In this issue of Blood, Alhasan et al1 report the existence of circular RNAs (circRNAs) in circulating human platelets, thereby revealing yet another facet of their already diverse transcriptome.
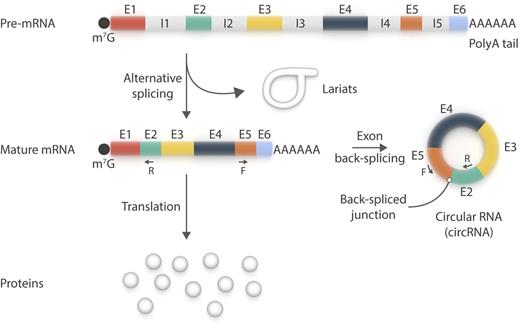
circRNA is a new member of the platelet transcriptome. circRNA species primarily use canonical splice sites and may be amplified by reverse transcriptase polymerase chain reaction using pairs of oligonucleotides (F+R) designed to be divergent on the linear messenger RNA (mRNA) species, but convergent on circRNAs. The existence of lariats in platelets remains to be confirmed. E, exon; F, forward; I, intron; m7G, 7-methylguanylate cap; R, reverse. Professional illustration by Somersault18:24.
circRNA is a new member of the platelet transcriptome. circRNA species primarily use canonical splice sites and may be amplified by reverse transcriptase polymerase chain reaction using pairs of oligonucleotides (F+R) designed to be divergent on the linear messenger RNA (mRNA) species, but convergent on circRNAs. The existence of lariats in platelets remains to be confirmed. E, exon; F, forward; I, intron; m7G, 7-methylguanylate cap; R, reverse. Professional illustration by Somersault18:24.
Platelets are short-lived (∼8-11 days), translationally competent, and capable of premature messenger RNA (pre-mRNA) splicing into mature mRNAs.2 They do this using a process expected to generate lariats, which are formed of normally spliced out, circularized introns. Alhasan et al1 now report that platelet mRNAs may also undergo exon back-splicing to generate circRNAs (see figure). To detect circRNAs, the authors analyzed RNA sequencing datasets from human platelets and searched for “back-splice” exon junctions, inconsistent with the underlying genomic DNA, by using specialized bioinformatic tools. They then validated some of the circular reads by reverse transcriptase polymerase chain reaction. Next, they enriched the circRNA content of their samples by selectively digesting linear RNA with ribonuclease R, to which circRNAs are resistant and that has been used to define them.
Using this approach, the authors observed marked circRNA enrichment in platelets (and erythrocytes) relative to nucleated cells, mainly as a result of differential decay of linear RNAs. Conversely, circRNA enrichment in platelets vs cultured megakaryocytes, in the absence of de novo gene transcription, is consistent with circRNAs being produced in platelets rather than being inherited from their precursor cells. Two circRNA junctions (E4-E2 and E5-E2; E, exon) of one gene, MAN1A2, are even more abundant in platelets (and erythrocytes) than the linear mRNA. Based on their findings, the authors proposed that mRNA translation in platelets occurs against the backdrop of a rapidly degrading transcriptome, which would explain the relatively weak association between the human platelet transcriptome and proteome.3
At the interface of the platelet transcriptome and proteome is an unusually abundant and diverse array of microRNAs (miRNAs),4 which are key regulators of gene expression. Considering that (1) platelet mRNAs harbor 3′ untranslated regions (3′UTRs) twice as long as those of nucleated cells (1047 vs 492 nucleotide),5 (2) 3′UTRs are known to contain microRNA (miRNA) binding sites, and (3) 2 circRNAs from nucleated cells act as miRNA sponges,6 it would be interesting to determine whether platelet circRNAs are enriched in 3′UTR exons and miRNA binding sites, because this could explain the relative abundance of circRNAs and miRNAs in platelets.
To gain further insights into the biology of circRNAs in platelets, we may want to know if platelets contain RNA binding protein Quaking, an alternative splicing factor regulating the production of more than one-third of abundant circRNAs during human epithelial-mesenchymal transition.7 The biological relevance of platelet circRNAs, if any, remains unclear: do they play a role inside of platelets, or are they simply byproducts of platelet mRNA degradation? The latter assumption does not necessarily preclude them from mediating biological effects outside of platelets. Indeed, circRNAs may still play a role in other cells, as shown previously for mRNAs and miRNAs,8 provided that they are expelled within exosomes or larger microparticles, under basal conditions or upon activation, which remains to be seen. Because exon-intron circRNAs regulate transcription in nucleated cells,9 it is tempting to speculate that circRNAs that accumulate in platelets may end up regulating gene expression and function of other recipient cells of the circulatory system.
The possible clinical implications of platelet circRNAs may soon be studied and revealed, because we will want to know if circRNA abundance and/or sequences vary depending on the race, gender, age, or health status of the subjects. Are circRNAs present and conserved in the platelets of small animal models, such as the mouse? If so, very informative in vivo studies could be performed. The suggestion that platelet lifespan may be determined by translational competence support a possible relationship between circRNA levels and platelet half-life or aging. Could circRNAs be used as biomarkers of platelet-related diseases, or signal shorter platelet lifespan or premature platelet aging? Could these conditions be unveiled simply by measuring the level of specific circRNAs by quantitative PCR?
Produced upon cytoplasmic fragmentation of bone marrow megakaryocytes, circulating platelets contain an impressive array of RNA species (messenger RNA, transfer RNA, microRNA, long noncoding RNA)10 that now includes circRNAs. These recently described RNA species were identified by Alhasan et al1 among gigabytes of platelet RNA sequencing data simply by using publicly available bioinformatic tools, as one would experience by looking at the full moon with a telescope instead of binoculars. Has anyone thought that, by analyzing the transcriptome of an anucleate element of the blood, we would go round (and round) in circles?
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal