In this issue of Blood, studies by Lavallée et al and Maxson et al provide a powerful example of functional and genomic data integration to reveal unexpected correlations between genotype and phenotype in acute myeloid leukemia (AML).1,2
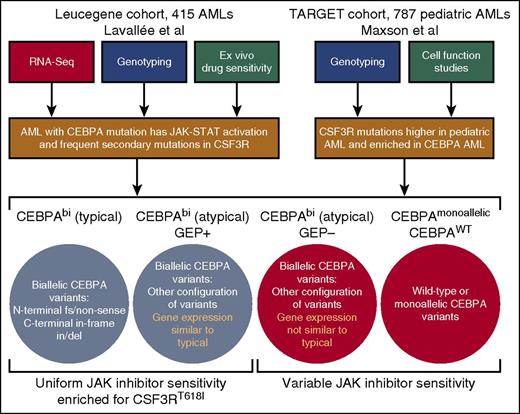
Workflow of studies and summary of findings. By using a combination of RNA-Seq, genotyping, and ex vivo drug sensitivity studies, Lavallée and colleagues identified a gene signature of JAK-STAT pathway activation in patients with biallelic CEBPA mutations, which corresponds with enrichment of secondary mutations in JAK-STAT regulating genes, most notably CSF3R, as well as uniform hypersensitivity to JAK kinase inhibitors. Maxson et al performed sequencing on a large cohort of pediatric AML cases with validation of variants in cell transformation assays. They found CSF3R mutations to be higher in this pediatric cohort than in any adult AML cohorts and, consistent with the findings of Lavallée et al, found a significant enrichment of CSF3R mutations within the CEBPA mutated subset.
Workflow of studies and summary of findings. By using a combination of RNA-Seq, genotyping, and ex vivo drug sensitivity studies, Lavallée and colleagues identified a gene signature of JAK-STAT pathway activation in patients with biallelic CEBPA mutations, which corresponds with enrichment of secondary mutations in JAK-STAT regulating genes, most notably CSF3R, as well as uniform hypersensitivity to JAK kinase inhibitors. Maxson et al performed sequencing on a large cohort of pediatric AML cases with validation of variants in cell transformation assays. They found CSF3R mutations to be higher in this pediatric cohort than in any adult AML cohorts and, consistent with the findings of Lavallée et al, found a significant enrichment of CSF3R mutations within the CEBPA mutated subset.
The landscape of recurrent genetic events in AML has been elegantly unraveled in recent years3 ; however, for most disease subsets this has not yet led to facile deployment of therapeutics targeting aberrant genetic events. The resulting gap in our ability to decipher genetic drivers versus our capacity to harness this knowledge for therapeutic advantage will require the integration of genotype information with transcriptomic, proteomic, and drug response data. The studies by Lavallée et al and Maxson et al conclude that AML patients with biallelic mutation of CCAAT/enhancer binding protein α (CEBPA) exhibit dysregulation of and dependence upon Janus kinase–signal transducer and activator of transcription (JAK-STAT) signaling.
In the first study, Lavallée et al show that JAK-STAT pathways are dysregulated more frequently than average in biallelic CEBPA (CEBPAbi)–mutated AML, which correlates with sensitivity to JAK kinase inhibitors. Both this study by Lavallée et al and another study by Maxson et al show that CEBPA mutations also frequently coincide with secondary mutations in JAK-STAT pathway regulators, especially granulocyte colony-stimulating factor 3 receptor (CSF3R). These findings yield the exciting conclusion that JAK kinase inhibitors could be an effective component of improved therapeutic regimens for AML patients with CEBPAbi mutation, a hypothesis that can be tested with prospective clinical trials targeted to this patient population.
CEBPA is a transcription factor that is crucial for regulating myeloid lineage development. In particular, it has been shown to regulate expression of CSF3R, thereby promoting neutrophil differentiation.4 Mutations in CEBPA are observed in 5% to 15% of AML patients,3,5,6 and those mutations can be classified into the following 3 categories: (1) monoallelic mutation, (2) typical CEBPAbi AML (ie, biallelic mutation with one allele harboring an N-terminal frameshift or nonsense mutation and the other allele having an inframe insertion or deletion of the C terminus), and (3) atypical CEBPAbi AML (ie, biallelic mutation with other configurations of mutations on each allele [eg, point mutations, two N-terminal mutations]). Prior studies have shown that CEBPAbi AML patients exhibit distinct gene expression patterns compared with those who have wild-type (WT) or monoallelic CEBPA AML, and only CEBPAbi mutation confers improved prognosis.7 However, the significance of these CEBPAbi AML gene expression signatures for disease pathogenesis or design of targeted therapies has remained unclear.
CSF3R is a cell surface receptor that binds the ligand CSF3 and activates downstream signaling cascades, most notably JAK-STAT and SRC-family kinase pathways. CSF3R mutations were originally reported in a fraction of patients with severe congenital neutropenia (SCN), and these patients exhibited increased risk of developing AML.8 The variants observed in SCN always took the form of frameshift or nonsense mutations that truncated the cytoplasmic tail of the receptor. Subsequently, it was found that mutation of CSF3R is quite frequent in Philadelphia-negative neutrophilic leukemia with a majority of chronic neutrophilic leukemia (CNL) patients and a minority of atypical chronic myeloid leukemia (aCML) patients exhibiting CSF3R variants.9 The vast majority of mutations seen in CNL/aCML are point mutations proximal to the transmembrane that result in increased receptor dimerization and ligand-independent signaling, although some patients also exhibit truncating mutations similar to those seen in SCN patients. CSF3R mutations have rarely been observed in AML3,9,10 and, until now, had not been observed to cluster into specific disease subsets.
Lavallée et al examined RNA-Seq and genotype data from the Leucegene cohort of 415 primary AML patient specimens to identify a gene signature of JAK-STAT pathway activation in patients with CEBPAbi mutations (see figure). Typical CEBPAbi AML patients uniformly exhibited the JAK-STAT activation signature, whereas a portion of the atypical CEBPAbi AML patients showed the same signature (termed “atypical CEBPAbi AML GEP+”). Genotyping data showed that a high percentage of these patients also harbor mutations in genes that regulate JAK-STAT signaling, most notably the gain-of-function CSF3RT618I variant that is commonly observed in CNL. Examination of ex vivo drug sensitivity revealed that all typical CEBPAbi AML patients as well as those with atypical CEBPAbi AML with the JAK-STAT gene expression signature (GEP+) were more sensitive on average to JAK kinase inhibitors than CEBPAWT AML or atypical CEBPAbi AML patients without the JAK-STAT gene expression signature (GEP–). Sensitivity to other drugs lacking activity against JAK kinases was not similarly biased between patients with wild-type or mutant CEBPA.
Maxson et al focused on a large cohort of pediatric patients with AML; whole-exome sequencing was performed on 186 patients and a targeted sequencing panel of ∼200 genes was performed on an additional 601 patients (787 patients total). They found CSF3R mutations to be higher in this pediatric cohort than in any reported adult AML cohort and, consistent with the findings of Lavallée et al, they found a significant enrichment of CSF3R mutations within the CEBPA-mutated subset.
Collectively, these two studies suggest that typical CEBPAbi AML patients uniformly exhibit JAK-STAT pathway activation and sensitivity to JAK kinase inhibitors, and a proportion of atypical CEBPAbi AML patients, as marked by a gene expression signature, also display JAK-STAT dysregulation and dependence. In numerous patients, this JAK-STAT pathway activation appears to be at least partially driven by mutations in JAK-STAT–regulating genes such as CSF3R or STAT5B, but the etiology of JAK activation in the remaining patients remains to be determined.
From a translational standpoint, these studies suggest the exciting possibility that CEBPAbi AML patients may benefit from treatment with JAK kinase inhibitors. These studies certainly provide a strong rationale for testing this hypothesis in prospective clinical trials. The authors of both articles are to be commended for the scope, strategy, and potential impact of their studies, and for their elegant work in establishing complex connections between genotype and phenotype. Much more work of this caliber is needed to improve outcomes for patients with AML.
Conflict-of-interest disclosure: J.W.T. received research support from Agios Pharmaceuticals, Array BioPharma, Aptose Biosciences, AstraZeneca, Constellation Pharmaceuticals, Genentech, Incyte, Janssen Pharmaceuticals, Seattle Genetics, and Takeda Pharmaceuticals and is a consultant for Leap Oncology.