Key Points
We report a cell-based pharmacologic screening strategy to identify new therapeutic targets in mutant NRAS transformed leukemia cells.
The screen and mechanistic analysis identified a previously unknown synergy between germinal center kinase and ACK1/AKT in mutant NRAS transformed cells.
Abstract
Oncogenic forms of NRAS are frequently associated with hematologic malignancies and other cancers, making them important therapeutic targets. Inhibition of individual downstream effector molecules (eg, RAF kinase) have been complicated by the rapid development of resistance or activation of bypass pathways. For the purpose of identifying novel targets in NRAS-transformed cells, we performed a chemical screen using mutant NRAS transformed Ba/F3 cells to identify compounds with selective cytotoxicity. One of the compounds identified, GNF-7, potently and selectively inhibited NRAS-dependent cells in preclinical models of acute myelogenous leukemia and acute lymphoblastic leukemia. Mechanistic analysis revealed that its effects were mediated in part through combined inhibition of ACK1/AKT and of mitogen-activated protein kinase kinase kinase kinase 2 (germinal center kinase). Similar to genetic synthetic lethal approaches, these results suggest that small molecule screens can be used to identity novel therapeutic targets in cells addicted to RAS oncogenes.
Introduction
The RAS family (H-, K-, and NRAS) of guanosine triphosphate–binding proteins is a frequent target of oncogenic mutation in human cancers. RAS mutations, especially in NRAS, are common in acute myelogenous leukemia (AML), identified in >10% of patients. The mutations lead to constitutive RAS activation and subsequent activation of multiple signaling pathways, including mitogen-activated protein kinase, PI3K-AKT, and others.1 Many efforts to directly target RAS itself or posttranslational modifications of RAS with small molecules have been unsuccessful.1,2 Current efforts to develop covalent binders of G12C KRAS and allosteric inhibitors are still in early stages of development.3 Given this challenge, efforts have been directed at targeting canonical downstream effectors of RAS, such as RAF, mitogen-activated protein kinase kinase (MEK), and others. However, efficacy has been limited by the complexity of RAS signaling, including redundancy and activation of compensatory pathways.4 For example, treatment with RAF inhibitors increase proliferation of RAS-transformed cells by paradoxically increasing activation of MEK. Similarly, in a recent clinical trial with the MEK inhibitor, selumetinib (AZD6244), none of 3 patients with AML with NRAS mutation responded to the therapy in this early study.5 Large-scale RNA interference screens have identified TBK1,6 STK33,7 and GATA2,8 as synthetically lethal genes9 for KRAS mutations. However, thus far, generally these results have not translated into clinical successes.10,11 Recognizing the redundancy, lineage specificity, and complexity of RAS signaling, we hypothesized that small molecule chemical screens could complement genetic screens by potentially inhibiting multiple signaling pathways simultaneously.
Here, we used a cell-based screen to identify a multitargeted kinase inhibitor, N-(4-methyl-3-(1-methyl-7-(6-methylpyridin-3-ylamino)-2-oxo-1,2-dihydropyrimido[4,5-d]pyrimidin-3(4H)-yl)phenyl)-3-(trifluoromethyl)benzamide (GNF-7),12 as an inhibitor of RAS signaling in preclinical leukemia models. By utilizing this as a tool compound, mechanistic studies suggest that inhibition of AKT/ACK1 phosphorylation and germinal center kinase (GCK) (genome nomenclature: mitogen-activated protein kinase kinase kinase kinase 2 [MAP4K2]) are relevant targets of GNF-7 in this context.
Methods
Chemical screen
Ba/F3-NRAS-G12D cells were plated in 384 well plates (2 × 103 cells/well) in a volume of 50 μL of media in the presence or absence of recombinant murine IL-3 (mIL-3). Dimethylsulfoxide (DMSO) or a compound (0.3 μM final concentration) from a kinase inhibitor library (mainly consisting of 450 multitargeted kinase inhibitors) was added. Cells were incubated for approximately 2 days and cell growth was measured by the addition of CellTiter Glo.
In situ kinase selectivity profiling (KiNativ profiling)
OCI-AML3 cells were treated with DMSO or 0.3 μM or 1 μM of GNF-7 or 1 μM of YHJ0557 for 2 hours and cell lysate was made. Then biotinylated acyl phosphates of adenosine triphosphate (ATP) and adenosine 5′-diphosphate (5 μM) that react with protein kinases on conserved lysine residues in the ATP-binding pocket were added. Samples were analyzed by liquid chromatography-mass spectrometry and liquid chromatography-tandem mass spectrometry on a linear ion trap mass spectrometer. Characteristic fragment ions for each kinase peptide-probe conjugate were used to extract signals and a comparison of inhibitor-treated to DMSO-treated lysates enabled determination of the percentage of inhibition.13
In vitro kinase assay
Kinase/substrate pairs and required cofactors of a panel of kinases14 were prepared in reaction buffer.14 Compounds were first added to the reaction, and ATP and 33P ATP were added to a final concentration of 10 μM. Two hours later, the reactions were spotted onto P81 ion exchange filter paper. After subtraction of background derived from control reactions, kinase activity data were expressed as the percent residual kinase activity in test samples as compared with vehicle (DMSO) reactions.
Gene expression profiling
OCI-AML3 cells and KO52 cells were treated with 1.0 μM of GNF-7 for 8 hours in biologic triplicate and HuGene 1.0 ST chip was used for global transcriptional profiling. The data were normalized using robust multi-array average and Gene Set Enrichment Analysis was performed by comparing the samples using gene signatures of C2 curated gene sets in MSigDB (Molecular Signatures Database) version 3 by GenePattern.
In vivo xenotranplantation study
The 2.5 × 106 MOLT-3 luc+ cells were administered by tail-vein injection to 6-week-old NSG mice. Cohorts of mice were treated with oral administration of a vehicle, (7.5 mg/kg GNF-7 per day or 15 mg/kg GNF-7 per day). Tumor burden was assessed by serial bioluminescence imaging, and statistical significance was determined by the Student t test.
AML patient samples
Cryopreserved bone marrow samples from 5 AML patients identified as harboring mutated NRAS, wild-type (WT) KRAS, and WT FLT3 were obtained from the tissue bank of the Dana Farber Cancer Institute.
Additional methods can be found in the supplemental Methods, available on the Blood Web site.
Results
Identification of GNF-7 as an inhibitor of leukemia cells with mutant RAS
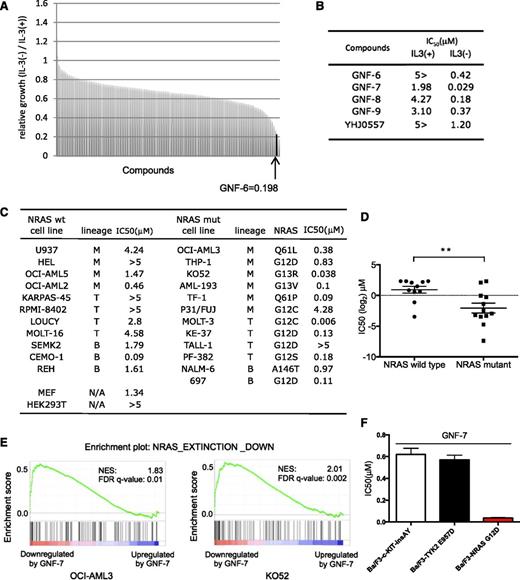
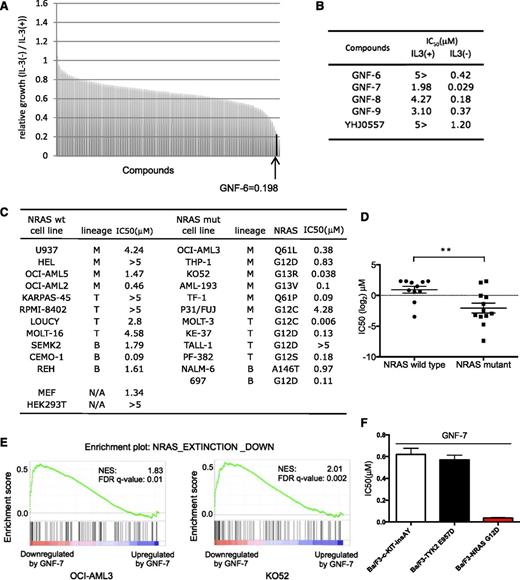
Murine Ba/F3 cells are not dependent on N-RAS when cultured with IL-3,15 but can be readily transformed by NRAS-G12D (Ba/F3-NRAS-G12D cells), resulting in IL-3-independence. Growth of Ba/F3-NRAS-G12D cells was suppressed >95% by short hairpin RNA (shRNA) for NRAS, which could be rescued upon addition of IL-3 (supplemental Figure 1A). A chemical screen was performed using a diverse library of 450 multitargeted kinases for compounds that were selectively cytotoxic to Ba/F3-NRAS-G12D cells. This screen resulted in the identification of GNF-6 (Figure 1A-B; supplemental Figure 1B), which was originally developed as a kinase inhibitor capable of inhibiting BCR-ABL.12 We further explored 10 analogs of GNF-6, including GNF-7, -8, and -9,12 and YHJ0557 (supplemental Figure 1B). GNF-7 displayed the highest antiproliferative potency against Ba/F3-NRAS-G12D cells (Figure 1B). YHJ0557, a close structural analog of GNF-7 showed 40-fold less potency against Ba/F3-NRAS-G12D cells relative to GNF-7, which was therefore selected as a “negative” control for subsequent target identification efforts. GNF-7 is 65-fold more potent against Ba/F3-NRAS-G12D cells cultured in the absence of IL-3 (IC50 = 0.029 μM) vs in its presence (IC50=1.98 μM), demonstrating its selective differential cytotoxicity (Figure 1B; supplemental Figure 1C).
Identification of GNF-7 as an inhibitor of leukemia cells with mutant RAS. Using Ba/F3-NRAS-G12D cells, a chemical screen was performed in the presence or absence of IL-3. A unique and diverse library of 450 multitargeted kinase inhibitors was screened. (A) Relative growth of Ba/F3-NRAS-G12D cells in the screen was cultured in the absence of IL-3 vs the presence of IL-3. GNF-6 was identified as a hit in the screen. (B) IC50s of GNF-6 and its analogs tested against Ba/F3-NRAS-G12D cells in the absence of IL-3 vs the presence of IL-3. (C) IC50 of GNF-7 tested against a panel of human leukemia cell lines expressing WT or mutated NRAS gene. Cell lines expressing mutated FLT-3 were excluded. (D) A comparison was made of IC50 for GNF-7 as it was tested against human leukemia cell lines with or without NRAS mutation. Data are represented as mean ± standard error of the mean. (E) Enrichment plot of a gene set NRAS EXTINCTION DOWN (as described in “Results”) and its size, NES (normalized enrichment score), and FDR (false discovery rate) q-value for OCI-AML3 cells and KO52 cells. (F) IC50 of GNF-7 was tested against Ba/F3-TYK2 E957D, Ba/F3-c-KIT insAY, and Ba/F3-NRAS-G12D cells. wt, wild type. **P < .01.
Identification of GNF-7 as an inhibitor of leukemia cells with mutant RAS. Using Ba/F3-NRAS-G12D cells, a chemical screen was performed in the presence or absence of IL-3. A unique and diverse library of 450 multitargeted kinase inhibitors was screened. (A) Relative growth of Ba/F3-NRAS-G12D cells in the screen was cultured in the absence of IL-3 vs the presence of IL-3. GNF-6 was identified as a hit in the screen. (B) IC50s of GNF-6 and its analogs tested against Ba/F3-NRAS-G12D cells in the absence of IL-3 vs the presence of IL-3. (C) IC50 of GNF-7 tested against a panel of human leukemia cell lines expressing WT or mutated NRAS gene. Cell lines expressing mutated FLT-3 were excluded. (D) A comparison was made of IC50 for GNF-7 as it was tested against human leukemia cell lines with or without NRAS mutation. Data are represented as mean ± standard error of the mean. (E) Enrichment plot of a gene set NRAS EXTINCTION DOWN (as described in “Results”) and its size, NES (normalized enrichment score), and FDR (false discovery rate) q-value for OCI-AML3 cells and KO52 cells. (F) IC50 of GNF-7 was tested against Ba/F3-TYK2 E957D, Ba/F3-c-KIT insAY, and Ba/F3-NRAS-G12D cells. wt, wild type. **P < .01.
Next, the potency of GNF-7 was compared with those of inhibitors targeting known downstream RAS effectors, including inhibitors of MEK and PI3K/AKT/mammalian target of rapamycin (mTOR). GNF-7 most potently suppressed growth of Ba/F3-NRAS-G12D cells among the inhibitors tested (supplemental Figure 1D). When tested on a panel of established human leukemia cell lines, GNF-7 was more active against leukemia cell lines expressing mutant NRAS compared with those expressing WT NRAS (Figure 1C-D). Cell lines expressing mutated FLT3 were excluded because GNF-7 directly inhibits these kinases (supplemental Figure 1E).
Mutant NRAS is known to impact a large number of genes and pathways.1 We performed global transcriptional profiling using 2 NRAS mutant AML cell lines (OCI-AML3 and K052) and compared genes downregulated by GNF-7 to genes reported to be downregulated after genetic ablation of NRAS.16 By Gene Set Enrichment Analysis, the 99 genes that were most downregulated in the reported mouse model (P < .005), “NRAS EXTINCTION DOWN” (supplemental Table 1) were found to be enriched among genes downregulated by GNF-7 (Figure 1E).
To further explore the selectivity of GNF-7, we investigated antiproliferative activities of GNF-7 for Ba/F3 cells transformed by oncogenes other than RAS. Ba/F3 cells transformed by oncogenic c-KIT-502-503 insAY17 or TYK2-E957D18 showed IC50 of 0.62 μM and 0.57 μM, respectively, which are around 20 times less effective compared with mutant NRAS (Figure 1F). Also a previous study reported that the IC50 of GNF-7 against NPM-ALK or TPR-c-MET- transformed Ba/F3 cells is 1.47 and 3.40 μM, respectively.12
These results suggest that treatment with GNF-7 leads to suppression of an NRAS-regulated gene network/signaling pathway, which argues that GNF-7 may inhibit proliferation by suppressing NRAS signaling, and not via a nonspecific toxic effect.
Effect of GNF-7 on the RAS signaling pathways and identification of AKT signaling as a functional target
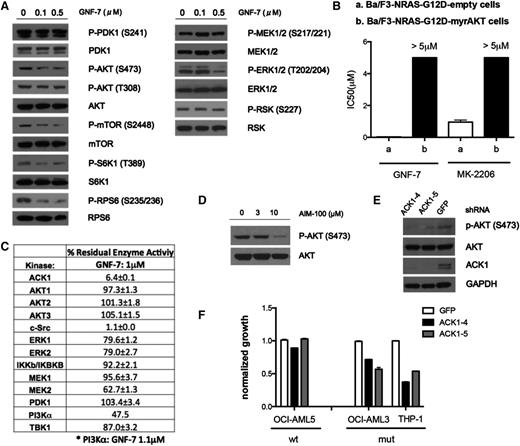
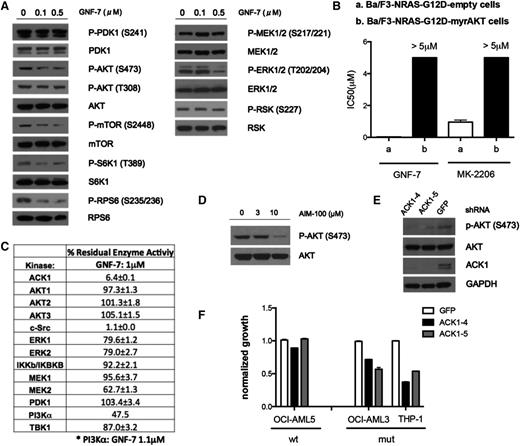
To elucidate the mechanism underlying the inhibitory effect of GNF-7 against cancer cells with the NRAS G12D mutation, NRAS–guanosine triphosphate levels were examined before and after GNF-7 exposure. As anticipated, GNF-7 did not affect NRAS–guanosine triphosphate levels (supplemental Figure 2A). Next, the effect of GNF-7 on major components of RAS signaling was assessed biochemically. Interestingly, treatment of Ba/F3-NRAS-G12D cells with concentrations of GNF-7 up to 0.1 μM (IC50 = 0.029 μM) failed to reduce phospho-MEK or phospho-ERK (Figure 2A). In contrast, we observed suppression of the AKT/mTOR signaling pathway (Figure 2A). Similar results were observed in multiple human leukemia cell lines (supplemental Figure 2B).
Effect of GNF-7 on the RAS signaling pathways, and identification of AKT signaling as a functional target. (A) Ba/F3-NRAS-G12D cells were treated with various concentrations of GNF-7 for 2 hours. Changes in phosphorylation of major kinases mediating the RAS signaling pathway were investigated by immunoblot analysis. (B) IC50 of GNF-7 tested against Ba/F3-NRAS-G12D cells and Ba/F3-NRAS-myrAKT cells. MK-2206, an allosteric AKT inhibitor, was also tested against Ba/F3-NRAS-G12D cells. Data are represented as mean ± standard error of the mean (SEM). (C) In vitro kinase assays for kinases related to the RAS signaling pathway were performed as described in “Methods”. Residual kinase activity after treatment with 1.0 μM of GNF-7 of each kinase is shown. Data are represented as mean ± SEM. (D) Ba/F3-NRAS-G12D cells were treated with indicated concentrations of AIM-100 and change of phosphorylation of AKT was immunoblotted. (E) Validation of the KD efficiency of ACK1 by shRNAs and its effect on phospho-AKT. (F) Proliferation of AML cell lines characterized by KD of ACK1 by shRNA. Data are represented as mean ± SEM.
Effect of GNF-7 on the RAS signaling pathways, and identification of AKT signaling as a functional target. (A) Ba/F3-NRAS-G12D cells were treated with various concentrations of GNF-7 for 2 hours. Changes in phosphorylation of major kinases mediating the RAS signaling pathway were investigated by immunoblot analysis. (B) IC50 of GNF-7 tested against Ba/F3-NRAS-G12D cells and Ba/F3-NRAS-myrAKT cells. MK-2206, an allosteric AKT inhibitor, was also tested against Ba/F3-NRAS-G12D cells. Data are represented as mean ± standard error of the mean (SEM). (C) In vitro kinase assays for kinases related to the RAS signaling pathway were performed as described in “Methods”. Residual kinase activity after treatment with 1.0 μM of GNF-7 of each kinase is shown. Data are represented as mean ± SEM. (D) Ba/F3-NRAS-G12D cells were treated with indicated concentrations of AIM-100 and change of phosphorylation of AKT was immunoblotted. (E) Validation of the KD efficiency of ACK1 by shRNAs and its effect on phospho-AKT. (F) Proliferation of AML cell lines characterized by KD of ACK1 by shRNA. Data are represented as mean ± SEM.
The effects of GNF-7 on the MEK-ERK pathway were further evaluated by generating a Ba/F3 cell line transformed with a constitutively active MEK allele (MEK-DD)19 (supplemental Figure 2C). Treatment of these cells with an ATP noncompetitive MEK1/2 inhibitor, CI-1040, suppressed growth as expected, however, GNF-7 had no effect (supplemental Figure 2D). This result suggests that GNF-7 does not function by inhibition of MEK or its downstream pathways. This notion is further supported by the finding that growth of Ba/F3-NRAS-G12D cells in the presence of GNF-7 could be further suppressed by the addition of CI-1040 (supplemental Figure 2E).
To better understand the role of AKT, the effects of GNF-7 were compared with those of a known, potent allosteric AKT inhibitor, MK-2206.20 GNF-7 inhibited the proliferation of Ba/F3-NRAS-G12D cells more potently than MK-2206 (Figure 2B). Interestingly, overexpression of a constitutively active version of AKT, myr-AKT, in Ba/F3-NRAS-G12D cells (supplemental Figure 2C) conferred strong resistance to not only MK-2206, but also to GNF-7 (Figure 2B), suggesting that suppression of phospho-AKT is functionally necessary for growth suppression by GNF-7.
To determine if GNF-7 directly inhibits AKT, we performed an in vitro kinase assay with recombinant protein kinases in the PI3K-AKT and MEK/extracellular signal-regulated kinase signaling pathways. None of the kinases studied in the PI3K/AKT pathway (PI3Kα, PDK1, AKT1-3) were inhibited by GNF-7 (1 μM; Figure 2C). Although c-Src was strongly inhibited, treatment of Ba/F3-NRAS G12D cells with the selective c-Src inhibitor, saracatinib (AZD0530), failed to suppress growth (data not shown), suggesting that c-Src is not a functional target for suppression of NRAS signaling. Interestingly, activation of a PI3K-independent kinase, ACK1 (activated Cdc42-associated tyrosine kinase 1), which has been reported to activate AKT,21 was strongly suppressed (Figure 2C). Indeed, the reported ACK1 inhibitor, AIM-100, which does not directly inhibit AKT1-3, PI3Kα, or PI3Kβ, suppressed phospho-AKT (S473) in Ba/F3-NRAS-G12D cells at a concentration of 10 μM, similar to what has been previously reported22 (Figure 2D) and growth was suppressed by 40% (data not shown). In addition, the growth of NRAS-mutant cell lines (OCI-AML3 and THP-1) was more suppressed by AIM-100 than was that of an NRAS–wild cell line (OCI-AML5) (supplemental Figure 2F). Knockdown (KD) of ACK1 using shRNAs suppressed phospho-AKT (Figure 2E) and growth of the same cell lines (Figure 2F), again suggesting that ACK1 is a relevant target of GNF-7 in these cells.
Taken together, it is likely that suppression of the AKT pathway, at least in part through inhibition of ACK1, but not the MEK pathway, is important for the activity of GNF-7 on NRAS–mutant cell lines. However, direct AKT inhibition only partially recapitulated the antiproliferative effects of GNF-7 on mutant NRAS-expressing cells, suggesting that GNF-7 was likely to possess additional functional targets.
Identification of GCK as a functionally relevant target of GNF-7
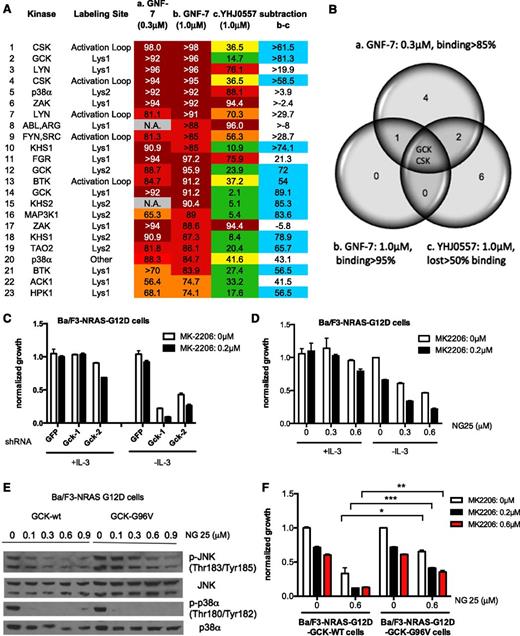
To expand the search for additional functional targets of GNF-7, we performed in situ kinase profiling of GNF-7 and the less potent analog YHJ0557 using the KiNativ method13 (supplemental Tables 2-4). The top 15 kinases whose probe labeling was inhibited by treatment of cells with 1 μM GNF-7 are shown in Figure 3A. Importantly, ACK1 (74.7%) was among the top hits.
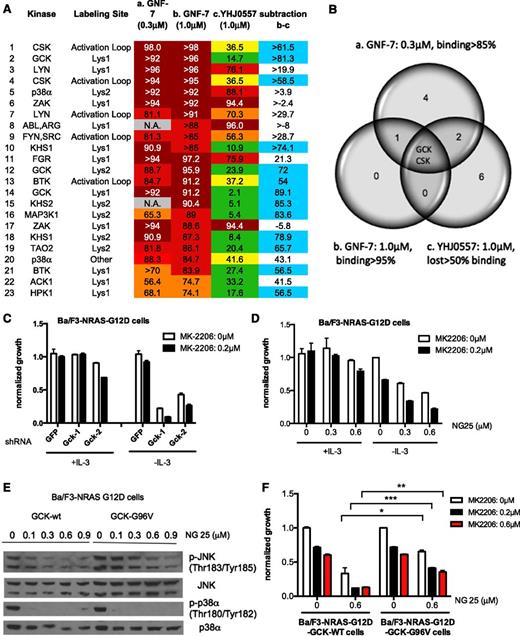
Identification of GCK as a functionally relevant target of GNF-7. OCI-AML3 cells (NRAS mutant) were treated with DMSO, 0.3 μM or 1 μM of GNF-7, or 1.0 μM of YHJ0557 for 2 hours and in situ kinase profiling by Kinativ method was performed as described in “Methods”. (A) Shown are the leading targets as revealed by the in situ kinase selectivity profiling. The difference between GNF-7 (1.0 μM) and inactive YHJ0557 in terms of inhibitory activity displayed against each target kinase was calculated by subtraction. (B) Venn diagram showing 3 groups of target kinases. a. Kinases inhibited more than 85% by 0.3 μM of GNF-7. b. Kinases inhibited more than 95% by 1.0 μM of GNF-7. c. Kinases that lost inhibitory activity more than 50% compared with group b after treatment with 1.0 μM of YHJ0557. (C) Proliferation of MK-2206-treated Ba/F3-NRAS-G12D cells in the absence or presence of IL-3 after KD of Gck by shRNA. Data are represented as mean ± SEM. (D) Proliferation of Ba/F3-NRAS-G12D cells treated with MK-2206 and the MAP4K2 inhibitor, NG25, in the absence or presence of IL-3. Data are represented as mean ± standard error of the mean. (E) Effect of overexpression of GCK-G96V in Ba/F3-NRAS-G12D on phosphor JNK, which is downstream of GCK, and phospho p38 as assessed by immunoblotting. (F) Rescue of growth suppression of MK-2206- and NG25-treated Ba/F3-NRAS-G12D cells by overexpression of GCK-G96V. Data are represented as mean ± SEM. Lys1, conserved Lysine 1; Lys2, conserved Lysine 2; activation loop, activation loop near DFG (Asp-Phe-Gly) motif peptide sequences detected are shown in supplemental Figure 3A. *P < .05; **P < .01; ***P < .001.
Identification of GCK as a functionally relevant target of GNF-7. OCI-AML3 cells (NRAS mutant) were treated with DMSO, 0.3 μM or 1 μM of GNF-7, or 1.0 μM of YHJ0557 for 2 hours and in situ kinase profiling by Kinativ method was performed as described in “Methods”. (A) Shown are the leading targets as revealed by the in situ kinase selectivity profiling. The difference between GNF-7 (1.0 μM) and inactive YHJ0557 in terms of inhibitory activity displayed against each target kinase was calculated by subtraction. (B) Venn diagram showing 3 groups of target kinases. a. Kinases inhibited more than 85% by 0.3 μM of GNF-7. b. Kinases inhibited more than 95% by 1.0 μM of GNF-7. c. Kinases that lost inhibitory activity more than 50% compared with group b after treatment with 1.0 μM of YHJ0557. (C) Proliferation of MK-2206-treated Ba/F3-NRAS-G12D cells in the absence or presence of IL-3 after KD of Gck by shRNA. Data are represented as mean ± SEM. (D) Proliferation of Ba/F3-NRAS-G12D cells treated with MK-2206 and the MAP4K2 inhibitor, NG25, in the absence or presence of IL-3. Data are represented as mean ± standard error of the mean. (E) Effect of overexpression of GCK-G96V in Ba/F3-NRAS-G12D on phosphor JNK, which is downstream of GCK, and phospho p38 as assessed by immunoblotting. (F) Rescue of growth suppression of MK-2206- and NG25-treated Ba/F3-NRAS-G12D cells by overexpression of GCK-G96V. Data are represented as mean ± SEM. Lys1, conserved Lysine 1; Lys2, conserved Lysine 2; activation loop, activation loop near DFG (Asp-Phe-Gly) motif peptide sequences detected are shown in supplemental Figure 3A. *P < .05; **P < .01; ***P < .001.
To identify functionally critical targets, we compared the targets of GNF-7 to those of YHJ0557 (Figure 3A and supplemental Table 4). This analysis (Figure 3B) showed that 2 kinases (ie, GCK and CSK) were most significantly different in terms of affinity with GNF-7 and YHJ0557. Kinase activity of these kinases was suppressed with 1 μM of GNF-7 by 97.7% and 96.9%, respectively, in a biochemical kinase assay.
To examine the potential role of GCK and CSK as functional GNF-7 targets, these genes were knocked down in Ba/F3-NRAS-G12D cells cultured ± IL-3, using shRNA. The cell growth was suppressed after KD of GCK in the absence of IL-3, but not in its presence, and the addition of MK-2206 resulted in additional suppression of proliferation (Figure 3C, supplemental Figure 3B). In contrast, KD of CSK did not affect cell growth (supplemental Figure 3C).
These results were validated by testing an alternative inhibitor of GCK, NG25,23 which has been previously shown using in situ kinase profiling (KiNativ method) to have high affinity for GCK (86.3% at 1 μM). We confirmed that NG25 inhibited GCK using an in vitro kinase assay with an IC50 of 21.7 nM (supplemental Figure 3D). Treatment of Ba/F3-NRAS-G12D cells with a combination of MK-2206 and NG25 suppressed cell growth in the absence of IL-3, but not in its presence (Figure 3D). To exclude the possibility that this suppression was due to inhibition of kinases other than GCK, we introduced a mutation (GCK-G96V) in the ATP-binding pocket of GCK predicted to block binding of NG25,24 and overexpressed either GCK-G96V or WT GCK in Ba/F3-NRAS-G12D cells (supplemental Figure 3E-F). The in vitro IC50 of NG25 against WT GCK or G96V GCK overexpressed in 293T cells was 0.11 μM and >21.3 μM, respectively. To confirm this mutation was indeed resistant to NG25 in cells, we assessed the phosphorylation of JNK, which is one of the established downstream targets of GCK.25 Suppression of phospho-JNK by NG25 was significantly rescued by overexpression of GCK-G96V, but not WT GCK; in contrast, suppression of phospho-p38, another target of NG25, was not rescued (Figure 3E). This suggests that this mutation at least partially blocks binding of NG25 to GCK. Next, Ba/F3-NRAS-G12D-GCK WT or G96V cells were treated with a combination of MK-2206 and NG25. Growth suppression was significantly rescued by overexpression of GCK-G96V compared with that of WT GCK, suggesting that NG25 suppresses the growth of Ba/F3-NRAS-G12D cells by suppression of GCK, at least in part (Figure 3F). Next, Ba/F3-NRAS-G12D cells were treated with MK-2206 and an inhibitor of p38 (VX-702) or TAK1 (5Z-7-Oxozeaenol), which are other known targets of NG25. The former resulted in minimal suppression of the cell growth and the latter suppressed the growth both in the presence and absence of IL-3 (supplemental Figure S3G-I), suggesting that p38 and TAK1 are not important targets of GNF-7 in this context. Overall, both the shRNA and pharmacologic studies support the notion that the combined inhibition of GCK and ACK1/AKT provides a means to selectively inhibit the proliferation of mutant NRAS-transformed cells.
Comparing the function and targets of GNF-7 in additional leukemia cell lines
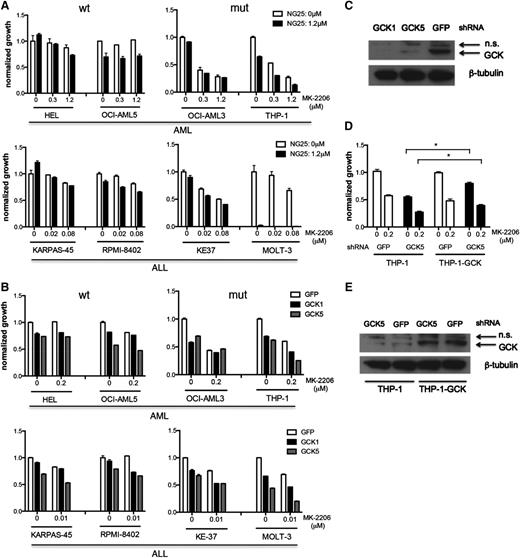
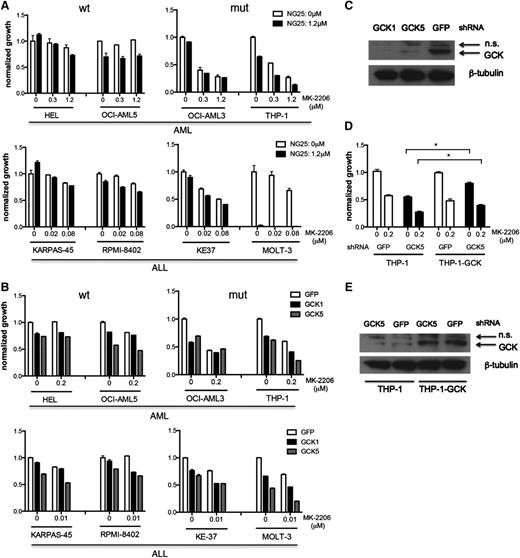
Four myeloid leukemia cell lines and 4 lymphoid leukemia cell lines expressing WT or mutant NRAS, as indicated in Figure 4A, were treated with NG25 and MK-2206 in an effort to simultaneously inhibit GCK and ACK/AKT signaling. Cell lines expressing mutated NRAS were more sensitive to GNF-7, or the combination of NG25 and MK-2206, than cells expressing WT NRAS (Figure 1D, 4A). These differences could not be explained by differences in the level of expression of GCK (supplemental Figure 4A). Similar results were obtained when these same cell lines were treated with MK-2206, and GCK was knocked down by 2 different shRNAs (Figure 4B-C). To exclude the possibility that the observed effects were due to off-target effects, we expressed empty vector or WT GCK in THP-1 cells, and knocked down GCK using shRNA (GCK5) that targets the GCK 3′ untranslated region in the presence of MK-2206. We found that forced expression of GCK partially rescued the cells, suggesting that the suppression of growth by the shRNA (GCK5) is an on-target effect of this hairpin RNA (Figure 4D-E). Taken together, these results suggest that GCK and ACK1/AKT are functionally important targets of GNF-7 in human leukemia lines expressing mutated NRAS.
Comparing the function and targets of GNF-7 in additional leukemia cell lines. (A) Proliferation of WT and mutant NRAS-expressing AML or ALL (acute lymphoblastic leukemia) cell lines treated with MK-2206 and NG25. Data are represented as mean ± standard error of the mean (SEM). (B) Proliferation of AML or ALL cell lines treated with MK-2206 and characterized by KD of GCK by shRNA. Data are represented as mean ± SEM. (C) Validation of the KD efficiency of GCK by shRNAs used (as shown in B). (D) Proliferation of THP-1 cells and THP1-GCK-WT cells treated with MK-2206 and characterized by KD of GCK by shRNA. Data are represented as mean ± SEM. (E) Validation of the KD efficiency of GCK by shRNA (used in D) in THP-1 cells and THP1-GCK-WT cells. GFP, green fluorescent protein; mut, mutant; n.s., nonsignificant; wt, wild type. *P < .05.
Comparing the function and targets of GNF-7 in additional leukemia cell lines. (A) Proliferation of WT and mutant NRAS-expressing AML or ALL (acute lymphoblastic leukemia) cell lines treated with MK-2206 and NG25. Data are represented as mean ± standard error of the mean (SEM). (B) Proliferation of AML or ALL cell lines treated with MK-2206 and characterized by KD of GCK by shRNA. Data are represented as mean ± SEM. (C) Validation of the KD efficiency of GCK by shRNAs used (as shown in B). (D) Proliferation of THP-1 cells and THP1-GCK-WT cells treated with MK-2206 and characterized by KD of GCK by shRNA. Data are represented as mean ± SEM. (E) Validation of the KD efficiency of GCK by shRNA (used in D) in THP-1 cells and THP1-GCK-WT cells. GFP, green fluorescent protein; mut, mutant; n.s., nonsignificant; wt, wild type. *P < .05.
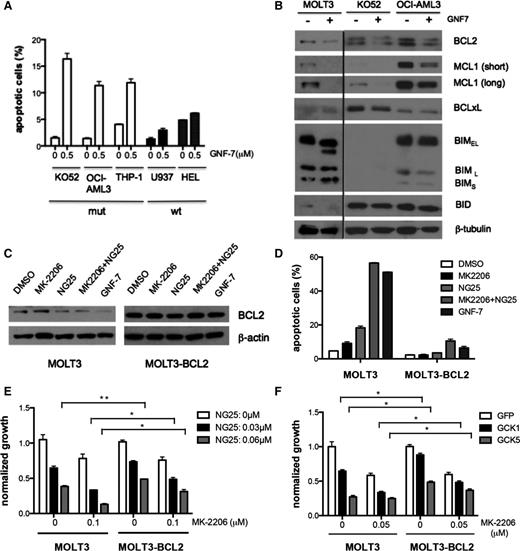
Effect of GNF-7 suppression of AKT and/or GCK on induction of apoptosis and cell cycle arrest
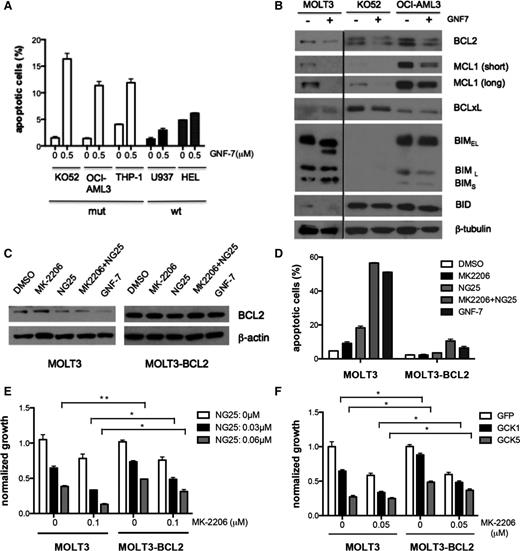
Next, the mechanism of growth suppression of NRAS-mutated cells by GNF-7 was explored. GNF-7 induced apoptosis in NRAS mutated cells (Figure 5A; supplemental Figure 5A). Importantly, apoptosis was partially reversible by the pan-caspase inhibitor Z-VAD-FMK (supplemental Figure 5B). Among proapoptois- and antiapoptosis-related proteins, BCL2 expression levels were found to be downregulated in mutant NRAS-expressing cell lines after treatment with GNF-7 (Figure 5B), and the effects of GNF-7 could be partially rescued by overexpression of BCL2 (supplemental Figure 5C-D). To explore suppression of which pathway is inducing suppression of BCL2, MOLT-3 cells were treated with MK2206 or NG25, or the combination. Suppression of BCL2 and apoptosis was strongly induced by NG25, and its effect on apoptosis and growth suppression was potentiated by MK-2206 treatment (Figure 5D-F). Overexpression of BCL2 effectively rescued cells from apoptosis and modestly enhanced cell growth. (Figure 5D-F). Next, GCK was knocked down by shRNAs in MOLT-3 or MOLT-3–BCL2 cells. GCK knockdown induced significant apoptosis and growth suppression, which were rescued by BCL2 (supplemental Figure 5E), suggesting that the suppressive effects by NG25 are mainly through GCK. GNF-7 also induced G1 arrest in NRAS–mutated cells as shown in supplemental Figure 5F. To determine if GCK inhibition was important, MOLT-3 cells were treated with MK2206, or NG25, or the combination. The arrest was mainly induced by NG25 (supplemental Figure 5G) and knockdown of GCK by shRNAs confirmed that inhibition of GCK was important (supplemental Figure 5H). These data show that suppression of growth of NRAS-mutated cells is due to both apoptosis and induction of cell cycle arrest, and that GCK is important in both these effects.
Effect of GNF-7 suppression of AKT and/or GCK on induction of apoptosis and cell cycle. (A) Indicated cell lines were treated with GNF-7 for 24 hours and percentage of annexin V-positive cells are shown. Data are represented as mean ± standard error of the mean (SEM). (B) Indicated cell lines were treated with 0.5 μM of GNF-7 (0.1 μM for MOLT3) for 24 hours and expression of proteins involved in apoptosis are shown. (C) Expression of BCL2 in MOLT3 cells and MOLT3-BCL2 cells 24 hours after treatment with DMSO or MK-2206 (0.2 μM) or NG25 (0.03 μM), or a combination of MK-2206 (0.2 μM) and NG25 (0.03 μM) or GNF-7 (0.1 μM). (D) Percentage of annexin V-positive cells after 24 hours in the same experiment (as shown in C). (E) Proliferation of MOLT3 cells and MOLT3-BCL2 cells with presence of MK-2206 and NG25. (F) Proliferation of MOLT3 cells and MOLT3-BCL2 cells characterized by KD of GCK by shRNA with presence of MK-2206. Data are represented as mean ± SEM. GFP, green fluorescent protein; mut, mutant; wt, wild type. *P < .05; **P < .01
Effect of GNF-7 suppression of AKT and/or GCK on induction of apoptosis and cell cycle. (A) Indicated cell lines were treated with GNF-7 for 24 hours and percentage of annexin V-positive cells are shown. Data are represented as mean ± standard error of the mean (SEM). (B) Indicated cell lines were treated with 0.5 μM of GNF-7 (0.1 μM for MOLT3) for 24 hours and expression of proteins involved in apoptosis are shown. (C) Expression of BCL2 in MOLT3 cells and MOLT3-BCL2 cells 24 hours after treatment with DMSO or MK-2206 (0.2 μM) or NG25 (0.03 μM), or a combination of MK-2206 (0.2 μM) and NG25 (0.03 μM) or GNF-7 (0.1 μM). (D) Percentage of annexin V-positive cells after 24 hours in the same experiment (as shown in C). (E) Proliferation of MOLT3 cells and MOLT3-BCL2 cells with presence of MK-2206 and NG25. (F) Proliferation of MOLT3 cells and MOLT3-BCL2 cells characterized by KD of GCK by shRNA with presence of MK-2206. Data are represented as mean ± SEM. GFP, green fluorescent protein; mut, mutant; wt, wild type. *P < .05; **P < .01
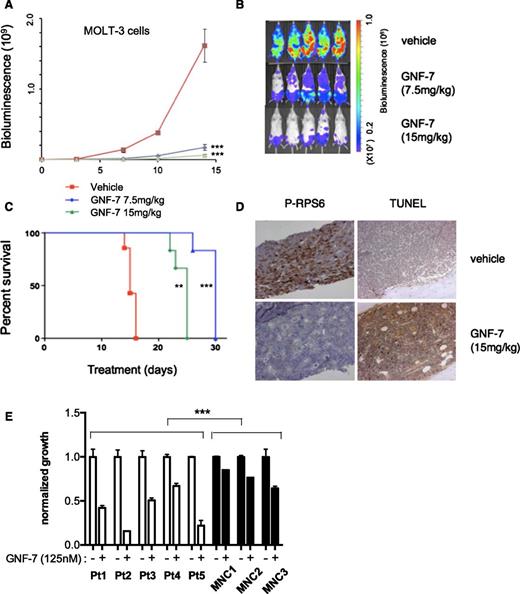
In vivo efficacy of GNF-7 in a xenotransplantation model and activity against primary AML patient samples
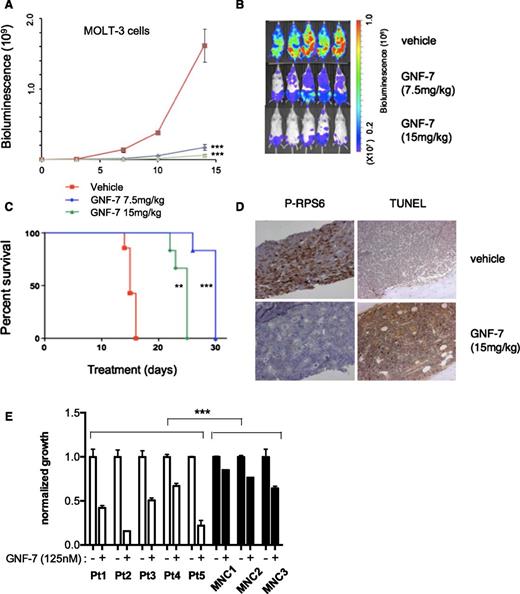
To evaluate the in vivo therapeutic efficacy of GNF-7, a xenotransplantation model in which bioluminescence of MOLT-3 cells was quantified as a measurement of disease burden. After injection of leukemia cells intravenously, mice were treated with GNF-7 (7.5 mg/kg daily or 15 mg/kg daily), or with the vehicle alone. Treatment with GNF-7 significantly decreased disease burden in mice and prolonged overall survival compared with vehicle controls (Figure 6A-C). Bone marrow cells from GNF-7–treated mice harvested on day 15 showed strong suppression of phospho-AKT and phospho-RPS6, suggesting that GNF-7 was effective in vivo (supplemental Figure 6A). Similarly, expression of phospho-RPS6 (by immunohistochemistry) was reduced by GNF-7 and there was an increase in apoptotic cells by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling staining (Figure 6D). GNF-7 treatment (15 mg/kg) did not result in appreciable body weight loss (supplemental Figure 6B) and did not alter gross microscopic morphology (supplemental Figure 6C) in major organs, suggesting the drug was well tolerated under these conditions without toxicity.
In vivo efficacy of GNF-7 in a xenotransplantation model and activity against primary AML patient samples. Bioluminescence was measured in untreated and GNF-7-treated NSG (NOD scid gamma) mice injected via tail-vein with human mutant NRAS-expressing MOLT-3-luc+ cells. (A) Bioluminescence values plotted for mice treated with vehicle or 7.5 mg/kg or 15 mg/kg of GNF-7 (once a day, per os). Data are represented as mean ± standard error of the mean. (B) Representative whole-body bioluminescence images of NSG mice treated with vehicle or 7.5 mg/kg or 15 mg/kg of GNF-7 on day10. (C) Survival curve (Kaplan-Meier). Treated groups show prolongation of overall survival compared with vehicle control. (D) On day 15, a femur of mice injected with MOLT-3-luc+ cells and treated with vehicle or 15 mg/kg of GNF-7 was harvested and sectioned. Bone marrow was stained by antiphospho RPS6 antibody and also terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining was performed. (E) Proliferation of primary AML patient samples expressing mutant NRAS and WT FLT3 and mononuclear cells of bone marrow from healthy donors treated with 0.125 μM of GNF-7 for 3 days. Bar represents 100 μm (D). MNC, mononuclear cells. *P < .05; **P < .01; ***P < .001.
In vivo efficacy of GNF-7 in a xenotransplantation model and activity against primary AML patient samples. Bioluminescence was measured in untreated and GNF-7-treated NSG (NOD scid gamma) mice injected via tail-vein with human mutant NRAS-expressing MOLT-3-luc+ cells. (A) Bioluminescence values plotted for mice treated with vehicle or 7.5 mg/kg or 15 mg/kg of GNF-7 (once a day, per os). Data are represented as mean ± standard error of the mean. (B) Representative whole-body bioluminescence images of NSG mice treated with vehicle or 7.5 mg/kg or 15 mg/kg of GNF-7 on day10. (C) Survival curve (Kaplan-Meier). Treated groups show prolongation of overall survival compared with vehicle control. (D) On day 15, a femur of mice injected with MOLT-3-luc+ cells and treated with vehicle or 15 mg/kg of GNF-7 was harvested and sectioned. Bone marrow was stained by antiphospho RPS6 antibody and also terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining was performed. (E) Proliferation of primary AML patient samples expressing mutant NRAS and WT FLT3 and mononuclear cells of bone marrow from healthy donors treated with 0.125 μM of GNF-7 for 3 days. Bar represents 100 μm (D). MNC, mononuclear cells. *P < .05; **P < .01; ***P < .001.
Finally, we tested the efficacy of GNF-7 in vitro using primary AML patient samples expressing mutant NRAS (supplemental Table 5), along with mononuclear bone marrow cells from healthy donors. The proliferation of patient-derived samples was significantly suppressed with 125 nM of GNF-7 (Figure 6E), with minimal toxicity observed on normal marrow cells.
Discussion
Two genes are synthetic lethal if mutation (dysfunction) of either alone is compatible with viability, but mutation of both leads to death. The concept that transformation by specific oncogenes, such as NRAS, may create a synthetically lethal dependence on a second gene is well established genetically.9 Indeed, large scale RNA interference studies have been conducted in RAS transformed cell lines to look for such genes.6-8 In this study, we performed a differential cytotoxicity screen of NRAS-transformed Ba/F3 cells, using a panel of small molecule kinase inhibitors to identify compounds and their targets that are required for NRAS–transformed cells. We identified GNF-6 and then GNF-7 as RAS pathway inhibitors against leukemia cells using preclinical models of AML and ALL. The finding of differential growth suppression between NRAS mutant and WT leukemia cell lines (Figure 1C), suppression of a known NRAS gene signature (Figure 1E), and reduced potency against Ba/F3 cells transformed by oncogenes other than RAS (Figure 1F) suggest it is unlikely that GNF-7 targets pathways that are generally important for cell growth. Previous studies have reported the complexity and exceptional difficulty of defining the mechanism of action of any multitargeted kinase inhibitor with certainty.26,27 RAS mutations are known to activate a significant number of downstream effector pathways that have relative importance changes according to cellular background. We have tackled these challenges using state-of-the-art methodologies including screening large panels of kinase binding and activity assays, as well as chemical proteomics approaches. In addition, to simplify the process, we compared the kinase profiling data of GNF-7 to that of a less active analog, YHJ0557, generated to test the hypothesis that one or more key targets may be significantly less inhibited by the less active analog. This integrated approach has led to the identification of 2 pathways (ACK1/AKT and GCK) that synergistically contribute to the growth-suppressing effects of GNF-7 on NRAS–mutated cells as summarized in the scheme shown in supplemental Figure 6D.
Interestingly, we found that suppression of AKT signaling by GNF-7 is critically important, but it is not likely to occur by direct inhibition of AKT. This was confirmed by analyzing GNF-7-resistant Ba/F3-NRAS-G12D cells obtained after long-term culture with GNF-7 (IC50 is 40 times higher). These cells were found to have acquired a nonsense mutation of Pten. Reexpression of Pten significantly reversed drug resistance (data not shown). Kinase selectivity profiling identified ACK1 as a target of GNF-7, with this being of interest because ACK1 is known to form a robust complex with AKT and it directly regulates recruitment of AKT to the plasma membrane, “shunting” the PI3K pathway.21 Furthermore, ACK1 has been reported to be required for survival of v-H-Ras–transformed mammalian cells.28 Overall, it seems likely that inhibition of ACK1 by GNF-7 contributes to the suppression of AKT signaling.
Most importantly, the serine/threonine kinase, GCK, was revealed to be another novel critical target of mutant NRAS signaling. GCK is homologous to the yeast Ste20 kinase and is reported to act as a MAP4K. Mammalian Ste20-related kinases have been implicated in cellular processes ranging from stress responses to cell growth, proliferation, and death.29 GCK is highly expressed in germinal center of B cells, but it is also expressed in many tissues including the lung, brain, and others.30 GCK can be activated by tumor necrosis factor α or IL-1, and is involved in the activation of MEKK1 and MLK3 (MAP3Ks) in the stress-activated protein kinase/c-Jun N-terminal kinase signaling pathway.25,29,31 It has also been reported that JNK, which is an established downstream target of GCK, is required for KRAS-initiated tumor formation.32 Despite this, treatment of Ba/F3-NRAS-G12D cells with MK2206 and JNK inhibitor, SP600125, did not show synergy (supplemental Figure 4B). This suggests that JNK by itself is not an important downstream target of mutant NRAS signaling in the context of cell growth in this model. GCK is also not known to be mutated in leukemia and a survey of 200 primary AML patient samples (Cancer Genome Atlas Research Network, 2013) did not reveal any copy number alteration or changes in messenger RNA expression levels of GCK. Thus, it appears that the dependency of NRAS–mutant cells on GCK may be due to synthetic lethal interaction9 rather than transcriptional or genomic alteration.
ACK1/AKT and GCK are important targets of GNF-7. However, it is possible that inhibition of additional targets is required to reach the full efficacy of suppression of mutant NRAS signaling to overcome complex feedback loops, bypass signaling, and redundancy.
In conclusion, we have identified GNF-7 as a multitargeted kinase inhibitor of cells transformed by NRAS by a chemical screen using the Ba/F3-NRAS-G12D cell line. By utilizing this as a tool compound, mechanistic analysis revealed that ACK1/AKT and GCK were functionally important novel therapeutic targets of acute leukemia with NRAS mutation. Development of more selective inhibitors of ACK1 and GCK is underway. Interestingly, while the AKT pathway is well known to be a mediator of canonical RAS signaling, GCK is not. Rather, GCK may well be important in the context of NRAS transformation in hematopoietic cells for reasons that will need further study. Our work is potentially impactful because we demonstrate that it is possible to devise efficient cell-based screening paradigms to identify targets that are important only in the context of mutant NRAS signaling, and are thus synthetically lethal. The fact that GNF-7 has more than one potentially important target in combination is of interest. This is not surprising given the fact that many kinase inhibitors have multiple known targets, and this is theoretically an advantage over some types of genetic screens where a hit is a single gene. Larger scale screens of the sort described here and these pharmacologic approaches would be expected to provide information that could not be obtained solely from large scale genetic screens.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Hwan Geun Choi for the synthesis of GNF-7 and Dr Nam Doo Kim for the molecular docking study.
This study is supported by the National Institutes of Health, National Cancer Institute grants CA66996 (J.D.G), CA130876-04 (N.S.G), and CA154303-01A1 (N.S.G), and HG005693-01 (N.S.G), a grant from the Kanae foundation for the promotion of medical science (A.N.), and grants from the Korea Institute of Science and Technology (T.S.), the creative/challenging research program of the National Research Foundation of Korea (NRF-2011-0028676) (T.S.), and the Korea Basic Science Institute (D33400, T.S.).
Authorship
Contribution: A.N. designed the study and experiments, analyzed the data, and prepared the manuscript; M.S. and E.W. provided support and prepared the manuscript; Q.L. and J.Z. provided support for the chemical screen and helpful discussion; M.P.P. performed in situ kinase selectivity profiling; A.L.C., A.M.S., N.E.K., and A.L.K. performed the in vivo xenotransplantation study; H.Y. and T.S. performed synthesis of GNF-7 and design and synthesis of the analogs of GNF-7 and also performed the in vitro kinase assay; and N.S.G. and J.D.G. designed the study and supervised the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Taebo Sim, 145 Anam-ro, Seongbuk-gu, Seoul, 136-713, Korea; e-mail: tbsim@kist.re.kr; and Nathanael S. Gray, 250 Longwood Ave, Seeley G. Mudd 628A, Boston, MA 02115; e-mail: nathanael_gray@dfci.harvard.edu.