Key Points
BCL2 mutations in FL correlate with activation-induced cytidine deaminase expression and frequently alter the amino acid sequence of the protein.
Mutations in the BCL2 coding sequence at diagnosis are associated with shortened time to transformation and earlier death due to lymphoma.
Abstract
Follicular lymphoma (FL), an indolent neoplasm caused by a t(14;18) chromosomal translocation that juxtaposes the BCL2 gene and immunoglobulin locus, has a variable clinical course and frequently undergoes transformation to an aggressive lymphoma. Although BCL2 mutations have been previously described, their relationship to FL progression remains unclear. In this study, we evaluated the frequency and nature of BCL2 mutations in 2 independent cohorts of grade 1 and 2 FLs, along with the correlation between BCL2 mutations, transformation risk, and survival. The prevalence of BCL2 coding sequence mutations was 12% in FL at diagnosis and 53% at transformation (P < .0001). The presence of these BCL2 mutations at diagnosis correlated with an increased risk of transformation (hazard ratio 3.6; 95% CI, 2.0-6.2; P < .0001) and increased risk of death due to lymphoma (median survival of 9.5 years with BCL2 mutations vs 20.4 years without; P = .012). In a multivariate analysis, BCL2 mutations and high FL international prognostic index were independent risk factors for transformation and death due to lymphoma. Some mutant Bcl-2 proteins exhibited enhanced antiapoptotic capacity in vitro. Accordingly, BCL2 mutations can affect antiapoptotic Bcl-2 function, are associated with increased activation-induced cytidine deaminase expression, and correlate with increased risk of transformation and death due to lymphoma.
Introduction
Follicular lymphoma (FL) has a highly variable clinical course.1-3 Although some patients do well for decades, often with limited therapy, at some point 30% to 50% of patients experience histologic transformation to a more aggressive lymphoma, usually diffuse large B-cell lymphoma (DLBCL).4-11 This transformation, which is thought to reflect the acquisition of new genetic abnormalities leading to further genomic instability,12-16 has generally been associated with a poor clinical outcome.17 Retrospective analyses from the prerituximab era have reported a median survival of only 1 to 2 years after transformation,18,19 although a recent prospective observational study suggests somewhat better survival after transformation in the rituximab era.20 At the present time, the FL international prognostic index (FLIPI), which integrates patient characteristics at diagnosis, is the gold standard for predicting FL clinical outcome.21,22 There is, however, considerable interest in identifying characteristics of the FL cells themselves that might also impact prognosis.22,23
The BCL2 gene is critical for FL pathogenesis.24,25 Originally identified because of its translocation to the immunoglobulin heavy chain (IGH) locus as a part of the t(14;18) translocation that typifies FL, BCL2 encodes a protein that inhibits apoptosis.26,27 In particular, Bcl-2 and related antiapoptotic proteins diminish apoptosis by binding and neutralizing activated proapoptotic Bcl-2 family members, including the mitochondrial outer membrane permeabilizers Bax and Bak, as well as the intracellular stress sensors Bim and Puma that activate Bax and Bak.27-29 Because the IGH locus is activated in B cells, the translocated BCL2 gene is overexpressed in FL cells25 and enhances their survival.30
Activation-induced cytidine deaminase (AID), which is highly expressed in germinal center B cells, introduces mutations into the variable and switch regions of B-cell receptor genes during the physiological process of secondary antibody diversification.31 After AID deaminates cytidine to uracil, the resulting uracil-guanine mismatches are replicated to produce a C→T transition, a hallmark of AID activity, or removed by uracil DNA glycosylase to create an abasic site that is recognized by nucleases and error prone polymerases, leading to a variety of other mutations or DNA double-strand breaks.32,33 In addition to its role in antibody diversification, AID is postulated to play a critical role in lymphomagenesis by mutating genes outside the immunoglobulin locus,32,34 including the proto-oncogenes BCL6, PIM1, RhoH/TTF, PAX5, and MYC.35,36 Moreover, AID contributes to genetic instability, including the acquisition of chromosomal abnormalities such as IGH translocations.37,38 Even though AID levels vary substantially among different FLs,34 the implications of this variation have been unclear.
At the time BCL2 was cloned, single nucleotide variants (SNVs) were observed in human lymphoma cell lines39 and a handful of clinical lymphomas40 compared with normal tissues. Because these BCL2 variants, like wild-type (WT) BCL2, inhibited cell death41 and facilitated lymphomagenesis,42 it was assumed that BCL2 mutations do not affect Bcl-2 protein function. Consistent with this possibility, previous studies showed that the majority of BCL2 mutations in DLBCL were silent43,44 and had no effect on survival.44
More recently, large-scale genomic studies have also identified frequent BCL2 mutations in FL.16,45 Whether these BCL2 mutations have a biological impact has been unclear. Our previous studies demonstrated that some mutant Bcl-2 proteins found in lymphoma cell lines exhibit enhanced affinity for proapoptotic Bcl-2 family members and increased the ability to protect from death stimuli.46,47 Accordingly, the present studies were designed to assess the occurrence, nature, and potential clinical importance of mutations in the BCL2 coding sequence in FL.
Methods
Patients
Patients with grades 1 and 2 FL who provided written consent for enrollment in the actively maintained Mayo Clinic Lymphoma Database were studied. After approval by the Mayo Clinic Institutional Review Board (Rochester, MN), slides from initial diagnostic biopsies and transformation were subjected to pathology review applying World Health Organization criteria. Patients with grade 3 FL were excluded. Three distinct cohorts were studied. Research was conducted in accordance with the Declaration of Helsinki.
Discovery cohort.
The discovery cohort consisted of 38 FL patients with frozen tissue samples stored in the Mayo Clinic Lymphoma Tissue Bank at diagnosis (n = 20) or during the course of the disease (n = 18) and available clinical data, including outcome (median follow-up, 10.5 years). Of the 18 patients biopsied during the course of the disease, 16 had received prior therapy with various regimens. The discovery cohort was used to explore the frequency of BCL2 mutations, as well as the relationship between BCL2 mutation status and clinical outcome.
Validation cohort.
The validation cohort consisted of 128 patients with newly diagnosed FL seen at the Mayo Clinic between May 1975 and July 2005 who were included in an actively maintained database, and had (1) formalin-fixed paraffin embedded (FFPE) samples available from biopsy at initial diagnosis and (2) tissue available from the time of transformation if transformation occurred. Visual scoring of hematoxylin and eosin slides from the same block by an experienced hematopathologist indicated that samples contained a median of 70% (range, 30% to 90%) neoplastic cells. The validation cohort was used to assess the frequency of mutations in the coding regions of BCL2 at diagnosis and transformation, as well as the relationship between BCL2 mutations at diagnosis and clinical outcome in FL patients with uniformly recorded clinical characteristics, long-term follow-up, and known outcome.
Paired FL/germline control.
Paired samples of peripheral blood DNA and lymph node biopsy from 11 FL patients at diagnosis were examined to evaluate the possibility that FL SNVs represent germline variants rather than somatic mutations. Because patients in this group had a relatively short follow-up (5.5 years), samples were used for tumor/germline comparison but were not included in outcome analysis.
Other B-cell lymphomas and normal controls.
To assess the frequency of BCL2 mutations in other lymphoma subtypes, DNA was isolated from 39 DLBCLs, 117 mucosa-associated, marginal zone, and lymphoplasmacytic (“postfollicular”) lymphomas, 36 acute lymphocytic leukemias (ALLs), and 90 chronic lymphocytic leukemias (CLLs), 40 without and 50 with IGH variable region somatic mutations. DNA from 300 normal individuals (100 Caucasians, 100 African American, and 100 Han Chinese) was purchased from the Coriell Institute (Camden, NJ).
Isolation of genomic DNA, sequencing, and identification of tumor-specific mutations
DNA was isolated using a QIAamp DNA Kit (Qiagen). For FFPE FL samples, an Illumina Infinium HD FFPE Restore Kit (Illumina) and QIAamp DNA FFPE Kit (Qiagen) were used. Polymerase chain reaction (PCR) was performed using primers (see supplemental Table 1, available on the Blood Web site) for exons 1 (discovery cohort only), 2, and 3 of BCL2 (NG_009361.1). PCR products were subjected to Sanger sequencing on both strands using ABI PRISM BigDye Terminator technology at the Mayo Clinic DNA Sequencing Core. All SNVs were detected in both 3′ and 5′ directions and confirmed in a separate sequencing reaction using independently amplified DNA. Mixing experiments demonstrated that this approach readily detects variants that are present in ≥10% of DNA molecules. Candidate somatic variants (tumor-specific) were identified after removing known polymorphisms present in NCBI dbSNP database, HapMap, and in our set of normal samples. The absence of the identified mutations in the COSMIC database and in the Cancer Gene Census database was verified. A similar approach was likewise used to sequence the exons of PIM1 using primers shown in supplemental Table 1.
RNA isolation
RNA was extracted from frozen FL samples using an RNeasy Mini Kit (Qiagen). Complementary DNA (cDNA) was synthesized with Superscript III, amplified with Promega master mix and primers (supplemental Table 1), and sequenced as described above.
AID immunostaining and messenger RNA (mRNA) detection
FL samples were stained with murine monoclonal anti-AID primary antibody (cat #39-2500; Life Technologies) and peroxidase-labeled secondary antibody. Samples were scored based on the percentage of tumor cells expressing AID, with <20%, 21% to 40%, 41% to 60%, 61% to 80%, and >80% being scored 1, 2, 3, 4, and 5, respectively, by a lymphoma pathologist blinded to outcome data. AID mRNA was assessed by semi-quantitative reverse transcription PCR (RT-PCR) using primers (5′-AGGCAAGAAG-ACACTCTGGACACC-3′ and 5′-TCAAAGTCCCAAAGTACGAAATGC-3′) with β-actin as a positive control.
Statistical analysis
Risk of transformation over time and time to transformation (TTT) were estimated as time from diagnosis to histologically proven transformation using Kaplan-Meier methods48 ; patients without transformation at the time of last follow-up or death were censored. The FL-specific survival was estimated as the time from diagnosis to lymphoma-associated death, or last follow-up, using Kaplan-Meier methods. Patients alive at last follow-up or with death not lymphoma related were censored. The log-rank test and Fisher’s exact test were used to test for association between variables as appropriate. The Cox proportional hazards model was used to evaluate the impact of prognostic variables on TTT and lymphoma-specific survival, and to adjust for other known prognostic variables in FLIPI (0 to 2 vs 3 to 4).49 All P values were two-sided and P < .05 was considered to be statistically significant. All analyses were performed using JMP 9.0 statistical discovery software by SAS.
Results
BCL2 mutation frequency in FL
Based on recent studies reporting frequent BCL2 mutations in FL,16,44,45 as well as our previous studies suggesting that certain Bcl-2 sequence variants found in lymphoma cell lines exhibit enhanced antiapoptotic properties,46,47 we examined the occurrence and significance of mutations in the BCL2 coding sequence in FL. In an initial prevalence study, BCL2 exons 1, 2, and 3 were sequenced in cryopreserved FL specimens from 38 FL patients (termed the “discovery” cohort) who had a median age of 64 years, median follow-up of 10.5 years, and median survival of 13.2 years. DNA from 300 normal individuals served as a control. We identified lymphoma-specific variants (supplemental Table 3) by removing polymorphisms present in the control samples, NCBI dbSNP and HapMap.
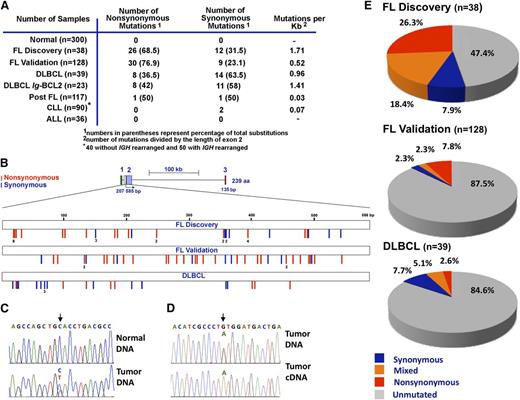
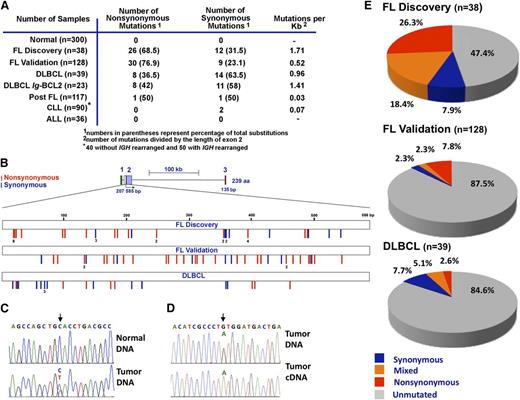
A total of 38 SNVs were observed in 20 of the 38 discovery cohort samples (Figure 1A; supplemental Table 3). These SNVs were located exclusively within exon 2 and not within the silent exon 1 or within exon 3 that encodes the Bcl-2 transmembrane domain (Figure 1B). To distinguish between rare germline variants and somatic mutations, we sequenced paired DNA samples from peripheral blood mononuclear cells and lymph nodes of 11 FL patients. As illustrated in Figure 1C for 1 case, 8 of these 11 FLs had SNVs in BCL2 exon 2 that were not present in the corresponding normal cells, indicating that the BCL2 SNVs represent somatic mutations.
Distribution of BCL2 somatic mutations in FL and non-Hodgkin lymphoma subtypes. (A) Summary of somatic sequence mutations for BCL2 in FL and other non-Hodgkin lymphoma subtypes analyzed by Sanger sequencing in normal samples, a discovery (prevalence) cohort of 38 FLs, a validation cohort of 128 FLs collected exclusively at diagnosis, and cohorts of 117 post-FLs, 39 DLBCLs, 90 CLLs (50 with IGH rearrangement and 40 without), and 36 ALLs. (B) Schematic diagram of the BCL2 gene (top) and somatic mutation distribution along the BCL2 gene in the FL discovery cohort (n = 38), FL validation cohort (n = 128), and DLBCL cohort (n = 39). Exons 1, 2, and 3 are coded in green, purple, and red, respectively. Blue and red bars indicate synonymous and nonsynonymous mutations, respectively. (C-D) Sequencing chromatograms of representative mutated FL samples (lymph node) and paired normal (peripheral blood) DNA (C), or FL DNA and corresponding cDNA (D). Arrows point to the positions of nucleotide change, with the variant nucleotide labeled in red. (E) Overall frequency of BCL2 somatic mutations in FL and DLBCL. Within each group, samples without mutation are depicted in gray, samples with only synonymous mutations in blue, samples with mixed mutations in orange, and samples with nonsynonymous mutations in red.
Distribution of BCL2 somatic mutations in FL and non-Hodgkin lymphoma subtypes. (A) Summary of somatic sequence mutations for BCL2 in FL and other non-Hodgkin lymphoma subtypes analyzed by Sanger sequencing in normal samples, a discovery (prevalence) cohort of 38 FLs, a validation cohort of 128 FLs collected exclusively at diagnosis, and cohorts of 117 post-FLs, 39 DLBCLs, 90 CLLs (50 with IGH rearrangement and 40 without), and 36 ALLs. (B) Schematic diagram of the BCL2 gene (top) and somatic mutation distribution along the BCL2 gene in the FL discovery cohort (n = 38), FL validation cohort (n = 128), and DLBCL cohort (n = 39). Exons 1, 2, and 3 are coded in green, purple, and red, respectively. Blue and red bars indicate synonymous and nonsynonymous mutations, respectively. (C-D) Sequencing chromatograms of representative mutated FL samples (lymph node) and paired normal (peripheral blood) DNA (C), or FL DNA and corresponding cDNA (D). Arrows point to the positions of nucleotide change, with the variant nucleotide labeled in red. (E) Overall frequency of BCL2 somatic mutations in FL and DLBCL. Within each group, samples without mutation are depicted in gray, samples with only synonymous mutations in blue, samples with mixed mutations in orange, and samples with nonsynonymous mutations in red.
To assess whether BCL2 mutations occurred in the translocated alleles, we sequenced cDNA prepared from discovery cohort samples with BCL2 mutations. The mutated message was vastly more abundant than WT in all 20 samples containing BCL2 mutations (Figure 1D), suggesting that the translocated allele is mutated.
To provide a basis for further examination of BCL2 mutations, we also sequenced the PIM1 gene, which is thought (like BCL2) to be a target of AID-induced mutagenesis. In the discovery cohort, only 5 of the 39 samples (3 of the 20 BCL2-mutant samples) harbored PIM1 mutations.
The discovery cohort had 1.71 × 10−3 mutations/bp in BCL2 exon 2, a frequency that approaches the IGH mutation rate (5.8 × 10−3 mutations/bp) during B-cell differentiation (Figure 1A).44,50 The mutation rate was much lower in other B-cell neoplasms, with BCL2 SNVs in 2 of 117 mucosa-associated, marginal zone, and lymphoplasmacytic (“postfollicular”) lymphomas, 0 of 36 ALLs and 2 of 90 CLLs (Figure 1A). Moreover, BCL2 was mutated twice as often in FL as in DLBCL (Figure 1A-B,E; supplemental Table 3), consistent with earlier results.44,50 Of the 39 DLBCLs examined, 23 were positive for Ig-BCL2 rearrangements involving the major breakpoint region (21) or minor cluster region (2), respectively. Nineteen BCL2 mutations occurred in BCL2-rearranged cases and 3 mutations occurred in nonrearranged cases (supplemental Table 3).
Presence of BCL2 mutations in FL at diagnosis
The discovery cohort contained samples collected at diagnosis and at relapse after chemotherapy. To rule out the possibility that BCL2 mutations were chemotherapy induced, as well as provide an opportunity for analyzing the relationship between BCL2 mutations at diagnosis and outcome, BCL2 sequencing was performed on a second, independent cohort collected exclusively at diagnosis. This validation cohort consisted of 128 patients with grades 1 or 2 FL and a median follow-up of 8.4 years (range, 0 to 30). Patient characteristics and survival are summarized in Table 1 and supplemental Figure 1. Fifty-eight (44%) of these patients had histologic evidence of transformation during follow-up, with a median TTT of 14 years, consistent with earlier findings.6,10,11 Sequencing revealed a total of 39 BCL2 mutations in 16 specimens at diagnosis in this cohort (Figure 1A-B).
Nature and patterns of BCL2 mutations in FL
Further analysis indicated that 26 of 38 mutations in the discovery cohort and 30 of 39 mutations in the validation cohort were amino acid altering (nonsynonymous; Figure 1B,E), providing an overall ratio of 2.7:1 for nonsynonymous vs synonymous mutations in FL. In contrast, the nonsynonymous:synonymous ratio was 0.57:1 in DLBCL (Figure 1E), in agreement with recent reports.43,44 Furthermore, some FLs had multiple nonsynonymous mutations (n = 2 in 6 samples, n = 3 in 2 samples, and n = 4, 5, or 10 in 1 sample each). Importantly, 90% of FL synonymous mutations occurred in combination with nonsynonymous SNVs (supplemental Table 3). This high frequency of nonsynonymous mutations, along with the absence of truncating mutations, raised the possibility that BCL2 mutations might confer an advantage to FL cells.
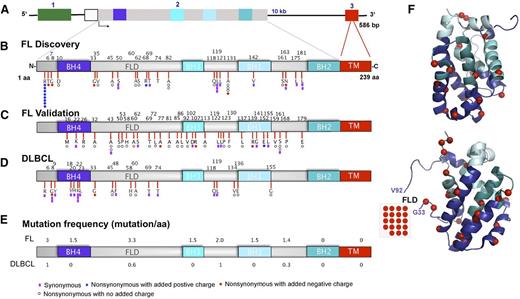
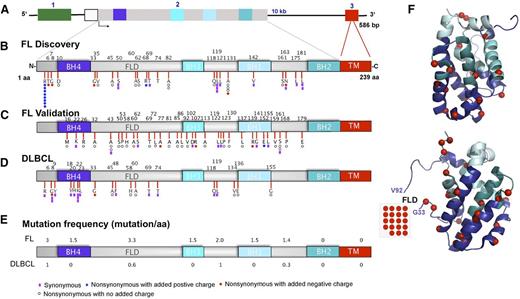
As indicated in Figure 2A-C, nonsynonymous mutations in FL were spread along the protein, including the N-terminus (10 of 77 mutations), flexible loop domain (FLD) (20 of 77), and conserved BH4, BH3, and BH1 domains (31 of 77). The distribution of mutations in DLBCL is shown in Figure 2D for comparison. In FL, the highest mutation frequency (defined as mutation load per 10 bp) occurred in the unstructured FLD (Figure 2E-F and supplemental Figure 2), which was previously implicated in binding p53 and c-myc as well as regulating the affinity of Bcl-2 for proapoptotic Bcl-2 family members.51-53
Distribution of mutations in the Bcl-2 protein. (A) Schematic diagram of the BCL2 gene. Exons 1, 2, and 3 are color-coded in green, gray, and red, respectively. (B-D) Linear diagram of Bcl-2 protein showing Bcl-2 homology domains and distribution of amino acid alterations in (B) FL discovery (n = 38), (C) FL validation (n = 128), and (D) DLBCL (n = 39) cohorts. Color-coded symbols depict distinct types of alterations, with purple for synonymous, white for nonsynonymous with no charge introduction, red for nonsynonymous with introduction of a negative charge, and blue for nonsynonymous with introduction of a positive charge. (E) Overall BCL2 coding mutation frequency normalized for an interval of 10 amino acids in FL and DLBCL cohorts. (F) Two different projections of Bcl-2 are presented showing coding somatic mutations (red spheres) in the 2 FL cohorts mapped together onto the Bcl-2 3D structure (PDB 2O21). BH, Bcl-2 homology domain; FLD, flexible loop domain; TM, transmembrane domain.
Distribution of mutations in the Bcl-2 protein. (A) Schematic diagram of the BCL2 gene. Exons 1, 2, and 3 are color-coded in green, gray, and red, respectively. (B-D) Linear diagram of Bcl-2 protein showing Bcl-2 homology domains and distribution of amino acid alterations in (B) FL discovery (n = 38), (C) FL validation (n = 128), and (D) DLBCL (n = 39) cohorts. Color-coded symbols depict distinct types of alterations, with purple for synonymous, white for nonsynonymous with no charge introduction, red for nonsynonymous with introduction of a negative charge, and blue for nonsynonymous with introduction of a positive charge. (E) Overall BCL2 coding mutation frequency normalized for an interval of 10 amino acids in FL and DLBCL cohorts. (F) Two different projections of Bcl-2 are presented showing coding somatic mutations (red spheres) in the 2 FL cohorts mapped together onto the Bcl-2 3D structure (PDB 2O21). BH, Bcl-2 homology domain; FLD, flexible loop domain; TM, transmembrane domain.
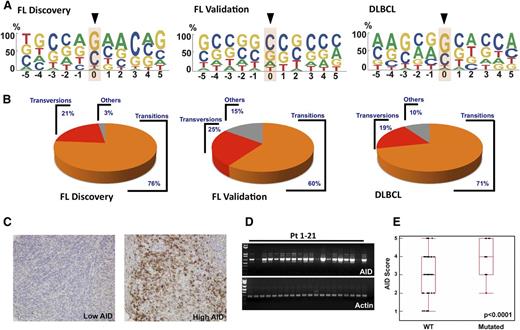
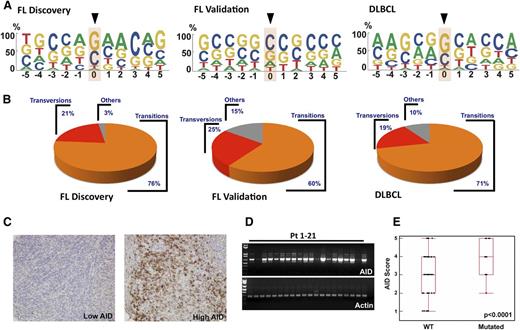
Some 95% of SNVs in the discovery cohort and 71% in the validation cohort occurred at Gs and Cs (Figure 3A-B). Moreover, 76% and 60% of mutations in the discovery and validation cohorts, respectively, were transitions (C to T or G to A) (Figure 3B). This predilection for Gs and Cs, as well as a high transition/transversion ratio, has been observed during AID-induced somatic hypermutation in normal B cells,31 suggesting that AID might be responsible for the observed BCL2 mutations. Consistent with this possibility, ∼40% of FL SNVs occurred at preferred AID target sequences (GCT/A or A/TGC) (Figure 3A and supplemental Table 4), even though AID can also mutate cytidines in other sequence contexts. Moreover, AID sites were not randomly mutated. Instead, mutations were only observed in exon 2, even though 39 preferred AID sites were present in exons 1 and 3 compared with 12 in exon 2 (supplemental Table 4).
C/G predominance of BCL2 mutations and AID expression in FL. (A) Depiction of bp preference of mutations in the FL discovery (left), FL validation (middle), and DLBCL (right) cohorts. The size of letters corresponds to the frequency with which that nucleotide is mutated in the BCL2 coding strand. (B) Transition and transversion load in FL discovery, FL validation, and DLBCL cohorts. Within each group, transitions are depicted in orange, transversions in red, and other changes in gray. (C) Images of FL validation cohort lymph node biopsies from diagnosis showing low (left) and high (right) staining after reaction with anti-AID primary antibody and peroxidase-labeled secondary antibodies. (D) RT-PCR was performed in 21 FL patient samples to detect AID expression differences at the mRNA level. Actin was used as an RNA loading control. (E) AID score (calculated as described in “Methods”) from 128 pretreatment FL validation cohort diagnostic samples depicted as a function of BCL2 mutation status (P < .0001).
C/G predominance of BCL2 mutations and AID expression in FL. (A) Depiction of bp preference of mutations in the FL discovery (left), FL validation (middle), and DLBCL (right) cohorts. The size of letters corresponds to the frequency with which that nucleotide is mutated in the BCL2 coding strand. (B) Transition and transversion load in FL discovery, FL validation, and DLBCL cohorts. Within each group, transitions are depicted in orange, transversions in red, and other changes in gray. (C) Images of FL validation cohort lymph node biopsies from diagnosis showing low (left) and high (right) staining after reaction with anti-AID primary antibody and peroxidase-labeled secondary antibodies. (D) RT-PCR was performed in 21 FL patient samples to detect AID expression differences at the mRNA level. Actin was used as an RNA loading control. (E) AID score (calculated as described in “Methods”) from 128 pretreatment FL validation cohort diagnostic samples depicted as a function of BCL2 mutation status (P < .0001).
Relationship between AID expression and BCL2 mutations in FL
When AID expression in the FL validation cohort at diagnosis was examined, immunohistochemistry demonstrated substantial intersample variability (Figure 3C). Similar variability was observed when AID was assessed by RT-PCR (Figure 3D). Importantly, AID expression was higher in samples with BCL2 mutations than in those without (Figure 3E) (P < .0001).
Increased BCL2 mutations at transformation
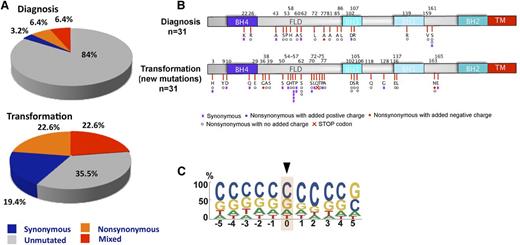
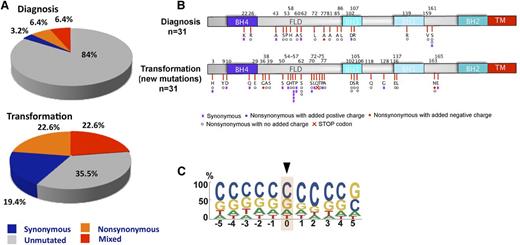
The presence of the mutagenic AID enzyme in many FLs raised the possibility that BCL2 mutations might continue to accumulate during the course of disease. To assess this possibility, we sequenced BCL2 in 31 available FL pairs, the first collected at diagnosis (part of the validation cohort) and the second at median of 4.5 (range, 0.5 to 14.5) years later at transformation. In these 31 pairs, the prevalence of BCL2 mutations was 16% at diagnosis and 65% at transformation (Figure 4A). Like mutations at diagnosis, those at transformation were distributed throughout the protein, with a predilection for the FLD (Figure 4B) and a strong preference for Gs and Cs (79%) (Figure 4C). Nonetheless, the mutation signature at transformation was distinct from that at diagnosis (compare Figure 3A with Figure 4C), perhaps because 26 of the 31 patients had been treated with radiation or chemotherapy between diagnosis and transformation. Of the 5 FL patients not treated with a DNA damaging therapy prior to transformation, only 2 had BCL2 mutations, a number that is too small to allow derivation of a therapy independent mutation signature at transformation.
BCL2 mutations at transformation in FL. (A) Overall frequency of BCL2 somatic mutations in FL validation cohort at diagnosis and transformation. Gray, blue, orange, and red depict samples without mutations, with only synonymous mutations, with mixed synonymous and nonsynomymous mutations, and with only nonsynonymous mutations, respectively. (B) Schematic diagram of the Bcl-2 protein showing Bcl-2 homology domains and distribution of new amino acid alterations for FL validation samples (n = 31) at transformation. Color-coded symbols depict distinct types of alterations, with purple for synonymous, white for nonsynonymous with no charge introduction, red for nonsynonymous with introduction of a negative charge, and blue for nonsynonymous with introduction of a positive charge. Red cross identifies a stop codon. (C) Depiction of bp preference of mutations in the FL validation at diagnosis and transformation (n = 31). The size of letters corresponds to the frequency with which that nucleotide is mutated in the BCL2 coding strand. bp, base pair; TM, transmembrane domain.
BCL2 mutations at transformation in FL. (A) Overall frequency of BCL2 somatic mutations in FL validation cohort at diagnosis and transformation. Gray, blue, orange, and red depict samples without mutations, with only synonymous mutations, with mixed synonymous and nonsynomymous mutations, and with only nonsynonymous mutations, respectively. (B) Schematic diagram of the Bcl-2 protein showing Bcl-2 homology domains and distribution of new amino acid alterations for FL validation samples (n = 31) at transformation. Color-coded symbols depict distinct types of alterations, with purple for synonymous, white for nonsynonymous with no charge introduction, red for nonsynonymous with introduction of a negative charge, and blue for nonsynonymous with introduction of a positive charge. Red cross identifies a stop codon. (C) Depiction of bp preference of mutations in the FL validation at diagnosis and transformation (n = 31). The size of letters corresponds to the frequency with which that nucleotide is mutated in the BCL2 coding strand. bp, base pair; TM, transmembrane domain.
Some FL BCL2 mutations exhibit gain-of-function
Based on the nonrandom distribution and nonsynonymous nature of FL BCL2 mutations, we began examining the effects of these mutations on Bcl-2 protein function. As illustrated in Figure 5 for the Bcl-2 G33R and A43T variants, products of some mutant BCL2 alleles displayed increased affinity for immobilized Bim and Puma BH3 peptides in vitro (Figure 5A-B), as well as increased binding to these proapoptotic proteins in a cellular context (Figure 5C), consistent with BCL2 gain-of-function mutations reported in some lymphoid cell lines.46,47 Furthermore, in cell-based assays, these Bcl-2 variants offered more protection from apoptosis induced by BimEL overexpression (Figure 5D). The increased affinity for proapoptotic BH3 domains and enhanced protection was not, however, seen with all variant Bcl-2 proteins, as exemplified by Bcl-2 A82G.
Some Bcl-2 amino acid substitutions increase affinity for proapoptotic family members. (A) Binding between immobilized human Bim BH3 peptide and 300 nM WT Bcl-2 or the G33R variant was analyzed by surface plasmon resonance. (B) Summary of dissociation constants for complexes of WT or mutant Bcl-2 with Bim BH3 peptide, Puma BH3 peptide, or BakΔTM protein showing that Bim and Puma BH3 domains are bound more tightly to Bcl-2 G33R or A43T than to WT. Error bars, SD of 3 independent experiments using different chips and protein preparations. *P < .03 vs WT sequence. (C) After cDNA encoding WT or variant Bcl-2 fused to S-peptide was expressed for 24 hours in Jurkat cells, S-peptide pulldowns (top) or whole cell lysates (bottom) were probed for S-peptide or the indicated Bcl-2 binding partners. (D) Nalm6 or Jurkat cells were transfected with plasmid encoding enhanced green fluorescent protein (EGFP) or EGFP fused to BimEL,59 along with empty vector or plasmid encoding the indicated Bcl-2 variant. Cells were cultured for 24 hours in the presence of the broad spectrum caspase inhibitor Q-VD-OPh,60 and subjected to immunoblotting to assess expression (left panels). Alternatively, after culture for 24 hours in the absence of Q-VD-OPh for 24 hours, cells were stained with APC-conjugated Annexin V and subjected to 2-color flow cytometry. Right panels show summary of EGFP-positive/Annexin V-negative cells (as a percentage of total cells) after transfection of Nalm6 cells (upper panel) or Jurkat cells (lower panel). Error bars, ± SD of 3 independent experiments. Procedures used for these in vitro studies were previously published46,47,53,61 and are described in the supplemental Methods.
Some Bcl-2 amino acid substitutions increase affinity for proapoptotic family members. (A) Binding between immobilized human Bim BH3 peptide and 300 nM WT Bcl-2 or the G33R variant was analyzed by surface plasmon resonance. (B) Summary of dissociation constants for complexes of WT or mutant Bcl-2 with Bim BH3 peptide, Puma BH3 peptide, or BakΔTM protein showing that Bim and Puma BH3 domains are bound more tightly to Bcl-2 G33R or A43T than to WT. Error bars, SD of 3 independent experiments using different chips and protein preparations. *P < .03 vs WT sequence. (C) After cDNA encoding WT or variant Bcl-2 fused to S-peptide was expressed for 24 hours in Jurkat cells, S-peptide pulldowns (top) or whole cell lysates (bottom) were probed for S-peptide or the indicated Bcl-2 binding partners. (D) Nalm6 or Jurkat cells were transfected with plasmid encoding enhanced green fluorescent protein (EGFP) or EGFP fused to BimEL,59 along with empty vector or plasmid encoding the indicated Bcl-2 variant. Cells were cultured for 24 hours in the presence of the broad spectrum caspase inhibitor Q-VD-OPh,60 and subjected to immunoblotting to assess expression (left panels). Alternatively, after culture for 24 hours in the absence of Q-VD-OPh for 24 hours, cells were stained with APC-conjugated Annexin V and subjected to 2-color flow cytometry. Right panels show summary of EGFP-positive/Annexin V-negative cells (as a percentage of total cells) after transfection of Nalm6 cells (upper panel) or Jurkat cells (lower panel). Error bars, ± SD of 3 independent experiments. Procedures used for these in vitro studies were previously published46,47,53,61 and are described in the supplemental Methods.
BCL2 mutations correlate with FL clinical course
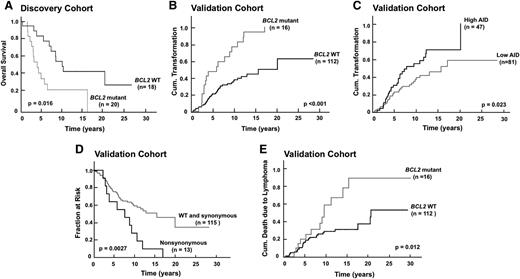
Examination of patient outcome in the FL discovery cohort revealed a striking decrease in overall survival from diagnosis if a BCL2 mutation was present (Figure 6A). Further analysis examined the relationship between BCL2 mutations and outcome in the validation cohort. As shown in Figure 6B, the presence of a BCL2 mutation at diagnosis was associated with a marked increase in transformation risk compared with the absence of a BCL2 mutation (P < .0001 by log-rank test; hazard ratio [HR] 3.56; 95% CI, 1.98-6.17). This association was even stronger than the relationship between AID expression and transformation risk (HR 1.70; 95% CI, 1.06-3.00; Figure 6C). These effects were most prominent in FLs with nonsynonymous mutations (Figure 6D). There was also a striking acceleration of death due to lymphoma (Figure 6E) in cases with BCL2 mutations at diagnosis vs those without (median, 9.6 vs 20.4 years; P = .012 by log-rank test). In multivariate analysis, FLIPI and presence of BCL2 mutations were independent risk factors for transformation, with HRs of 3.43 (95% CI, 1.67-6.66; P = .0013) and 3.61 (95% CI, 2.02-6.24; P < .0001), respectively. FLIPI and the presence of BCL2 mutations were also independent risk factors for dying from lymphoma in multivariate analysis, with HRs of 2.67 (95% CI, 1.17-5.57; P = .021) and 2.28 (95% CI, 1.14-4.33; P = .021), respectively.
BCL2 mutations correlate with transformation risk and disease-specific death in FL. (A) Kaplan-Meier plots showing impact of BCL2 mutations on overall survival in FL discovery cohort. (B-C) Kaplan-Meier plots showing impact of BCL2 mutations (B) or AID expression (C) on transformation risk in FL validation cohort. (D) Kaplan-Meier plots showing impact of BCL2 nonsynonymous mutations on TTT in FL validation cohort. (E) Kaplan-Meier plots showing impact of BCL2 mutations on death due to lymphoma in FL validation cohort.
BCL2 mutations correlate with transformation risk and disease-specific death in FL. (A) Kaplan-Meier plots showing impact of BCL2 mutations on overall survival in FL discovery cohort. (B-C) Kaplan-Meier plots showing impact of BCL2 mutations (B) or AID expression (C) on transformation risk in FL validation cohort. (D) Kaplan-Meier plots showing impact of BCL2 nonsynonymous mutations on TTT in FL validation cohort. (E) Kaplan-Meier plots showing impact of BCL2 mutations on death due to lymphoma in FL validation cohort.
Discussion
In the present study, we report for the first time an association between BCL2 mutations at diagnosis in FL, high AID expression, early disease progression, and poorer survival. These observations are potentially important for understanding FL transformation to clinically aggressive disease and the development of future therapeutic strategies.
Even though BCL2 mutations have been previously reported in FL,16,40,45,54,55 their correlation with clinical outcome has not been systematically explored. Several observations in the present study suggest that BCL2 mutations might be functionally important. First, in our study, mutations were detected only in exon 2 and not in exons 1 or 3 (Figures 1B and 2A-C) even though the former would be closer to the IGH locus and more prone to AID-induced mutagenesis. Second, in marked contrast to DLBCL,43 there was an excess of nonsynonymous mutations in FL (Figure 1E). Indeed, the vast majority of synonymous BCL2 mutations in FL occur in association with nonsynonymous alterations (supplemental Table 3). Third, some of the mutant Bcl-2 proteins (eg, Bcl-2 G33R and Bcl-2 A43T), bound and neutralized proapoptotic Bcl-2 family members more effectively (Figure 5).
Although we have observed functional consequences of the BCL2 mutations found in certain lymphoma cell lines46,47 and in FL (eg, this study), it is important to emphasize that not all of the BCL2 mutations are functionally significant. In particular, we have observed that the conservative A82G mutation has little impact on Bcl-2 function (Figure 5). Thus, the situation with BCL2 mutations might be somewhat analogous to BRCA1 or BRCA2 missense mutations, where some are functionally important and others are not.56
Several additional observations suggest that AID might be involved in the generation of the BCL2 SNVs. First, the vast majority of BCL2 mutations occur at Cs and Gs, consistent with AID-induced cytidine deamination. Second, BCL2 mutations exhibit a preference for AID target sequences rather than CpG sequences that would be preferred if spontaneous chemical deamination were responsible. Third, higher AID expression was observed in samples with BCL2 mutations at diagnosis (Figure 3E).
When the relationship between AID, BCL2 mutations, and survival was examined, transformation correlated with high AID expression (Figure 6C) and even more strongly with the presence of BCL2 mutations. Initially observed in a prevalence study (Figure 6A), this relationship between BCL2 mutations and earlier transformation was independently replicated in a larger independent study of FL samples, uniformly collected at diagnosis (Figure 6B). In short, the presence of BCL2 mutations at diagnosis correlated with decreased TTT of FL and increased risk of death due to lymphoma (Figure 6B,E).
We also observed more BCL2 mutations at transformation than at diagnosis (Figure 4), possibly reflecting ongoing mutagenic processes in FL. Comparison of the mutations at diagnosis and transformation nonetheless revealed potentially important differences. In particular, those acquired at transformation had a lower frequency of obvious AID signatures (15% at transformation vs 40% at diagnosis (supplemental Table 4), perhaps reflecting additional effects of intervening therapy or increased AID action at noncanonical sequences.57 Moreover, some mutations at transformation were clearly nonfunctional (eg, a BCL2 variant with a premature stop codon at transformation), again emphasizing that not all BCL2 mutations enhance Bcl-2 protein function.
Despite the strong correlation between BCL2 mutations at diagnosis and the poor outcome in 2 separate cohorts, we are aware of several limitations to the present study. First, the 2 cohorts differed from each other in important ways. The discovery cohort consisted of FLs with multiple cryopreserved vials decades after diagnosis. Consistent with the possibility that this cohort might have had bulkier disease, twice as many patients in the discovery cohort had FLIPI scores of 3 or 4 at diagnosis (Table 1). The frequency of mutations at diagnosis in this cohort also appeared higher, although the sample size was small.
Second, in order to have sufficient follow-up, the present study used cohorts treated almost exclusively in the prerituximab era. Accordingly, it will be important in the future to assess whether a similar correlation between BCL2 mutation and poor outcome exists in patients receiving chemoimmunotherapy.
Third, because of our focus on sequence-associated changes in function of the Bcl-2 protein,46,47 the present study differs from other recent efforts to catalog FL mutations. Sanger sequencing, a gold standard for detecting sequence variation, was used here. Moreover, the present study focused on BCL2 exons, especially protein encoding exons 2 and 3. This more focused approach likely underestimates the total number of BCL2 mutations compared with studies that used much more sensitive sequencing techniques and/or examined more noncoding BCL2 sequence in an effort to identify all mutations in the BCL2 locus.16,44,45
Finally, the present study does not address the question of whether BCL2 mutations cause FL transformation. In our cohort, over 50% of transformed FLs have synonymous mutations or no mutations at all (Figure 4A), indicating that BCL2 mutations are not required for FL transformation but leaving open the question of their contribution in transformed FLs that harbor them. In particular, we cannot rule out the possibility that BCL2 mutations are simply a surrogate for AID-induced mutagenesis. AID has previously been implicated in mutating BCL2,43 as well as BCL6, MYC, PIM1, and PAX5.35,36 In the present study, PIM1 mutations were less frequent than BCL2 mutations and did not correlate with early transformation. Moreover, transformation correlated more strongly with BCL2 mutation status (Figure 6B) than with AID expression (Figure 6C). Nonetheless, further evaluation of AID expression, BCL2 mutation status, and disease progression in additional patient cohorts is required to further address this issue. Moreover, even if BCL2 mutations are simply a reflection of AID activity, they might represent an objective and readily quantifiable measure of AID mutagenesis.
The observation that BCL2 mutation status at diagnosis is an independent prognostic factor in FL that supplements the FLIPI has potentially important implications. A high FLIPI score has been associated with transformation in multiple studies.2,17,20,22,23,58 If poor outcome in patients with BCL2-mutant FL is also widely replicated, this might serve as the foundation for further studies in FL. In particular, the assessment of BCL2 mutation status along with FLIPI could allow better stratification of FL patients in future clinical trials. In addition, it might also be reasonable to assess the effects of earlier therapeutic intervention, use of agents that bypass the mitochondrial apoptotic pathway, or treatment with Bcl-2 antagonists in this subgroup of FL patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Richard Weinshilboum for his encouragement, anonymous reviewers for helpful suggestions, and past and present members of the Mayo Clinic Lymphoma Group (who cared for the patients in this study) for their contributions.
This study was supported by grants from the Leukemia & Lymphoma Society (6125-10), National Institutes of Health, National Cancer Institute (P50 CA097274, R01 CA95241, and R01 CA166741), the Mayo Clinic Center for Individualized Medicine, and the Predolin Foundation.
Authorship
Contribution: S.H.K., H. Dai, G.S.N., M.J.M., and J.R.C. conceptualized and designed the study; A.J.N., J.R.C., S.L.S., T.M.H., J.E.K., S.D.G., N.E.K., and T.E.W. provided study material or patients; C.C., P.A.S., H. Dai, A.L.F., H. Ding, X.W.M., A.D., A.K.C., X.W., W.R.M., and T.M.H. collected and assembled the data; C.C., M.J.M., D.F.J., G.S.N., and S.H.K. analyzed and interpreted the data; S.H.K., G.S.N., and T.E.W. provided financial support for the study; and all authors wrote and gave final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Scott H. Kaufmann, Division of Oncology Research, Gonda 19-212, Mayo Clinic, 200 First St S.W., Rochester, MN 55905; e-mail: kaufmann.scott@mayo.edu.
References
Author notes
C.C. and P.A.S. contributed equally as cofirst authors of this study.
G.S.N. and S.H.K. contributed equally as cosenior authors.