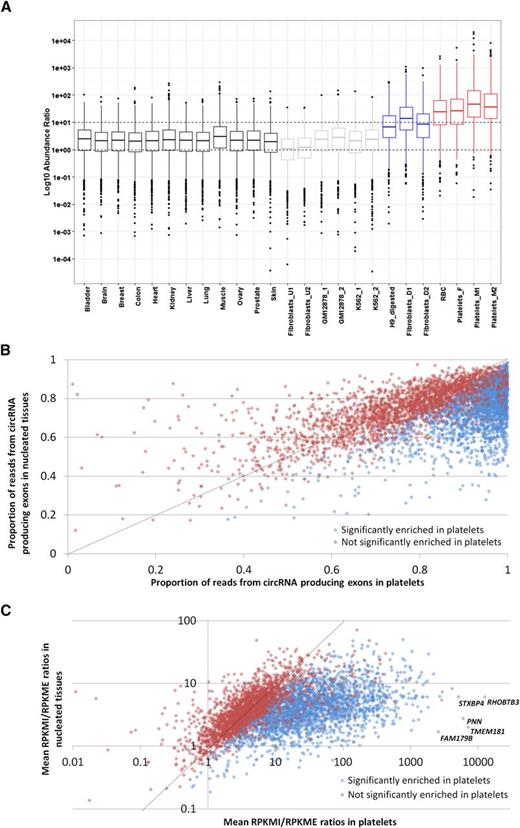
The abundance of individual circRNAs relative to mRNAs from the same loci has previously been estimated in silico by comparing back-splice junction read counts to flanking canonical junction counts.20,21,44 However, because most circRNA producing genes generate multiple overlapping structures (Figure 2B; supplemental Table S3), the canonical junctions flanking one circRNA are often present within others. We therefore performed a transcriptome-wide analysis where we calculated for each gene the reads per kilobase of transcript per million mapped reads (RPKM) across all exons internal to all known circRNA structures (RPKMI), and the RPKM across exons external to all known circRNAs (RPKME). The former is a measure of both linear and circRNA expression, the latter is linear only. Box and whisker plots showing RPKMI/RPKME ratios for all circRNA producing genes in all samples are presented in Figure 4A. The ratios from individual genes extend over 6 to 8 orders of magnitude for each sample, but what is most striking is that the median RPKMI/RPKME ratios of the anucleate samples are ∼12 times higher than nucleated tissues and ∼5 times higher than samples digested with RNAse R (Figure 4A). The distribution of RNAseq reads therefore reflects the pattern of circRNA structures inferred from exon-junction read counts alone (Figures 1 and 2) and provides further evidence that back-splice junctions are predominantly derived from circRNAs.
Differential read depth defines genes with significant circRNA enrichment in platelets. (A) Box and whisker plots showing the ratio of RPKM from circRNA producing exons (RPKMI) to RPKM from exons that do not produce circRNAs (RPKME) for all genes in each sample. The median and upper and lower quartiles are shown, with outliers as solid circles. (B) The proportion of reads from circRNA producing exons averaged across all nucleated samples (y-axis) and platelets (x-axis). (C) Fold enrichment of reads from circRNA producing exons in platelets relative to nucleated tissues. All genes with an average RPKMI >1 in platelets and expressed in 8 or more nucleated tissues are shown. Blue, genes significantly enriched in platelets; red, genes not significantly enriched in platelets. The data points corresponding to the 5 most enriched genes are indicated. The slope x = y is shown as a dashed line.
Differential read depth defines genes with significant circRNA enrichment in platelets. (A) Box and whisker plots showing the ratio of RPKM from circRNA producing exons (RPKMI) to RPKM from exons that do not produce circRNAs (RPKME) for all genes in each sample. The median and upper and lower quartiles are shown, with outliers as solid circles. (B) The proportion of reads from circRNA producing exons averaged across all nucleated samples (y-axis) and platelets (x-axis). (C) Fold enrichment of reads from circRNA producing exons in platelets relative to nucleated tissues. All genes with an average RPKMI >1 in platelets and expressed in 8 or more nucleated tissues are shown. Blue, genes significantly enriched in platelets; red, genes not significantly enriched in platelets. The data points corresponding to the 5 most enriched genes are indicated. The slope x = y is shown as a dashed line.
To identify genes significantly enriched for circRNAs in platelets, we then estimated the contribution of circRNA producing exons to total transcription for each gene by calculating RPKMI/(RPKMI + RPKME) ratios and compared ratios in the platelet samples to ratios in the 12 nucleated tissues using the Wilcoxon rank test (P = .05, Benjamin and Hochberg false discovery rate = 0.01). Of 8041 circRNA producing genes analyzed, 3162 showed significantly enriched transcription from circRNA-producing exons in platelets (supplemental Table S5). When the ratios from all genes with an average RPKM > 1 in platelets are plotted (Figure 4B), it can be seen that for the vast majority of enriched genes the contribution of circRNA exons to total transcription is >0.6 in nucleated tissues but >0.8 in platelets. Furthermore, for 457 genes, transcription from circRNA-producing exons exceeds an average of 99% in the platelet samples, and for 15 genes, it is 100% in all 3 samples (supplemental Table S5). As this assay is not dependent on exon junction counts, which require long sequence reads for accurate detection, we applied it to one of the SOLiD platelet RNAseq datasets15 and confirmed the presence of similar patterns of circRNA exon enrichment (supplemental Figure S4; supplemental Data File 1).
Reads from circRNA producing exons are enriched in platelets by up to 3000-fold
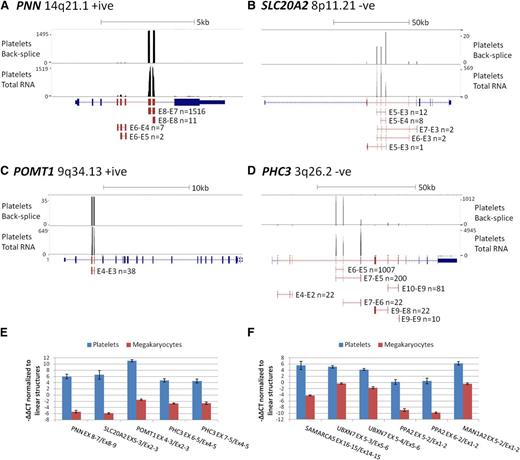
To estimate the magnitude of circRNA exon enrichment for each gene in platelets relative to other tissues, we further analyzed genes with an average RPKMI >1 by normalizing their mean platelet RPKMI/RPKME ratio against the mean RPKMI/RPKME ratio from all 12 nucleated samples. The average enrichment was 12.7 times for all genes and 22 times for the 3162 significantly enriched genes, with the highest enrichment being 3590 times for TMEM181 (Figure 4C; supplemental Table S5). To illustrate these extreme levels of enrichment, RNAseq read counts and circRNA junction read counts for 4 enriched genes from a single platelet sample are shown in Figure 5A-D. The vast majority of platelet RNAseq reads from PNN (enrichment 2189 times) map to exons 7 and 8, which correspond to the most abundant circRNA (Figure 5A); the majority of reads from SLC20A2 (enrichment 152 times) map to the abundant circRNAs involving exons 3 to 5 (Figure 5B); all reads from POMT1 (enrichment 127 times) map to exons within its only circRNA (E4-E3; Figure 5C), whereas for PHC3 (enrichment 240 times), reads map largely to the abundant E6-E5 and E7-E5 structures (Figure 5D).
circRNA enrichment occurs in platelets but not megakaryocytes. (A-D) Adapted University of California Santa Cruz screen shots for 4 genes with extreme circRNA enrichment in platelets. Each panel shows RNAseq read abundance in a single platelet total RNA sample (F1), together with the position and abundance of back-splice exon junctions in the same sample. The intron-exon structure is shown below, with exons known to contribute to circRNAs shown in red. CircRNA structures present in the sample together with back-splice frequencies are also shown (for PHC3, only structures with ≥10 back-splice reads are shown). Above each panel, the gene names, cytogenetic locations, coding strand, and scale in kilobases are indicated. (E-F) Expression levels of circRNAs relative to linear RNAs from the same loci in platelets and cultured megakaryocytes, normalized to the housekeeping pool (−∆∆CT values).
circRNA enrichment occurs in platelets but not megakaryocytes. (A-D) Adapted University of California Santa Cruz screen shots for 4 genes with extreme circRNA enrichment in platelets. Each panel shows RNAseq read abundance in a single platelet total RNA sample (F1), together with the position and abundance of back-splice exon junctions in the same sample. The intron-exon structure is shown below, with exons known to contribute to circRNAs shown in red. CircRNA structures present in the sample together with back-splice frequencies are also shown (for PHC3, only structures with ≥10 back-splice reads are shown). Above each panel, the gene names, cytogenetic locations, coding strand, and scale in kilobases are indicated. (E-F) Expression levels of circRNAs relative to linear RNAs from the same loci in platelets and cultured megakaryocytes, normalized to the housekeeping pool (−∆∆CT values).
Although these in silico analyses point to enrichment specifically in anucleate cells, no total RNAseq data have been generated from the direct progenitors of these cells. For all genes with assays available, we therefore also analyzed the relative levels of linear and circRNAs in megakaryocytes cultured from cord blood. These were harvested after 10 days, when >70% of cells were positive for CD41 and CD42 (supplemental Methods). The results, presented in Figure 5E-F, establish that relative levels of linear and circRNA in megakaryocytes are similar to other nucleated cell types (Figure 2B); all circRNAs are expressed at a much lower level relative to linear RNAs than in platelets (P = 1.4 × 10−6, Wilcoxon paired rank sum test across all structures). This is consistent with circRNA enrichment after platelet formation.
Confirmation of differential decay of linear and circRNAs in platelet preparations
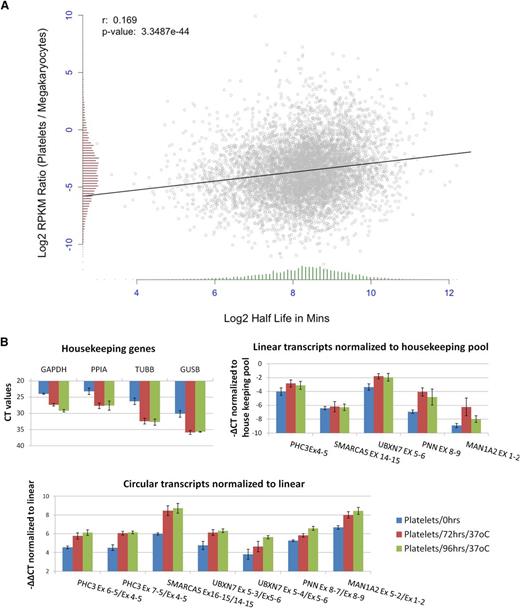
Human mRNAs are subject to a wide variety of decay pathways45 and exhibit extensive variation in decay rates.46-48 To investigate decay in platelets, we first correlated the change in transcript abundance between megakaryocyte PolyA+ RNA and platelet PolyA+ RNA with published estimates of mRNA half-life.46 This identified a weak but highly significant correlation, despite the difference in cell types between datasets, with short half-life genes showing a more pronounced reduction in transcript abundance (r = 0.17, P = 3.35e−44; Figure 6A). This is consistent with a transcriptome-wide role for mRNA decay in platelet expression levels.
Degradation of platelet RNA. (A) Correlation of the change in gene expression between megakaryocte polyA+ RNA and platelet polyA+ RNA (y-axis), and mRNA half-life estimates (x-axis). Distributions of both are shown as histograms on each axis. (B) qPCR analysis of differential decay of linear and circRNAs in platelets following incubation at 37°C for 0, 72, and 96 hours. Data from 3 biological replicates are shown. (Top left) Expression levels of housekeeping genes (CT values). (Top right) Expression levels of linear structures from 5 circRNA-producing genes relative to the housekeeping pool (−∆CT values). (Bottom) Expression levels of 7 circRNAs relative to linear transcripts from the same loci, both normalized to the housekeeping pool (−∆∆CT values).
Degradation of platelet RNA. (A) Correlation of the change in gene expression between megakaryocte polyA+ RNA and platelet polyA+ RNA (y-axis), and mRNA half-life estimates (x-axis). Distributions of both are shown as histograms on each axis. (B) qPCR analysis of differential decay of linear and circRNAs in platelets following incubation at 37°C for 0, 72, and 96 hours. Data from 3 biological replicates are shown. (Top left) Expression levels of housekeeping genes (CT values). (Top right) Expression levels of linear structures from 5 circRNA-producing genes relative to the housekeeping pool (−∆CT values). (Bottom) Expression levels of 7 circRNAs relative to linear transcripts from the same loci, both normalized to the housekeeping pool (−∆∆CT values).
To experimentally investigate differential decay, we then used real-time PCR to analyze changes in linear and circRNA abundance in PRP over time (see “Materials and methods”). Consistent with previous analyses,16 initial experiments suggested that all transcript levels remained stable at 4°C but that degradation of mRNA could be detected when incubation temperature was elevated (data not shown). To assess decay at a physiologically relevant temperature, we assayed the relative abundance of linear and circRNAs in PRP from 3 individuals, before and after incubation at 37°C for 72 and 96 hours (Figure 6B). The levels of all 4 housekeeping genes were reduced by incubation, with CT values increasing by 4 to 6 cycles (Figure 6B, top left panel). Reduction was also observed when genes which are highly abundant in platelets were analyzed (supplemental Figure S5). Normalization of linear transcripts from circRNA producing genes against the housekeeping pool (Figure 6B, top right panel) suggests that there are differences in decay rates between genes, with UBXN7, PNN, and MAN1A2 transcripts being enriched relative to control housekeeping RNAs. Strikingly, however, circRNAs show significant enrichment (2- to 6-fold) relative to linear RNAs after incubation (Figure 6B, lower panel; P < 2 × 10−3 for both day 0 vs 72 hours and day 0 v 96 hours, Wilcoxon rank sum test across all structures). This confirms that linear structures decay more rapidly than circRNAs in platelets.
Discussion
Platelets are anucleate yet retain machinery required for splicing and translation, and there is extensive evidence that genes are translated in activated platelets.5-7,49-51 As a result, recent platelet transcriptome analyses have generated considerable interest.8,17,18 However, here we showed that the platelet transcriptome is significantly enriched for circRNAs as assayed using back-splice junction counts, real-time PCR, and an RNAseq comparative read depth approach. Back-splice reads are enriched 17- to 188-fold relative to nucleated tissues, and >3000 genes have a statistically significant enrichment of reads from circRNA-producing exons. Furthermore, for some genes, only RNAseq reads derived from circRNA-producing exons are detected within platelet samples, highlighting the impact this enrichment will have on RNAseq-based estimates of expression levels. We also established that circRNAs are not enriched in cultured megakaryocytes compared with other nucleated cell types.
Although we cannot formally rule out the possibility that some circRNA enrichment occurs at the transcriptional level late in megakaryocyte maturation, collectively our results suggest that enrichment occurs specifically in platelets, after the loss of nuclear influence. The similarly high levels of enrichment observed in anucleate RBCs are consistent with this, whereas the observed reduction of linear RNAs relative to circRNAs in platelets incubated at 37°C confirms that differential decay occurs. The overall level of enrichment of RNAseq reads from circRNA-producing exons is consistent with platelet mRNA levels being ≥90% lower than within nucleated tissues (average enrichment, 12.7 times for genes with RPKMI >1). Although this is a rough and indirect estimate, it is likely to be conservative as it assumes no degradation of circRNAs. Furthermore, the fraction of translatable RNAs is likely to be even lower, as our analysis has only considered RNA abundance and not structural integrity. This is consistent with the suggestion that platelet lifespan may be determined by translational competence.52
Degradation has been assumed to be a feature of the platelet transcriptome based purely on their anucleate nature.52 Mitochondrially encoded genes are among the most highly expressed,53 levels of individual mRNAs have been shown to fall when platelet preparations are incubated at ambient temperature,16 and there is some evidence that translation is reduced in aged platelets relative to the whole population.52 Consideration of mRNA half lives alone would suggest that mRNA degradation is likely to be substantial, unless platelet-specific mechanisms exist to counteract it. If we assume the average age of circulating platelets is 4 days, then mRNA half-lives46 would predict gene specific reductions in expression ranging from 1.2 to 720 times, with a mean reduction of 14 times. Our estimate of 12.7 times based on RNAseq read counts is broadly consistent with this, despite the fact that RPKMI/(RPKMI + RPKME) ratios will vary between genes due to a variety of factors that are unrelated to circularity, including differential isoform abundance, overlapping untranslated regions between neighboring genes, and the reduction in RNAseq read coverage close to transcript termini.
Extensive mRNA degradation provides a simple explanation for conflicting reports concerning the correlation between the platelet transcriptome and proteome12,13,15,54 and for the presence of proteins in platelets without corresponding mRNAs.15,51 It can also explain why many genes shown to be spliced and/or translated in platelets, such as tissue factor (F3),7,55 IL1B,56 SLC23A2,57 FGA, FGB, FGG, ALB,58 and BCL3,59 are present at very low levels or not at all in some RNAseq datasets.10,11 Collectively, our results indicate that translation in platelets occurs against the backdrop of a rapidly degrading transcriptome, that platelets will differ in their translational potential in an age-dependent manner, and that RNAseq analyses targeted toward intact species that are capped15,18 or polysome bound will be required to analyze translational potential in these cells effectively.
circRNAs have been identified from RNAseq data by the back-splice exon junctions they contain, and such junctions have been taken prima facie as evidence of circularity.24,33,41,60 Our exon-partitioned analysis of read-depth provides additional in silico evidence of circularity, as trans-splicing of independent linear pre-mRNA20,43 would affect exons both up- and downstream of the back-spliced exons. The identification of higher numbers of circRNAs in platelets than in total RNA digested with RNAse R is, however, surprising and indicates that cessation of transcription in platelets leads to more extreme enrichment of circular RNA through differential decay in vivo than enzymatic digestion of linear species in vitro. This could be due both to limitations of RNAse R specificity and the lability of RNA after extraction. The significant enrichment of A nucleotides within back-splice transcripts present within platelet polyA+ RNA is also noteworthy as it means their presence within oligo-dT selected RNA fractions does not necessarily imply polyadenylation, as has been suggested previously,20,43 and may adversely affect the accuracy of RNAseq-based mRNA expression estimates of the genes involved.
It has recently been suggested that up to 50% of RNAs with back-splice exon junctions are RT artifacts. This was extrapolated from an analysis of 13 back-splice RNAs where the only 6 RNAs that could be independently validated were those that could be amplified using both AMV and MMLV RTs.43 However, among the 7 RNAs that could not be validated, 4 were intragenic back-splice structures, and all are present within platelets: The back-splice counts of 3 (PHC3 E6-E5, CNTLN E5-E3, RERE E3-E3) are enriched relative to other tissues (∼1000, ∼100, and ∼4 times), and read counts in exons between each back-splice are also enriched (240, ∼30, and 500 times) indicative of circRNAs. Furthermore, the PHC3 E6-E5 structure degrades more slowly than linear RNA, is resistant to RNAse R digestion,23 and in our hands can be amplified from cDNA generated using either MMLV or AMV (supplemental Figure S3). These diverse lines of evidence suggest that there may not be a straightforward relationship between RT specificity and artifacts as previously suggested.43
Finally, cellular levels of circRNAs are known to be low in proliferating and neoplastic human cells44,60 but to increase during differentiation of ES cells26 and during Drosophila development,61 suggesting that they may be a biomarker for aging. Among nucleated tissues, they are highest in both human and Drosophila neural tissues44,61 and are particularly enriched in synapses.44 They are also enriched in exosomes.62 These observations, together with the presence of microRNA binding sites within circRNAs61 and the dynamic expression patterns of some,44 implies that they may be of functional importance. circRNA levels can be modulated by intronic sequence identity23,27,28 and splicing factors such as Quaking31 and Muscleblind.29 However, our qPCR analysis of transcripts in megakaryocytes suggests that the extreme enrichment in platelets reported here is not due to modulation at the pre-mRNA level within progenitor cells. With no transcription in platelets it can, therefore, only be accounted for by differential degradation/decay. The very high levels of enrichment reported here suggests that, even if the relative levels of linear and circRNAs from each gene remain constant at the level of transcription, subtle changes in expression level, rates of mRNA export, changes in the cell cycle,60 changes in nuclear and cytoplasmic volumes, and distance from the nucleus, could all alter relative abundance over time. Many reported differences in linear and circRNA levels could, therefore, be due to differences in stability alone.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The financial support from the Leverhulme Trust (grant RPG-2012-795 to M.S.J., M.S.-K., and D.J.E.), Biotechnology and Biological Sciences Research Council (studentship BB/J014516/1 to O.G.I.), and the Wellcome Trust (grants WT089225MA and WT089225/Z/09/Z to D.J.E.) is gratefully acknowledged.
Authorship
Contributions: O.G.I. performed the bioinformatic analyses of RNAseq data, circRNA abundance, circRNA exon enrichment, and RNA decay; A.A.A. performed all cell purification and isolation, RNA extraction, real-time PCR analyses, and platelet time course analyses; H.H.A.-B. developed all real-time PCR assays; J.S.S. provided assistance with the analysis of PolyA+ transcripts; J.C.M. and L.M. provided the erythrocytes used to generate RNAseq data; A.E., M.C., and C.G. isolated human haematopoietic progenitors from cord blood, followed by differentiation to megakaryocytes and sorting to purity; M.S.J., M.S.-K., and D.J.E. conceived the project; and M.S.J. drafted the manuscript with input from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael S. Jackson, Institute of Genetic Medicine, International Centre for Life, Central Parkway, Newcastle upon Tyne NE1 3BZ, United Kingdom; e-mail: michael.jackson@newcastle.ac.uk.
References
Author notes
A.A.A. and O.G.I. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal