Key Points
Mutations in SETBP1 can be detected using droplet digital polymerase chain reaction in at least 30% of patients with JMML and are associated with a dismal prognosis.
Patients harboring rare cells with mutant SETBP1 at diagnosis should be considered candidates for swift hematopoietic stem cell transplant.
Abstract
Juvenile myelomonocytic leukemia (JMML) is an aggressive myeloproliferative neoplasm of childhood associated with a poor prognosis. Recently, massively parallel sequencing has identified recurrent mutations in the SKI domain of SETBP1 in a variety of myeloid disorders. These lesions were detected in nearly 10% of patients with JMML and have been characterized as secondary events. We hypothesized that rare subclones with SETBP1 mutations are present at diagnosis in a large portion of patients who relapse, but are below the limits of detection for conventional deep sequencing platforms. Using droplet digital polymerase chain reaction, we identified SETBP1 mutations in 17/56 (30%) of patients who were treated in the Children’s Oncology Group sponsored clinical trial, AAML0122. Five-year event-free survival in patients with SETBP1 mutations was 18% ± 9% compared with 51% ± 8% for those without mutations (P = .006).
Introduction
Juvenile myelomonocytic leukemia (JMML) is an aggressive myeloproliferative neoplasm of childhood with overlapping myeloproliferative and myelodysplastic properties.1,2 Curative therapy uses hematopoietic stem cell transplantation (HSCT), leading to a 5-year event-free survival of 52%, with relapse being the most common reason for treatment failure.3,4 Although 85% to 90% of patients harbor mutations in NF1, NRAS, KRAS, PTPN11, or CBL at diagnosis, these mutations are not reliably associated with differences in outcome.5-10 Although JMML frequently manifests as an aggressive disease requiring swift treatment, there are rare patients who spontaneously resolve with little to no intervention.11,12 Importantly, there are no reliable biomarkers capable of distinguishing those patients who can be safely observed from those with an aggressive disease course.13,14
Sakaguchi et al recently reported that SETBP1 mutations were detected in 7/92 (8%) patients with JMML and were associated with a poor prognosis.15 Given that these mutations had a lower allelic frequency compared with other diagnostic Ras pathway mutations, they hypothesized that SETBP1 mutations contribute to disease progression rather than initiation. Similarly, Shiba et al identified SETBP1 mutations in 2/42 (4.8%) patients with JMML.16 Makishima et al reported that SETBP1 mutations were seen in 17% of secondary acute myeloid leukemias and 15% of chronic myelomonocytic leukemias.17 By analyzing serial samples, they demonstrated that SETBP1 mutations were acquired during leukemic evolution and likely represented secondary events.
After identifying an increase in the allelic fraction of a SETBP1 mutation in a patient at diagnosis who subsequently relapsed, we hypothesized that subclonal mutations might be present in a greater number of patients than previously reported, and that their presence would confer a poor prognosis.
To detect such rare subclonal events we applied a technology capable of detecting events as rare as 0.001%.18 Droplet digital polymerase chain reaction (ddPCR) and the QX100 system partitions template molecules into uniform, nanoliter-sized droplets before amplification. This approach is particularly useful for rare mutation detection in a high background of wild-type alleles because the vast majority of background alleles are partitioned away from the mutant, facilitating the detection of rare mutants relative to other genomic approaches.19-21
Methods
Patient samples
DNA was extracted using standard methods from bone marrow or peripheral blood mononuclear cells obtained at diagnosis from 78 patients diagnosed with JMML. Fifty-six of these patients were enrolled in clinical trial AAML0122 and had outcome data available. Five patients also had material at relapse time points. Approvals for these studies were obtained from the University of California San Francisco (UCSF) Committee on Human Research. All participants/guardians provided informed consent in accordance with the Declaration of Helsinki.
Sanger sequencing
Specimens from all 78 patients with JMML were screened for SETBP1 mutations by Sanger sequencing. We performed Sanger sequencing of SETBP1 exon 4 (NM_015559) using Hot Start Polymerase (Promega) and the following primers: 5′ CTTCACCAGCAGCTATGCA and 5′ CGGTGGGAGATTCTGAACAC. PCR conditions were 95°C for 2 minutes; 35 cycles of 94°C for 45 seconds, 60°C for 30 seconds, and 72°C for 40 seconds; and 1 cycle at 75°C for 5 minutes. Sequences were aligned using CLC software (CLC Bio, Aarhus, Denmark).
Digital PCR
The ddPCR assays used to identify mutations in the SETBP1 gene were designed using Beacon Designer 8.0 (Premier Biosoft, Palo Alto, CA). The PCR primer sequences used to amplify the SETBP1 mutational hotspot were: 5′ CACAGTGAGGAGACGATC and 5′ GTACCTCCTTCGGGATTC. Hydrolysis probes were designed to the following mutations. The mutation in each probe is noted in a lower case letter. The wild-type probe was labeled with HEX and all of the mutant probes were labeled with 6-carboxyfluorescein (6-FAM).
Wild-type: 5′ HEX CAGCGACAGCGGCATT 3′ NFQ
D868N: 5′ 6-FAM CAGCaACAGCGGCATT 3′ NFQ
G870S: 5′ 6-FAM CAGCGACAGCaGCATT 3′ NFQ
G870D: 5′ 6-FAM CAGCGACAGCGaCATT 3′ NFQ
I871T: 5′ 6-FAM CAGCGGCAcTGGGACA 3′ NFQ
Primers and probes were obtained from Bio-Rad Laboratories, Hercules, CA. Each assay was ordered as 9 μM PCR primers and 5 μM probes as 20× stock solutions. For each assay duplex (wild-type/mutant), the final concentration of primers was 900 nM and each probe was 250 nM into ddPCR. A multiplex assay was prepared using wild-type (HEX), D868N (FAM), G870S (FAM), G870D (FAM), and I871T (FAM) for a final assay concentration of 2250 nM of primers and 250 nM of each probe.
The digital PCR reactions were prepared using the QX100 reagents and consumables from Bio-Rad Laboratories: 2× ddPCR supermix for probes no deoxyuridine triphosphate (#186-3024), droplet generator cartridges and gaskets (#186-4007), and probes droplet generation oil (#186-3005). EcoRI HF (#R3101S, New England Bio-Labs) was added directly into the ddPCR mastermix at 2 U/well. A total of 100 ng genomic DNA was added to each well and the reaction was thoroughly mixed before droplet generation. After droplet generation, the droplets were transferred to an Eppendorf TwinTec PCR plate (#951020362) with a heat-activated foil seal (#181-4040).
Initially, a temperature gradient was performed (55 to 65°C) to determine the optimal annealing temperature for the SEPBP1 assays to give optimal specificity and separation of clusters. The droplets underwent 40 thermal cycles using the following 2-step parameters on a Bio-Rad C1000 thermal cycler: 95°C for 10 minutes, 94°C for 30 seconds, and 57°C for 1 minute × 40 cycles, 98°C for 10 minutes, and a 12°C hold.
The droplets were prepared and analyzed using the Bio-Rad QX100 ddPCR instrument at the UCSF Center for Advanced Technologies facility. The data were analyzed using QuantaSoft 1.5. Thresholds were set manually using the clustering tool for the 2-dimensional plot view, and a global threshold was determined for each assay on a particular plate based on the location of clusters in the positive control wells. For patient samples, the requirement for inclusion as a SETBP1-positive sample was the presence of at least 1 FAM-positive droplet in the upper left quadrant of the 2-dimensional plot (>3000 fluorescence units in FAM; <3000 fluorescence units in HEX). Using QuantaSoft, the Poisson concentrations for each well in addition to the 95% confidence interval (CI) were determined by using the Poisson formula: c (copies per droplet) = −ln(1 − p), in which p = the fraction of positive droplets. The droplet size is approximately 0.85 nL. Direct measurement across a large dynamic range (1 to 100 000 copies/well) is feasible because the 95% CI is reported for each well measurement.
Colony picks
Mononuclear cells were isolated from fresh bone marrow or peripheral blood samples and resuspended in Iscove modified Dulbecco medium + 2% fetal bovine serum. Cells were suspended at a concentration of 200 000 cells/mL. A total of 154 μL of the cell suspension was added to a tube with the following: 1.2 mL human methylcellulose complete media (R&D Systems, catalog no. HSC003), 15 μL of 100× penicillin/streptomycin, diluted in water and Iscove modified Dulbecco medium to complete the tube volume to 1.54 mL. The solution was vortexed for 15 seconds and rested for 15 minutes. A total of 1.1 mL was plated into a 35 × 10 mm Petri dish (BD Falcon, catalog no. 351008), placed into a 150 × 15 mm Petri dish (BD Falcon, catalog no. 351058) with another dish containing sterile water, and placed into an incubator at 37°C, 5% CO2. After 14 days, plates were removed from the incubator, and 2 μL of colonies (clusters of 50 cells or more) were plucked under a microscope at ×40 magnification and resuspended in 20 μL of sterile water. PCR amplification was then carried out for SETBP1 exon 4 using 1 μL of the vortexed colony suspension.
Single droplet sorting
Fabrication of microfluidic devices.
Poly-di-methyl-siloxane (PDMS) microfluidic devices are constructed using standard soft lithographic methods22 including SU8-on-Si wafer masters and PDMS-on-glass devices. Electrodes are designed as channels in the PDMS device and are filled with a 5 M solution of NaCl. To implement saltwater electrodes, syringes are filled with the 5 M NaCl solution and inserted into custom acrylic clamps. The acrylic clamps apply a constant pressure to the salt solution via a screw pushing on the syringe plunger. The syringes are connected to the device electrode via a 27-gauge needle and polyethylene tubing. Alligator clips connected to an exposed section of the syringe needle provide electrical contact. The driving bias is generated by a high-voltage amplifier (Trek, 609E-6). Two electrodes are incorporated into the device, 1 for the high-voltage signal and 1 for a ground. The grounded “moat” channel surrounds both the high-voltage electrode and the other inlets to prevent stray fields from inadvertently merging or otherwise affecting droplets at other locations on the device.
Droplet generation device.
The aqueous phase consists of 100 ng genomic DNA, Platinum Multiplex PCR Mix, Taqman probes and primers, restriction enzymes, and polyethylene glycol (molecular weight 6000) at 5% wt/vol to a final volume of 100 μL. Novec 7500 fluorinated oil (HFE) containing 2% of a triblock surfactant is used as the dispersed phase. Drops are generated in a flow-focusing geometry using a 70-μm nozzle with oil flow rates of 1500 μL/h and aqueous flow rates of 750 μL/h. Drops are collected in PCR tubes and thermal cycled using temperature ramp rates of 2°C/s with the following program: 95°C for 10 minutes, 40 cycles of 94°C for 30 seconds followed by 55°C for 1 minute, 1 cycle of 98°C for 10 minutes, and a final hold at 4°C.
Sorting device.
Droplets are periodically reinjected into the sorting channel by a flow-focus junction, where more oil is added to space the drops.23 Drops flow to the asymmetric “Y”-sorting junction, where they can take one of 2 paths. In the absence of sorting, drops preferentially flow to the waste channel because it has a lower fluidic resistance. When a drop passes the laser, its fluorescence is collected by a microscope objective and focused on a photomultiplier tube (Hamamatsu) connected to a computer running a custom LabVIEW program on a real-time field-programmable gate array card (National Instruments). When a drop is bright enough to cross the voltage threshold set in the program, the software sends several cycles of a 20-kHz single-ended square wave to the sorting electrodes after being amplified by a factor of 1000 by a high-voltage amplifier (Trek). The sorted drops move up the field gradient created by the electrodes by dielectrophoresis and are pulled into the keep channel.
TA subcloning
DNA was collected after lysis of the pooled FAM-positive droplets. Subcloning of FAM-positive droplet DNA using a TA cloning kit with pCR 2.1 vector (Invitrogen) was followed by directed sequencing of individually transformed colonies.
Progenitor sorting
Previously cryopreserved bone marrow samples were thawed and sorted into the following fractions as described:24 Lin− (CD2, CD3, CD4, CD4, CD7, CD8, CD10, CD11b, CD14, C19, CD20, CD56, CD235a), CD34+CD38−CD45RA−CD90+ (hematopoietic stem cells), Lin−CD34+CD38−CD45RA−CD90− (multipotent progenitors), Lin−CD34+CD38+CD45RA−CD90− (common myeloid progenitors), and Lin−CD34+CD38+CD45RA+CD90− (granulocyte-monocyte progenitors). DNA from each fraction was extracted using DNeasy Blood and Tissue Kit from Qiagen.
Deep sequencing
A customized TruSeq amplicon kit was designed by using the online DesignStudio pipeline (http://designstudio.illumina.com; Illumina) targeting the entire coding regions of 6 genes—PTPN11, NRAS, KRAS, CBL, NF1, and SETBP1—and covering 22 721 bp with 90 amplicon regions. Libraries prepared from DNA were subjected to 250 bp paired-end sequencing. Twelve patients with either clonal (n = 2) or subclonal (n = 10) SETBP1 mutations who had sufficient DNA for next-generation sequencing were analyzed. These 12 samples were run on a Mi-Seq platform that generated an average depth of coverage above 175× across all targeted regions. After quality trimming, the readings of each sample were aligned independently to the human genome reference sequence (GRC37/hg19) as well as sequences of designed regions and followed with variant calling using CLC Genomic Workbench (v7.5, CLC Bio, Denmark). An in-house tool was then used to filter through the variant list, and the consequence of protein sequence changes resulting from detected DNA sequence changes were predicted using the functional annotation tool ANNOVAR (http://www.openbioinformatics.org/annovar/).25
Statistical analyses
Data from AAML0122 were current as of June 8, 2010. The significance of observed differences in proportions was tested using the χ2 test and Fisher’s exact test when data were sparse. The Mann-Whitney test was used to determine the differences in medians. The Kaplan-Meier method26 was used to estimate probabilities of overall survival (OS) and event-free survival (EFS). OS was defined as time from study entry to death; EFS was defined as time from study entry to relapse, death, or graft failure. Estimates of EFS and OS are reported with a 1 Greenwood standard error. Cox proportional hazards models27 were used to estimate the hazard ratio (HR) for defined groups of patients in univariate and multivariate analyses of OS and EFS.
Results
Mutant SETBP1 clones increase over time
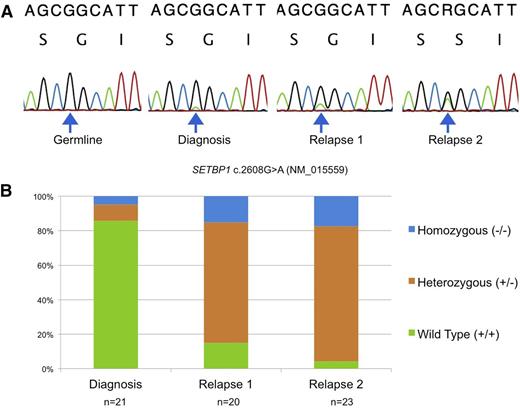
Using peripheral blood or bone marrow–derived mononuclear cell DNA, we identified SETBP1 mutations by Sanger sequencing in 3 patients at relapse. Interestingly, 2 patients who relapsed had serially acquired samples demonstrating SETBP1 mutations that were barely detectable at diagnosis using Sanger sequencing and increased in allelic fraction over time (Figure 1A). Differentiating these low allelic fraction mutations from a PCR artifact was only possible in retrospect after observing the clonal outgrowth at relapse. In addition, when cryopreserved samples from serial time points were plated in complete methylcellulose, the number of individual cells that were heterozygous or homozygous for SETBP1 increased at each time point despite intensive treatment, suggesting these cells are resistant to traditional cytotoxic therapy (Figure 1B).
Disease progression over serial time points. (A) Sanger sequencing from 1 patient over 4 time points reveals a serial increase in the allelic fraction of mutant SETBP1 c.2608G>A (p.G870S). (B) Sequencing of individual colonies from the same patient using defrosted, cryopreserved cells grown in complete methylcellulose at diagnosis, relapse 1, and relapse 2 reveals that the number of cells that contain heterozygous and homozygous mutant SETBP1 increase despite intensive treatment. n = number of colonies sequenced at each time point.
Disease progression over serial time points. (A) Sanger sequencing from 1 patient over 4 time points reveals a serial increase in the allelic fraction of mutant SETBP1 c.2608G>A (p.G870S). (B) Sequencing of individual colonies from the same patient using defrosted, cryopreserved cells grown in complete methylcellulose at diagnosis, relapse 1, and relapse 2 reveals that the number of cells that contain heterozygous and homozygous mutant SETBP1 increase despite intensive treatment. n = number of colonies sequenced at each time point.
Droplet digital PCR is a sensitive platform for detecting SETBP1 mutations
Primers and probes were designed for the 4 most common mutations in SETBP1 (D868N, G870S, G870D, and I871T). Determination of the sensitivity and specificity of the assay was carried out in both multiplex and singleplex reactions. The 4 mutant probes and 1 wild-type probe were first combined in a multiplex reaction. Any sample that was positive in the multiplex assay was repeated in singleplex to determine the exact nucleotide variant of interest (see supplemental Figure 1 on the Blood Web site). In singleplex, a dilution series for the most common single nucleotide variant, c.2602G>A (p.D868N), was performed. Using a heterozygous patient sample (sequenced using Sanger methods), an estimated 5000 copies/µL of genomic DNA based on NanoDrop measurement was serially diluted (1:10) 6 times to 0.005 copies/μL and placed in triplicate wells. The QX100 platform reliably detected levels as low as 1 mutant allele per well (supplemental Figure 2). In addition, no false positives were present in any of the wild-type or negative controls (Figure 2). In total, 23/66 (35%) JMML patients screened by ddPCR harbored SETBP1 mutations at diagnosis.
Representative ddPCR experiment. (A) Wild-type DNA reveals only HEX-positive droplets. (B) Positive control DNA from a patient with a SETBP1 p.G870D mutation reveals a mixed population of HEX- and FAM-positive droplets. Orange droplets indicate the presence of mutant and wild-type DNA within 1 droplet. (C) When no DNA is added, no template control wells contain only empty droplets. No HEX- or FAM-positive droplets are identified. (D) DNA from sample J435 reveals the presence of subclonal SETBP1 p.G870D mutations in 3 droplets, representing an allelic fraction of 0.09%.
Representative ddPCR experiment. (A) Wild-type DNA reveals only HEX-positive droplets. (B) Positive control DNA from a patient with a SETBP1 p.G870D mutation reveals a mixed population of HEX- and FAM-positive droplets. Orange droplets indicate the presence of mutant and wild-type DNA within 1 droplet. (C) When no DNA is added, no template control wells contain only empty droplets. No HEX- or FAM-positive droplets are identified. (D) DNA from sample J435 reveals the presence of subclonal SETBP1 p.G870D mutations in 3 droplets, representing an allelic fraction of 0.09%.
Sorting of individual droplets allows direct confirmation of mutant DNA
To confirm that the subclonal SETBP1 mutations detected by ddPCR represented true mutant alleles and not PCR artifacts, we sorted rare positive droplets using a microfluidic device with fluorescence-based detection capabilities (Abate Laboratory, UCSF). DNA was extracted from FAM-positive droplets from a patient with a subclonal mutation (0.45% allelic fraction). Pooled FAM-positive DNA was cloned and transformed into Escherichia coli, and DNA from individual bacterial colonies was then sequenced. Fifty percent of these colonies contained the mutant SETBP1 allele, confirming the existence of mutant SETBP1 DNA molecules in our FAM-positive droplets (Figure 3A). This patient also experienced a clinical relapse 226 days after HSCT, at which point a clonal mutation in SETBP1 (allelic fraction 38%) was detected by both ddPCR and Sanger sequencing (Figure 3B).
Confirmation of subclonal SETBP1. (A) Mutant droplets were sorted, collected, and lysed for DNA. A Sanger tracing of sorted mutant droplet DNA from diagnosis revealed the SETBP1 c.2608G>A (p.D868N) mutation. (B) ddPCR at diagnosis using pooled bone marrow–derived DNA from the same patient showed a subclonal mutation at diagnosis not detected by Sanger sequencing. FAM (blue) droplets indicate the presence of mutant SETBP1 DNA; HEX (green) droplets indicate the presence of wild-type DNA; orange droplets indicate the presence of both mutant and wild-type DNA. (C) At relapse, a clonal SETBP1 mutation is clearly visible on ddPCR and Sanger sequencing. Percentages indicate the allelic fraction of mutant SETBP1.
Confirmation of subclonal SETBP1. (A) Mutant droplets were sorted, collected, and lysed for DNA. A Sanger tracing of sorted mutant droplet DNA from diagnosis revealed the SETBP1 c.2608G>A (p.D868N) mutation. (B) ddPCR at diagnosis using pooled bone marrow–derived DNA from the same patient showed a subclonal mutation at diagnosis not detected by Sanger sequencing. FAM (blue) droplets indicate the presence of mutant SETBP1 DNA; HEX (green) droplets indicate the presence of wild-type DNA; orange droplets indicate the presence of both mutant and wild-type DNA. (C) At relapse, a clonal SETBP1 mutation is clearly visible on ddPCR and Sanger sequencing. Percentages indicate the allelic fraction of mutant SETBP1.
SETBP1 mutations confer a dismal prognosis
AAML0122 was a Children’s Oncology Group–sponsored clinical trial to assess the safety and efficacy of a farnesyl transferase inhibitor and chemotherapy in patients with JMML.4 The trial enrolled 85 patients between June 2001 and October 2006. Fifty-six patients had sufficient material for SETBP1 ddPCR analysis, had outcomes data available from the trial, and were therefore eligible for analysis. Of note, there were no significant differences in the baseline characteristics of the 56 patients included in the analysis when comparing those with SETBP1 mutations with those without (supplemental Table 1). In addition, EFS at 5 years for these 56 patients was no different than the 29 patients without sufficient material for analysis (41% ± 7% vs 40% ± 9%, P = .718).
Clonal mutations were defined as those clearly visible on Sanger chromatograms, whereas subclonal mutations were defined as those below the threshold detectable using Sanger sequencing (approximately 15%). Of 56 patients, 17 had SETBP1 mutations at diagnosis; 13 were subclonal and 4 were clonal. EFS at 5 years in patients with any SETBP1 mutation (n = 17) was 18% ± 9% compared with 51% ± 8% in those with wild-type SETBP1 (n = 39) (P = .006) (Figure 4A). When including only patients with subclonal mutations (n = 13) compared with those without mutations (n = 39), the difference in EFS remained significant (P = .004) (Figure 4B). However, 5-year OS differed between patients with SETBP1 mutations (44% ± 12%) vs those without (64% ± 8%), but did not reach statistical significance (Figure 4C-D). Of note, 6/17 (35.3%) patients with SETBP1 mutations received an HSCT after meeting criteria for progressive disease compared with 3/39 (7.7%) patients without a SETBP1 mutation (P = .017).
SETBP1 status at diagnosis affects outcome. (A) EFS in patients with a SETBP1 mutation was significantly lower than those without a mutation in the AAML0122 clinical trial (P = .006). (B) EFS in patients with a subclonal SETBP1 mutation that was only detectable by ddPCR was significantly lower than in those without a mutation (P = .004). (C) OS in patients with a SETBP1 mutation compared with those without a mutation (P = .258). (D) OS in patients with a subclonal mutation compared with those without any mutation (P = .507).
SETBP1 status at diagnosis affects outcome. (A) EFS in patients with a SETBP1 mutation was significantly lower than those without a mutation in the AAML0122 clinical trial (P = .006). (B) EFS in patients with a subclonal SETBP1 mutation that was only detectable by ddPCR was significantly lower than in those without a mutation (P = .004). (C) OS in patients with a SETBP1 mutation compared with those without a mutation (P = .258). (D) OS in patients with a subclonal mutation compared with those without any mutation (P = .507).
Deep sequencing of canonical mutations
PTPN11, KRAS, NRAS, CBL, and NF1 were sequenced in 12 patient samples that were identified by ddPCR to contain SETBP1 mutations and had sufficient DNA for analysis (supplemental Table 2). Canonical mutations were detected in proportion to what has been previously described.15 However, none of the subclonal SETBP1 mutations identified by ddPCR were detected by deep sequencing.
SETBP1 is an independent risk factor for poor prognosis
Several clinical characteristics have been variably associated with outcome in JMML.11 In particular, older age, higher white blood cell count (WBC), higher fetal hemoglobin, lower platelet count, PTPN11 mutation status, and male gender have all been previously associated with a poor prognosis.11,14,28,29 The only characteristic in univariate Cox analyses to reach statistical significance for OS on the AAML0122 study was age >2 years at diagnosis (HR 3.11, CI 1.34-7.21, P = .008) (supplemental Table 3). In a multivariate Cox analysis, SETBP1 mutation status remained an independent risk factor for poor prognosis (HR 2.54, CI 1.12-5.78, P = .026) after adjusting for PTPN11 mutation status, age, WBC, and platelets at diagnosis. Although HSCT conditioning received on the AAML0122 trial was not uniform, there was no statistically significant difference in the number of patients with a SETBP1 mutation (9/17, 53%) who received a protocol-compliant HSCT vs those without a SETBP1 mutation (23/39, 59%) (P = .675). When a protocol-compliant HSCT was included as a time-dependent variable in the multivariate analysis, SETBP1 remained an independent risk factor for poor prognosis (Table 1).
SETBP1 mutations occur in hematopoietic progenitor cells
To determine if the SETBP1 alteration occurs in a putative hematopoietic stem cell, 2 individual patient samples were sorted into early hematopoietic stem cells, multipotent progenitors, common myeloid progenitors, and common myeloid progenitors (supplemental Figure 3). Individual cells from a single compartment were then pooled together and DNA was extracted from pooled cells and analyzed by both Sanger methods and ddPCR. Using a diagnostic sample from 1 patient, mutant SETBP1 was detected in all 4 hematopoietic progenitor compartments using ddPCR with as little as 1 ng of input DNA (supplemental Figure 4). However, Sanger sequencing revealed only wild-type SETBP1. The same patient at relapse had an allelic fraction of SETBP1 of 50%, at which point sorting revealed clonal SETBP1 mutations in all 4 compartments using both Sanger and ddPCR methods. These data further support our hypothesis that SETBP1 mutations occur in early cancer-initiating cells and are highly resistant to intensive, albeit conventional, chemotherapy.
Discussion
JMML is an aggressive myeloproliferative neoplasm of childhood. One hallmark of JMML is aberrant signaling through the Ras pathway caused by mutations in NF1, NRAS, KRAS, PTPN11, and CBL. The current standard of care involves HSCT, but relapse is the most common reason for treatment failure. Robust predictors of response in this disease are lacking, yet they are needed to justify exposing these young patients to intensive and/or additional novel treatment strategies.
Germline mutations in SETBP1 cause Schinzel-Giedion syndrome and were first described by Hoischen et al.30 This disorder is characterized by severe mental retardation, short stature, predisposition to neuroepithelial malignancies, and death in the first decade of life. In 2013, Piazza et al identified heterozygous somatic mutations in a highly conserved 13-codon hotspot in the SKI domain of SETBP1 in patients with atypical chronic myelogenous leukemia.31 Interestingly, the identical c.2602G>A (p.D868N) and c.2608G>A (p.G870S) mutations that were previously described in Schinzel-Giedion syndrome as germline events were somatically acquired in atypical chronic myelogenous leukemia.
SET-related proteins have been previously reported to inhibit the tumor suppressor, protein phosphatase 2A (PP2A).32 In particular, SETBP1 is a 170-kDa nuclear protein that binds to SET and participates in the SET-PP2A complex. Previous work has demonstrated that the PP2A complex comprises a catalytic subunit (PP2Ac), scaffolding A subunit (PR65), and various B subunits.33 PP2A can both positively and negatively regulate the Ras–mitogen-activated protein kinase pathway via dephosphorylation of several individual substrates.34 For example, dephosphorylation of the kinase suppressor of RAS (KRS1) leads to localization of this scaffolding protein to the nucleus and activation of the Ras–mitogen-activated protein kinase pathway,35 whereas dephosphorylation of mitogen-activated protein kinase kinase and extracellular signal-regulated kinase leads to inhibition of the pathway.34 The critical interplay between the PP2A complex and the Ras pathway could explain the presence of SETBP1 mutations (which may inhibit PP2A) in the pathogenesis of JMML, which is largely considered a Ras-driven disease. Mutations in SETBP1 are predicted to be activating, and Cristobal et al reported that in a multivariate analysis, acute myeloid leukemia patients with overexpression of SETBP1 at diagnosis had a significantly shorter OS.32 Mutations in SETBP1 were previously shown to occur as secondary events in several myeloid malignancies and carry a poor prognosis.15,17,31,36,37
ddPCR proved capable of reliably identifying rare mutations with a lower limit of detection of 1 copy per well. Using this technology, we were able to identify patients harboring SETBP1 mutations at diagnosis with extremely low allelic fractions who had a worse prognosis compared with wild-type patients treated on the same clinical trial. Although EFS was statistically worse for patients with SETBP1 mutations compared with those without, OS was not statistically significant but did still carry an HR of 1.69 (CI 0.70-3.73). The most likely explanation for the discrepancy between EFS and OS is that a small sample size limited the power to detect a statistically significant difference. However, another potential explanation is that SETBP1 mutations did not impact OS because a significantly higher proportion of patients with SETBP1 mutations experienced early progressive disease followed by HSCT than patients without a mutation. AAML0122 allowed patients with JMML and progressive disease to proceed to transplant if there was an available donor. In addition, these patients may have gone on to receive a second transplant, as is commonly performed for patients who relapse after their first transplant.38 However, data on second transplants were not available on the AAML0122 study.
Deep sequencing results support that canonical mutations at diagnosis are clonal at presentation, whereas SETBP1 mutations can be either clonal or subclonal. Deep sequencing also failed to detect any of the subclonal mutations in SETBP1 that were identified by ddPCR. However, ddPCR permits detection of subclonal events because of the amplification of DNA within each of the 20 000 droplets per well, thereby partitioning rare mutant molecules into their own droplet before amplification. This allows for significantly reduced competition effects from wild-type molecules during amplification, allowing the mutant molecule to amplify.
Previous exome sequencing revealed that 7/92 (7.6%) patients with JMML harbored SETBP1 mutations.15 However, ddPCR in our cohort was able to detect rare SETBP1 mutations in 17/56 (30.3%) of patients with JMML that resulted in inferior outcomes. The presence of these subclonal mutations thus represents a biomarker for poor prognosis that will allow for improved risk stratification in patients with JMML and also justifies additional experiments to identify how somatically acquired SETBP1 alterations in hematopoietic cells cause such resistant phenotypes.
In summary, we have demonstrated that subclonal mutations in SETBP1 at diagnosis independently confer a dismal prognosis in JMML. In addition, these mutations are present in early progenitor cells and are resistant to currently available therapies. Understanding the mechanisms underpinning SETBP1-mediated resistance and relapse and identifying therapeutic vulnerabilities of these mutant proteins will be crucial to improve outcomes for patients with myeloid malignancies who possess SETBP1 mutations at diagnosis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for participating in the Children’s Oncology Group trial AAML0122, without which this research would not be possible. The authors also thank Scott Olsen from the Hartwell Center at St. Jude Children’s Research Hospital for his assistance with deep sequencing.
This work was supported by the St. Baldrick’s Foundation (E.S. and B.S.B.); the Leukemia and Lymphoma Society (grant number 6059-09) (M.L.L.); the American Society of Hematology (B.S.B.); the National Institutes of Health, National Cancer Institute grants T32 CA128583 (E.S.), R01CA173085 (B.S.B. and M.L.L.), R01 CA095621 (P.D.E.); the National Cancer Institute Cancer Center Support grant 5P30CA082103; the Frank A. Campini Foundation (E.S. and M.L.L.); Hyundai Hope on Wheels (M.L.L.); the Team Conner Foundation (M.L.L.); and a University of California San Francisco Dean's Commitment to the Center for Advanced Technologies facility.
Authorship
Contribution: E.S. and C.B.T designed the ddPCR experiments and analyzed the data; C.B.T. designed the ddPCR assays; E.S. performed the ddPCR and analyzed the data; E.S., K.B.Y., and L.C.G. performed Sanger sequencing; J.H. and A.A. carried out the droplet sorting; L.C.G., J.A., and B.S.B. optimized the progenitor sorting; A.N.T.-W., E.D.C., and Y.-D.W. analyzed the data; R.B.G. and T.A.A. performed the statistical analyses; K.B. assisted with sample processing; Y.L.L. and P.D.E. provided patient samples and clinical data; E.S. and M.L.L. wrote the manuscript; C.B.T., E.D.C., L.C.G., J.H., Y.L.L., R.B.G., T.A.A., and M.L.L. edited the manuscript; M.L.L. supervised the experiments; and all authors reviewed the literature, contributed to specific sections, and reviewed the final version of the manuscript.
Conflict-of-interest disclosure: C.B.T. was an employee of Bio-Rad Laboratories when the experiments were carried out. All other authors declare no competing financial interests.
The current affiliation for C.B.T. is Nanostring Technologies, Seattle, WA.
Correspondence: Elliot Stieglitz, Department of Pediatrics, Benioff Children's Hospital, University of California, San Francisco, 513 Parnassus Ave, HSE 302, San Francisco, CA 94143; e-mail: elliot.stieglitz@ucsf.edu; and Mignon Loh, Department of Pediatrics, Benioff Children's Hospital, University of California, San Francisco, 513 Parnassus Ave, HSE 302, San Francisco, CA 94143; e-mail: mignon.loh@ucsf.edu.